Abstract
Nucleoside reverse transcriptase inhibitors (NRTIs) are employed in first line therapies for the treatment of human immunodeficiency virus (HIV) infection. They generally lack a 3′-hydroxyl group, and thus when incorporated into the nascent DNA they prevent further elongation. In this report we show that 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA), a nucleoside analog that retains a 3′-hydroxyl moiety, inhibited HIV-1 replication in activated peripheral blood mononuclear cells with an EC50 of 0.05 nm, a potency several orders of magnitude better than any of the current clinically used NRTIs. This exceptional antiviral activity stems in part from a mechanism of action that is different from approved NRTIs. Reverse transcriptase (RT) can use EFdA-5′-triphosphate (EFdA-TP) as a substrate more efficiently than the natural substrate, dATP. Importantly, despite the presence of a 3′-hydroxyl, the incorporated EFdA monophosphate (EFdA-MP) acted mainly as a de facto terminator of further RT-catalyzed DNA synthesis because of the difficulty of RT translocation on the nucleic acid primer possessing 3′-terminal EFdA-MP. EFdA-TP is thus a translocation-defective RT inhibitor (TDRTI). This diminished translocation kept the primer 3′-terminal EFdA-MP ideally located to undergo phosphorolytic excision. However, net phosphorolysis was not substantially increased, because of the apparently facile reincorporation of the newly excised EFdA-TP. Our molecular modeling studies suggest that the 4′-ethynyl fits into a hydrophobic pocket defined by RT residues Ala-114, Tyr-115, Phe-160, and Met-184 and the aliphatic chain of Asp-185. These interactions, which contribute to both enhanced RT utilization of EFdA-TP and difficulty in the translocation of 3′-terminal EFdA-MP primers, underlie the mechanism of action of this potent antiviral nucleoside.
Introduction
Nucleoside reverse transcriptase inhibitors (NRTIs)4 are central components of first line regimens for treatment of HIV infections (1–6). Currently, there are eight clinically approved NRTIs: AZT, 3TC, FTC, ABC, ddI, ddC, d4T, and the nucleotide tenofovir (TFV; reviewed in Refs. 7 and 8). A structural hallmark of these NRTIs is the lack of a 3′-OH; it has long been considered that the absence of the 3′-OH is essential for antiviral activity. However, the absence of the 3′-OH in NRTIs also imparts detrimental properties to the inhibitor, including reduced affinity for RT compared with the analogous dNTP substrate, as well as reduced intracellular conversion to the active nucleoside triphosphate (9).
Previously we described a series of 4′-substituted NRTIs (10) that retain the 3′-OH group and have excellent antiviral properties and significantly improved selectivity indices (CC50/EC50) compared with the approved NRTIs. Furthermore, these NRTIs efficiently suppress various NRTI-resistant HIV. The most potent of these 4′-substituted NRTIs are the adenosine analogs that have an ethynyl group at the 4′ position of the ribose ring. Despite their high anti-HIV activity, 4′-substituted compounds are susceptible to degradation by adenosine deaminase (11), a property that limits the plasma and intracellular half-life of the drugs. To overcome the adenosine deaminase sensitivity of these 4′-ethynyl NRTIs, we developed a second generation of analogs substituted at the 2-position of the adenine ring (12). We recently reported that the 2-halogenated, 4′-ethynyl compounds have remarkably improved potency and selectivity indices (CC50/EC50) compared with the nonhalogenated analogs and significantly better ones compared with other approved NRTIs. These compounds are resistant to degradation by adenosine deamination (13). The most potent of these compounds is EFdA (Fig. 1A), which was recently shown not to inhibit human DNA polymerases α and β or mitochondrial DNA polymerase γ (12). Notably, clinically important drug-resistant HIVs (14, 15) are sensitive or hypersensitive to this compound (13).
FIGURE 1.
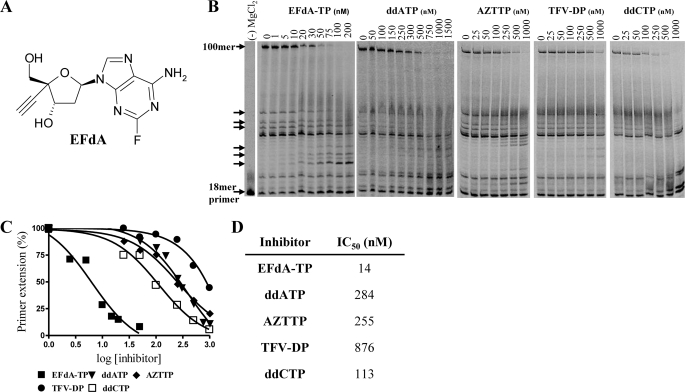
HIV RT inhibition by EFdA-TP and other NRTIs. A, structure of EFdA. B, primer extension by HIV-1 RT was observed in the presence of fixed concentrations of 4 dNTPs, Td100/Pd18, and MgCl2 and increasing concentrations of EFdA-TP, ddATP, AZTTP, TFV-DP, or ddCTP. The reactions were carried out for 15 min. The arrows denote stops of the elongating DNA chain where adenosine analogs (EFdA-TP, ddATP, or TFV-DP) were expected to be incorporated. The first lane is a negative control, where no MgCl2 was added; it shows the length of the 18-mer primer. C, the 100-mer products synthesized by HIV-1 RT were quantified and plotted against increasing concentrations of various inhibitors. The data points were fitted by GraphPad Prism 4. D, IC50 values of the nucleotide analogs were determined by quantifying the percent of full extension and fitting the data points to GraphPad Prism 4 using one-site competition nonlinear regression.
Despite its remarkable antiviral potency, the molecular mechanism by which EFdA and related compounds inhibit HIV is unknown. To elucidate this mechanism we carried out biochemical experiments that systematically decipher the effect of EFdA on each of the mechanistic steps of DNA synthesis by HIV RT. On the basis of these experiments we propose that EFdA-5′-triphosphate (EFdA-TP) inhibits RT by first being incorporated at the 3′-primer terminus, and after its incorporation it prevents further addition of nucleotides by blocking the translocation of the primer strand on the viral polymerase. We therefore termed EFdA a “translocation-defective reverse transcriptase inhibitor (TDRTI).” By understanding the molecular details of RT inhibition by a highly potent NRTI, we hope to gain insights into the design of even more efficacious inhibitors that may act via same or similar mechanisms.
EXPERIMENTAL PROCEDURES
Enzymes and Nucleic Acids
The RT genes coding for p66 and p51 subunits of BH10 HIV-1 were cloned in the pETDuet-1 vector (Novagen) using restriction sites NcoI and SacI for the p51 subunit and SacII and AvrII for the p66 subunit. The sequences coding for a hexahistidine tag and the 3C protease recognition sequence were added at the N terminus of the p51 subunit. RT was expressed in BL21 (Invitrogen) and purified by nickel affinity chromatography and MonoQ anion exchange chromatography (16). Oligonucleotides used in this study were synthesized chemically and purchased from Integrated DNA Technologies (Coralville, IA). Sequences of the DNA/RNA substrates are shown in Table 1. Deoxynucleotide triphosphates and dideoxynucleotide triphosphates were purchased from Fermentas (Glen Burnie, MD). EFdA was synthesized by Yamasa Corp. (Chiba, Japan) as described previously (12). Using EFdA as the starting material, the triphosphate form, EFdA-TP, was synthesized by TriLink BioTechnologies (San Diego, CA). Concentrations of nucleotides and EFdA-TP were calculated spectrophotometrically on the basis of absorption at 260 nm and their extinction coefficients. All nucleotides were treated with inorganic pyrophosphatase (Roche Diagnostics) as described previously (17) to remove traces of PPi contamination that might interfere with the rescue assay.
TABLE 1.
DNA and RNA sequences used in this study
The primers were fluorescently labeled at the 5′-end except for the footprinting experiments, in which the template was fluorescently labeled at the 5′-end.
| Polymerization experiments | |
| Td100 | 5′-TAG TGT GTG CCC GTC TGT TGT GTG ACT CTG GTA ACT AGA GAT CCC TCA GAC CCT TTT AGT CAG TGT GGA AAA TCT CTA GCA GTG GCG CCC GAA CAG GGA C |
| Pd18 | 5′-Cy3 GTC CCT GTT CGG GCG CCA |
| Td31 | 5′-CCA TAG CTA GCA TTG GTG CTC GAA CAG TGA C |
| Tr31 | 5′-CCA UAG CUA GCA UUG GUG CUC GAA CAG UGA C |
| Pd18 | 5′-Cy3 GTC ACT GTT CGA GCA CCA |
| Gel shift experiments | |
| Td43 | 5′-AAT CAG TGT AGA CAA TCC CTA GCA TTG GTG CTC GAA CAG TGA C |
| Pd18 | 5′-Cy3 GTC CCT GTT CGG GCG CCA |
| Footprinting experiments | |
| Pd20 | 5′-TTG TCA CTG TTC GAG CAC CA |
| Td43 | 5′-Cy3 CCA TAG CTA GCA TTG GTG CTC GAA CAG TGA CAA TCA GTG TAGA |
Cell-based HIV-1 Replication Assays
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donor buffy coats (purchased from the Central Blood Bank, Pittsburgh, PA) using Ficoll-Hypaque (Histopaque, Sigma-Aldrich) gradient centrifugation as described previously (18). PBMCs were stimulated with 5 μg/ml phytohemagglutinin (Sigma) in RPMI 1640 containing 10% fetal bovine serum for 48 h prior to exposure to drug and virus. After washing, the activated cells were resuspended in RPMI 1640/fetal bovine serum containing interleukin-2 (10 units/ml) and varying concentrations of the NRTIs and then were infected with HIV-1NL4-3 at a multiplicity of infection of 0.01. HIV-1 infection was assessed by measuring HIV-1 p24 antigen in cell-free culture supernatants obtained 7 days post-infection using an HIV-1 p24 antigen capture assay kit (SAIC, Frederick, MD).
Primer Extension Assays
Characterization of EFdA-TP as a Chain Terminator
DNA or RNA template was annealed to a 5′-Cy3-labeled DNA primer (3:1 molar ratio). To monitor the primer extension, the DNA/DNA or RNA/DNA hybrid (20 nm) was incubated at 37 °C with HIV-1 RT (20 nm) in a buffer containing 50 mm Tris (pH 7.8) and 50 mm NaCl (RT buffer). Subsequently, varying amounts of EFdA-TP or ddATP were added, and the reactions were initiated by the addition of 6 mm MgCl2 to a final volume of 20 μl. All dNTPs were present at a final concentration of 1 μm. The reactions were terminated after 15 min by adding an equal volume of 100% formamide containing traces of bromphenol blue. The products were resolved on a 15% polyacrylamide 7 m urea gel. In this and subsequent assays, the gels were scanned with a phosphorimaging device (FLA 5000, FujiFilm). The bands for fully extended product were quantified using Multi Gauge software (FujiFilm), and the results were plotted as percent full extension using GraphPad Prism 4 to determine the IC50 for EFdA-TP and other nucleotide analogs.
Steady State Kinetics
Steady state kinetic parameters, Km and kcat, for incorporation of EFdA-MP or dAMP were determined using single nucleotide incorporation in gel-based assays under saturating substrate conditions. The reactions were carried out in RT buffer with 6 mm MgCl2, 100 nm Td31/Pd18 or Tr31/Pd18, and 2.5 nm RT in a final volume of 20 μl and stopped at the indicated reaction times. The products were resolved and quantified as described above. Km and kcat were determined graphically using the Michaelis-Menten equation.
Incorporation of dNTP to the Template/Primer (T/P) Possessing Either EFdA-MP (T/PEFdA-MP) or ddAMP (T/PddAMP) at the 3′-Primer Terminus
T/PEFdA-MP and T/PddAMP were prepared by incubating 500 nm Td31/Pd18 with 1 μm HIV-1 RT in RT buffer and 6 mm MgCl2. EFdA-TP (1 μm) or ddATP (5 μm) was added into the reaction and the mixture was incubated at 37 °C for 1 h. After incorporation of the nucleotide analogs, the T/Panalog was purified using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA). Under these conditions, the extension of T/P to T/PEFdA-MP or T/PddAMP was complete. Purified T/PEFdA-MP (5 nm) was incubated with 20 nm HIV-1 RT in RT buffer and 6 mm MgCl2. The first incoming nucleotide was added at different concentrations (0–100 μm) in the presence of the other dNTPs (1 μm). The reactions were incubated at 37 °C for 15 or 60 min.
Gel Mobility Shift Assays
Formation of RT·DNA Binary Complex
T/PEFdA-MP and T/PddAMP were prepared using Td43/Pd18 as described above. Purified T/PEFdA-MP or T/PddAMP (20 nm) was incubated at room temperature for 10 min with different concentrations of HIV-1 RT in RT buffer and 6 mm MgCl2. RT was used at different concentrations to obtain RT/DNA ratios that ranged from 0.25 to 7.5. Four μl of 20% sucrose was added to each mixture in a final volume of 20 μl. The complexes were subsequently resolved on a native 6% polyacrylamide Tris borate gel and visualized as described above.
Formation of RT·DNAEFdA-MP·dTTP Ternary Complex
Purified T/PEFdA-MP or T/PddAMP (9 nm) was incubated at room temperature for 10 min with 100 nm HIV-1 RT, varying amounts of the next nucleotide (1–5000 μm) in RT buffer, and 6 mm MgCl2. Prior to the addition of sucrose, 150 ng/μl heparin was added, and finally the products were resolved on native 6% polyacrylamide Tris borate gels and visualized as described above.
Site-specific Fe2+ Footprinting Assay
Site-specific Fe2+ footprints were monitored on 5′-Cy3-labeled DNA templates. Td43/Pd20 (100 nm) was incubated with HIV-1 RT (600 nm) in a buffer containing 120 mm sodium cacodylate (pH 7), 20 mm NaCl, 6 mm MgCl2, and EFdA-TP (1 μm) to allow quantitative chain termination. Prior to treatment with Fe2+, complexes were preincubated for 7 min with increasing concentrations of the next nucleotide as indicated in Fig. 5A. The complexes were treated with ammonium iron sulfate (1 mm) as described previously (19). This reaction relies on autoxidation of Fe2+ (20) to create a local concentration of the hydroxyl radical, which cleaves the DNA at the nucleotide closest to the Fe2+ specifically bound to the RNase H active site.
FIGURE 5.
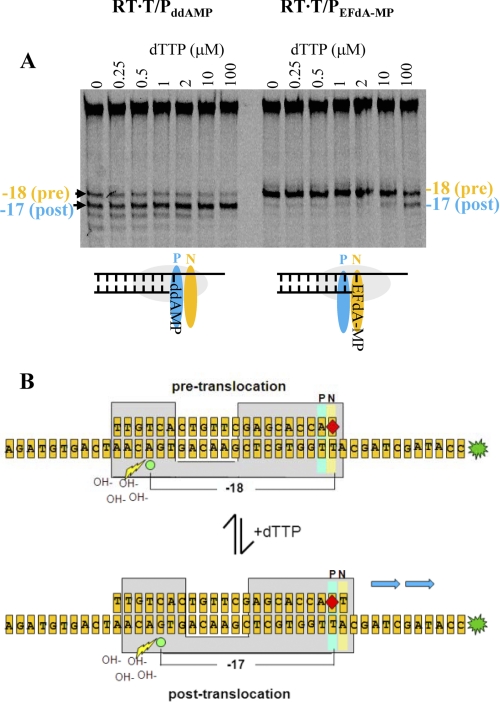
Determination of the translocation state of RT bound to T/PddAMP and T/PEFdA-MP template/primers. A, the translocation state of RT after EFdA-MP incorporation was determined using site-specific Fe2+ footprinting. T/PddAMP or T/PEFdA-MP (100 nm) with 5′-Cy3 label on the DNA template (see Fig. 5B) was incubated with HIV-1 RT (600 nm) and various concentrations of the next incoming nucleotide (dTTP) (as indicated). The complexes were treated for 5 min with ammonium iron sulfate (1 mm) and resolved on a polyacrylamide 7 m urea gel. An excision at position −18 indicates a pre-translocation complex, whereas the excision at position −17 represents a post-translocation complex. The scheme below the gel images indicates that in the absence of incoming dNTP, T/PddAMP is bound mostly in a post-translocation state, whereas T/PEFdA-MP is bound in a pre-translocation state, with EFdA-MP positioned at the N-site. B, schematic of the excision assay. Depending on whether the 3′-primer terminus is positioned at the pre-translocation (N-site) or post-translocation (P-site) site, cleavage is observed on the 5′-labeled template strand at positions −18 or −17, respectively. The addition of varying levels of the incoming complementary dNTP serves to force the 3′-primer terminus from the N-site to the P-site.
PPi- and ATP-dependent Excision and Rescue of T/PEFdA-MP and T/PddAMP
PPi-dependent Excision of T/PEFdA-MP and T/PddAMP
Purified T/PEFdA-MP and T/PddAMP (20 nm) were incubated at 37 °C with HIV-1 RT (60 nm) in the presence of 150 μm PPi in RT buffer and 6 mm MgCl2. Aliquots of the reaction were stopped at different times (0–30 min) and analyzed as described above.
PPi-dependent Rescue of T/PEFdA-MP and T/PddAMP
Purified T/PEFdA-MP and T/PddAMP (20 nm) were incubated with HIV-1 RT (60 nm) at various concentrations of PPi (0–150 μm) in RT buffer and 6 mm MgCl2. The assay was performed in the presence of a large excess of competing dATP (100 μm), which prevented reincorporation of EFdA-MP, 0.5 μm dTTP, and 10 μm ddGTP. After an incubation of 10 min at 37 °C, the reactions were stopped and analyzed as described above.
ATP-dependent Rescue of T/PEFdA-MP and T/PddAMP
Purified T/PEFdA-MP and T/PddAMP (20 nm) were incubated with HIV-1 RT (60 nm) in the presence of 3.5 mm ATP, 100 μm dATP, 0.5 μm dTTP, and 10 μm ddGTP in RT buffer and 10 mm MgCl2. Aliquots of the reaction were stopped at different time points (0–90 min) and analyzed as described above.
Molecular Modeling
Molecular models of two reaction intermediates that involve EFdA were built as follows. 1) A model of the ternary complex of HIV-1 RT·DNA·EFdA-TP (Fig. 7A) was built starting with the coordinates of the crystal structure of the HIV-1 RT·DNA·TFV-DP complex. The triphosphate of EFdA-TP was built using as a guide the corresponding atoms of TFV-DP in structure with PDB code 1T05 and of dTTP in PDB code 1RTD. The coordinates of the 4′-ethynyl sugar ring were from our NMR structure of EFdA5 showing that EFdA is in a North conformation similar to the sugar puckering observed in the crystal structure of 4′-ethynyl-2′-deoxycytidine (21). The structure of the EFdA-TP was assembled from its components using the sketch module of Sybyl 7.0 (Tripos Associates, St. Louis, MO), and minimized by the semiempirical quantum chemical method PM3 (22). The PM3 charges and the docking module of Sybyl 7.0 were used to dock the EFdA-TP at the RT dNTP-binding site (after removing the TFV-DP from 1T05) to generate the ternary complex HIV-1 RT·T/P·EFdA-TP. The final complex structure was minimized for 100 cycles using the AMBER force field with Coleman united charges on the protein and DNA molecules. 2) The model of the RT·T/PEFdA-MP binary complex with primer 3′-terminal EFdA-MP at the pre-translocation nucleotide-binding site (N-site) (or dNTP-binding site) (Fig. 7B) was built using as a starting model our crystal structure of the pre-translocation complex RT·T/PAZT-MP (PDB code 1N6Q). The structures of AZTMP and the base-pairing dA were replaced by EFdA-MP (built as in EFdA-TP above) and a dT, respectively, using the sketch module of Sybyl 7.0, and energy-minimized using the AMBER force field with Coleman united charges on the protein and DNA molecules.
FIGURE 7.
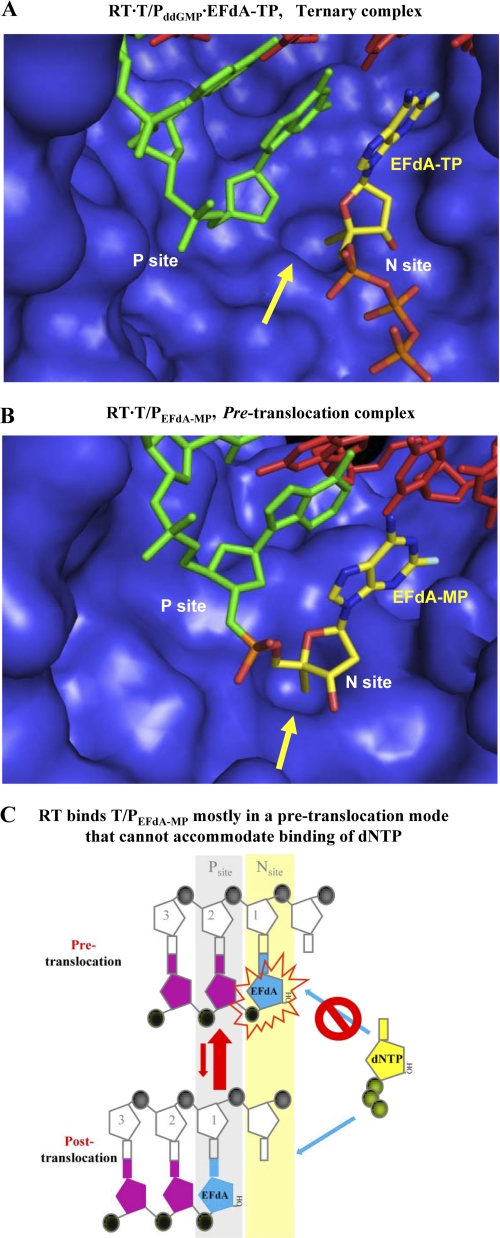
Molecular models representing intermediates of the DNA polymerization reaction. A, molecular model of a ternary complex among RT, DNA, and EFdA-TP. The primer is bound at the P-site, and the incoming EFdA-TP is bound at the N-site. The 4′-ethynyl group of EFdA-TP is bound at a hydrophobic pocket (shown by a yellow arrow) defined by residues Met-184, Ala-114, Tyr-115, and Phe-160 and the aliphatic chain of Asp-185. For purposes of clarity the p66 fingers subdomain is not shown. B, molecular model of RT bound to EFdA-MP-terminated T/P immediately after incorporation of the inhibitor at the primer terminus and before translocation. The EFdA-MP of the 3′-primer terminus is positioned at the N-site. C, schematic representation of RT inhibition by EFdA. After incorporation of EFdA-MP at the 3′-primer terminus RT remains bound to T/PEFdA mostly in a pre-translocation binding mode (top). In that binding mode the EFdA-MP at the 3′-primer terminus blocks binding of the incoming dNTP, thus inhibiting DNA polymerization.
RESULTS
EFdA-TP Is a Highly Potent Inhibitor of HIV-1 RT
EFdA inhibits HIV-1 replication in phytohemagglutinin-activated PBMCs with an EC50 of 50 pm (Table 2), consistent with previously published data obtained using T-cell lines (12, 13), and data published after completion of this work (23). The antiviral potency of EFdA is at least 4 orders of magnitude greater than the clinically used adenine nucleotide analog tenofovir and over 400-fold greater than that of AZT when assessed under the same conditions (Table 2). EFdA thus appears to be the most potent nucleoside inhibitor described to date of HIV-1 replication in primary cells. It is also interesting to note that EFdA is substantially more potent than analogs lacking a 3′-OH function (EFddA and EFd4A; Table 2). No cytotoxicity was noted at 10 μm EFdA (data not shown), the highest concentration tested, suggesting an in vitro selectivity index of over 200,000. To better understand the molecular basis for the exceptional antiviral potency of EFdA, we carried out a series of detailed in vitro evaluations of the impact of the active antiviral form of EFdA, namely EFdA-TP, on DNA synthesis catalyzed by purified HIV-1 RT.
TABLE 2.
Inhibition of HIV-1 replication in phytohemagglutinin-activated PBMCs by EFdA, EFdA analogs, and other NRTIs
| Compound | EC50a |
|---|---|
| nm | |
| EFdA | 0.05 ± 0.02 |
| EdA | 11 ± 7 |
| EFddA | 570 ± 92 |
| EFd4A | 14 ± 11 |
| Zidovudine (or AZT) | 22 ± 7 |
| Tenofovir | 3300 ± 1240 |
a Values are means ± S.D. of triplicate determinations and were determined by assessment of reduction in HIV-1 p24 antigen production in infected cells as described under “Experimental Procedures.”
We first compared the effect of EFdA-TP with other NRTI-TPs (ddATP, TFV-DP, AZTTP, and ddCTP) on RT-catalyzed DNA synthesis in in vitro primer extension assays using a nucleic acid T/P comprising a 100-nucleotide DNA template annealed to a Cy3–5′-labeled 18-nucleotide DNA primer (Table 1). As shown in Fig. 1B, EFdA-TP suppressed full-length DNA synthesis by RT in a dose-dependent manner. EFdA-TP was between ∼1 and 2 orders of magnitude more effective at inhibiting RT-catalyzed DNA synthesis than any of the NRTIs evaluated. The IC50 for suppression of full primer extension by EFdA-TP was 14 nm using the longer Td100/Pd18 (Fig. 1, C and D). To confirm the high efficiency at which RT uses EFdA, we performed single nucleotide incorporation assays under steady state conditions (using Td31/Pd18 or Tr31/Pd18 as a T/P, Table 1). Our results show that under these conditions the incorporation efficiency (kcat/Km) of EFdA-TP by RT is twice that for the natural dATP substrate and four times that for ddATP, primarily because of changes in the Km of RT to these substrates (Table 3). Moreover, we found that the increase in incorporation efficiency of EFdA-TP could be even higher at different nucleic acid substrate sequences, more than 10 times higher than dATP (data not shown).
TABLE 3.
Steady state kinetic parameters (Km and kcat) for EFdA-MP and dAMP incorporation by HIV-1 RT
Values are means ± S.D. of triplicate determinations and were determined from Michaelis-Menten equation using GraphPad Prism 4. ND, not determined.
| dNTP | T/P (DNA/DNA) |
T/P (RNA/DNA) |
||||||
|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Selectivitya | Km | kcat | kcat/Km | Selectivitya | |
| nm | min−1 | min−1·nm−1 | nm | min−1 | min−1·nm−1 | |||
| dATP | 73.11 ± 11 | 19.9 ± 0.7 | 0.272 | 1 | 21.3 ± 8 | 3.1 ± 0.2 | 0.145 | 1 |
| EFdA-TP | 39.2 ± 3 | 21.1 ± 0.4 | 0.538 | 2 | 24.1 ± 5 | 2.3 ± 0.1 | 0.095 | 0.7 |
| ddATP | 97.0 ± 9 | 15.4 ± 0.3 | 0.159 | 0.6 | ND | ND | ND | ND |
a Selectivity is the ratio of the incorporation efficiency (kcat/Km) of EFdA-MP or ddAMP over that of dAMP ([kcat/Km]EFdA-MP/[kcat/Km]dAMP or [kcat/Km]ddAMP/[kcat/Km]dAMP).
The EFdA-TP-mediated reduction in full-length DNA synthesis was accompanied by the concomitant appearance of products corresponding to the primer extension only at the length expected for the incorporation of adenosine nucleotides (indicated by arrows in Fig. 1B). Neither ddATP nor TFV-DP provided significant accumulation of this small DNA product.
EFdA-TP Inhibits DNA Synthesis Mainly at the Point of Incorporation
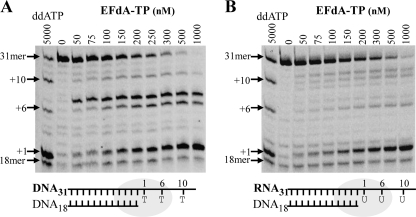
The stopping patterns of DNA synthesis were different in the presence of EFdA-TP compared with other dATP analogs such as ddATP and TFV-DP (marked by arrows in Fig. 1B). Hence, we used a shorter template (Td31/Pd18 (Table 1)), which allowed unambiguous identification of the stopping sites. As expected, the inhibitory potential of EFdA (and other NRTIs) appears lower in these shorter T/P (IC50 for suppression of full primer extension was 104 nm) in which there are fewer opportunities for incorporation (24). This substrate allows incorporation of dA, ddA, or EFdA at positions 1, 6, and 10 (Fig. 2). The results show that EFdA-TP causes major pauses at all possible points of incorporation ((Fig. 2A, positions 1, 6, and 10), suggesting that EFdA-TP inhibits RT mainly as an obligate chain terminator. Notably, there was a distinct difference at position +6 of Td31/Pd18, where we observed a strong stop not only at the point of incorporation but also at the position following (Fig. 2A, positions 6 and 7, respectively). These results suggest that in some cases EFdA-MP may also allow incorporation of an additional nucleotide depending upon the template sequence.
FIGURE 2.
Inhibition of DNA- and RNA-dependent DNA synthesis by EFdA-TP. A, Td31/Pd18 was incubated with HIV-1 RT for 15 min in the presence of 1 μm dNTPs and MgCl2 and increasing concentrations of EFdA-TP (0–1000 nm). The first lane (ddATP) shows the inhibition of primer extension by ddATP to identify points of adenosine analog (ddATP or EFdA-TP) incorporation (arrows: +1, +6, and +10). B, the primer extension under the same conditions with an RNA/DNA substrate containing an RNA template annealed to a DNA primer (Tr31/Pd18).
EFdA-TP inhibits both RNA- and DNA-dependent RT-catalyzed full-length DNA polymerization to comparable extents (Fig. 2). The selectivity for incorporation of EFdA-MP over dAMP ((kcat/Km)EFdA-TP/(kcat/Km)dATP) was slightly increased when we used DNA/DNA versus RNA/DNA template/primers (Table 3, 2 versus 0.7). Notably, the overall pausing pattern due to inhibition by EFdA-TP differs depending on whether the template is RNA or DNA. Inhibition differences based on type and sequence of template are currently under investigation.
Extension of EFdA-terminated T/P (T/PEFdA-MP)
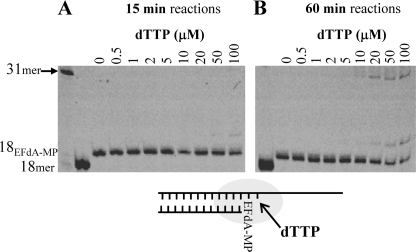
The data in Figs. 1 and 2 suggest that EFdA-TP can act as a terminator of RT-catalyzed DNA synthesis in a manner similar to that of other NRTI-TPs. We therefore examined the efficiency of nucleotide additions to primers that were synthesized to already possess a 3′-terminal EFdA-MP. Fig. 3 shows that the primer extension from EFdA-MP is very limited and is evident only at very high and nonphysiological concentrations (>50 μm) of the next nucleotide and with extended reaction times (Fig. 3B, 60-min incubation). It therefore appears that EFdA acts as a de facto chain terminator, despite the presence of a 3′-OH function.
FIGURE 3.
Incorporation of dNTP on EFdA-terminated template/primer (T/PEFdA-MP). EFdA-TP was first incorporated at Td31/Pd18 by HIV-1 RT and purified as described under “Experimental Procedures.” The incorporation of the next incoming nucleotide on T/PEFdA-MP was examined in the presence of HIV-1 RT and MgCl2 and increasing concentrations of dTTP. All other dNTPs were at a concentration of 1 μm. The reactions were stopped after 15 min (A) and 60 min (B).
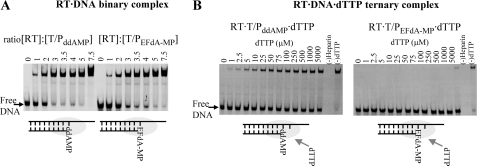
RT Binds T/PddAMP and T/PEFdA-MP at Similar Efficiencies
A possible reason for the inability of RT to efficiently extend the EFdA-MP-terminated primer is that it binds T/PEFdA-MP with less affinity than it does a T/P lacking a 3′-terminal EFdA-MP nucleotide. We therefore used gel mobility shift assays to compare the stabilities of the binary complexes of RT with T/P possessing either EFdA-MP (T/PEFdA-MP) or ddAMP (T/PddAMP) at the 3′-primer terminus (T/P chain terminated by EFdA-MP or ddAMP). As shown in Fig. 4A, RT binds T/PEFdA-MP with an apparent affinity comparable with that of the normal T/P (Kd for RT·T/PEFdA-MP = 51 nm; Kd for RT·T/PddA-MP = 42 nm). This observation suggests that RT is inhibited at a downstream step in the polymerization reaction.
FIGURE 4.
Effect of ddA or EFdA on formation of binary and ternary complexes. A, formation of a binary complex between RT and T/PddAMP or T/PEFdA-MP. Purified T/PddAMP or T/PEFdA-MP (20 nm) was incubated with HIV-1 RT at the indicated molar ratios and resolved by nondenaturing gel electrophoresis. B, formation of a ternary complex between RT and T/PddAMP or T/PEFdA-MP and incoming dTTP. The stability of the ternary complexes was analyzed by incubating 100 nm RT and 9 nm T/PddAMP or T/PEFdA-MP in the presence of increasing dTTP concentrations and heparin, which acted as an enzyme trap. In the absence of dTTP, the T/P·RT binary complex is unstable (lane 0), as RT dissociates from the T/P and is trapped by heparin.
RT Is Unable to Form a Stable Ternary Complex with T/PEFdA-MP and dNTP
The next step in the DNA polymerization mechanism is the binding of the next complementary dNTP to RT·DNA, thus forming the ternary complex that precedes catalysis. To determine whether EFdA exerts its inhibitory effect by interfering with the formation of a stable ternary complex with the incoming dNTP, we used a gel-based nondenaturing electrophoresis assay (25) that detects the ternary complex formed by RT·T/P and the next complementary dNTP (in this case, dTTP). In this assay, the stability of the ternary complex is assessed by the persistence of the RT·DNA·dNTP complex upon addition of a competing heparin trap (25). As seen in Fig. 4B (left panel), the ternary complex formed by RT with T/PddAMP and dTTP is quite stable. In contrast, no significant amount of ternary complex was noted in assays using T/PEFdA-MP even at very high dTTP concentrations (Fig. 4B, right panel).
Incorporation of EFdA-TP into DNA (T/PEFdA-MP) Decreases Translocation of RT
The inability of RT to form a stable ternary complex with T/PEFdA-MP and the next complementary dNTP could arise from several factors including (i) the inability of the 3′-terminal EFdA-MP primer to efficiently translocate from the N-site (which is also the pre-translocation site) to the post-translocation primer site (P-site), thereby preventing the next incoming nucleotide from binding; and (ii) the fact that T/PEFdA-MP can translocate, but the presence of unnatural substituents in the primer 3′-terminal EFdA-MP (4′-ethynyl and 2-fluoro) may alter geometric and electronic parameters at the primer end, thus preventing efficient incorporation of the next incoming dNTP such that catalysis cannot occur. These two different possibilities place the nucleic acid at different positions/registers with respect to a site-specific landmark in RT, the metal-binding ribonuclease H (RNase H) active site, as shown in the schematic of Fig. 5B. We therefore used a site-specific Fe2+ footprinting assay (19) to determine whether the primer 3′-terminal EFdA-MP of the RT·T/PEFdA-MP complex resides primarily in the pre- (N-site) or post-translocation (P-site) state in the absence and the presence of varying levels of the incoming complementary dNTP, which serves to “force” the 3′-primer terminus from the pre-translocation (N-site) to the post-translocation site (P-site). As shown on Fig. 5A (left panel), in the absence of dTTP (first lane), primers with a 3′-terminal ddAMP are located in both the pre- and post-translocation sites in approximately equal amounts, and the addition of the next complementary nucleotide (dTTP) forces the primer 3′-end almost entirely into the post-translocation site. In contrast, in the absence of dTTP, primers with a 3′-terminal EFdA- MP are located exclusively in the pre-translocation site (Fig. 5A, right panel, first lane). The next complementary nucleotide (dTTP) is unable to shift the position of the 3′-EFdA-MP except at very high (nonphysiological) concentrations.
Because it is physically impossible for the incoming dNTP to bind at the N-site (dNTP-binding site) when it is occupied by the 3′-primer terminus of the non-translocated T/PEFdA-MP, the present data demonstrate that the apparent termination of RT-catalyzed DNA synthesis upon incorporation of EFdA-MP arises from the inability of the 3′-EFdA-MP-terminated primer/template (T/PEFdA-MP) to efficiently translocate to the P-site and allow incorporation of the next dNTP. Moreover, the latter is unable to force translocation of the 3′-EFdA-MP-terminated primer strand. The inability of the 3′-EFdA-MP-terminated primer to translocate to allow binding of the next complementary dNTP effectively prevents continued elongation of the nascent viral DNA, despite the presence of the 3′-OH on EFdA. Therefore, we propose that EFdA acts as a translocation-defective reverse transcriptase inhibitor.
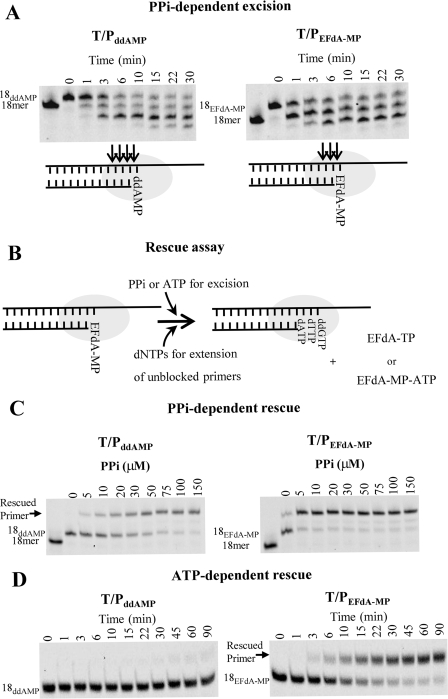
Phosphorolytic Excision of EFdA-MP
Two major mechanisms account for HIV resistance to NRTIs (26). One is based on NRTI discrimination, where the mutant RT preferentially incorporates the natural dNTP rather than the NRTI-TP. The other major resistance mechanism involves ATP-mediated phosphorolytic excision of the incorporated chain-terminating NRTI from the 3′-end of the primer (27, 28). We and others have previously shown that for excision to occur, the 3′-end of the primer must be positioned at the pre-translocation or N-site of RT (19, 29, 30). As we have already shown, the 3′-EFdA-MP-terminated primer strand binds predominantly in a pre-translocation mode. This suggests that EFdA-terminated primers might be especially susceptible to RT-catalyzed phosphorolytic removal of the terminating EFdA-MP. To assess this possibility we carried out primer unblocking experiments using nucleic acid substrates having at the 3′-primer terminus either EFdA-MP or ddAMP (T/PEFdA-MP or T/ PddAMP, respectively). The quantitation of results in Fig. 6A shows that the rate of hydrolysis of ddAMP- and EFdA-MP-terminated primers was 0.5 and 1.6 min−1, respectively. We also considered whether the EFdA-TP formed upon pyrophosphorolytic removal of the 3′-terminal EFdA-MP was promptly reincorporated. We tested this possibility using so-called phosphorolysis “rescue” assays, where in addition to the PPi that would react with the EFdA-MP from the 3′-primer terminus to produce EFdA-TP, we also included a high concentration of dATP that would compete with and prevent reincorporation of EFdA-TP. In the case of high excision activity, we expected to see higher bands corresponding to rescued and extended primers. Indeed, substantially more primer extension was noted in PPi- or ATP-mediated rescue assays using 3′-terminal EFdA-MP primers (Fig. 6, C and D, right panels) than in assays using primers with 3′-terminal ddAMP (Fig. 6, C and D, left panels). The rate for the ATP-dependent rescue of EFdA-MP-terminated primer was 0.063 min−1. The corresponding rate for the ATP-dependent rescue of the ddAMP-terminated primer could not be calculated because the reaction was very slow. Collectively, these data suggest that 3′-terminal EFdA-MP is phosphorolytically excised more efficiently than ddAMP, consistent with its preferential positioning at the phosphorolysis-susceptible pre-translocation N-site.
FIGURE 6.
PPi and ATP-dependent unblocking of ddAMP and EFdA-MP terminated primers. A, PPi-dependent unblocking of T/PddAMP and T/PEFdA-MP. Purified T/PddAMP or T/PEFdA-MP was incubated with HIV-1 RT in the presence of 6 mm MgCl2 and 150 μm PPi at 37 °C. Aliquots were removed and reactions stopped at the indicated time points (0–30 min). Cleavage sites are indicated with arrows in the schemes below the gels. B, schematic representation of PPi- and ATP-dependent rescue assay. The excision products of PPi- and ATP-dependent excision of EFdA-MP are EFdA-TP and the EFdA-MP-ATP dinucleoside tetraphosphate, respectively. C, PPi-dependent rescue of T/PddAMP and T/PEFdA-MP. Purified T/PEFdA-MP or T/PddAMP was incubated with HIV-1 RT in the presence of various amounts of PPi (0–150 μm), dATP (100 μm), dTTP (0.5 μm), or ddGTP (10 μm) and 10 mm MgCl2 at 37 °C. Aliquots of the reaction were stopped after 10 min. D, ATP-dependent rescue of T/PddAMP or T/PEFdA-MP. Purified T/PEFdA-MP or T/PddAMP was incubated with HIV-1 RT in the presence of ATP (3.5 mm), dATP (100 μm), dTTP (0.5 μm), or ddGTP (10 μm) and 10 mm MgCl2 at 37 °C. Aliquots of the reaction were stopped at the indicated time points (0–90 min).
Molecular Basis of RT Inhibition by TDRTIs
To understand the molecular basis of RT inhibition by EFdA-TP, we built molecular models of complexes that represent the following intermediates of the DNA polymerization reaction: 1) a precatalytic RT·DNA·EFdA-TP ternary complex and 2) a complex that corresponds to the product of EFdA-MP incorporation prior to translocation. Previously we had solved crystal structures representing both types of these intermediates for other NRTIs (29, 31), and the model building was guided by the structural characteristics of these complexes. Moreover, we built the EFdA sugar ring in both models in a 3′-endo (North) conformation based on our unpublished NMR experimental data, which clearly show that the equilibrium of the geometries of the EFdA sugar ring overwhelmingly favors the 3′-endo conformation (North). The RT·DNA·EFdA-TP ternary complex model was built using as a starting structure the coordinates of our RT·DNA·TFV-DP crystal structure (PDB code 1T05), not only because it is the highest resolution structure of an RT ternary complex but also because it is the only structure of RT in complex with an analog of deoxyadenosine triphosphate (31). The model of the RT·DNA·EFdA-TP complex represents the step of EFdA-TP binding to the preformed RT·DNA complex (Fig. 7A). It had no significant differences from the crystal structures of the ternary complexes of RT with DNA and TFV-DP (PDB code 1T05) or dTTP (PDB code 1RTD). It shows that the 4′-ethynyl of EFdA-TP is favorably positioned in a hydrophobic pocket formed by Ala-114, Tyr-115, Phe-160, and Met-184 and the aliphatic portion of Asp-185. These interactions are similar to those that have been proposed for binding of 4′-Ed4T, a related NRTI thymidine analog that also has a 4′-substitution but no 3′-OH group (32). These interactions are also consistent with the observed high efficiency of EFdA-MP incorporation by RT (Fig. 1D). Similar interactions stabilize the RT·DNAEFdA-MP pre-translocation binary complex, which has the primer 3′-terminal EFdA-MP positioned at the N-site (Fig. 7B, pre-translocation complex). These favorable interactions are also consistent with the enhanced binding of T/PEFdA-MP in a pre-translocated mode (Fig. 5).
DISCUSSION
The single most distinguishing feature of NRTIs used in HIV therapy is the absence of a 3′-OH. This property results in termination of further viral DNA synthesis upon incorporation of the inhibitor into the nascent viral DNA. We have shown (10, 13) that certain nucleoside analogs that retain the 3′-OH group can exert potent antiviral activity. One of these, EFdA, inhibits HIV-1 replication in PBMCs with a potency that is several orders of magnitude greater than that of any of the current clinically used NRTIs (Table 2 and Ref. 23), consistent with the inhibitory data obtained in transformed T-cell lines (13). The molecular basis for this exceptional antiviral activity of EFdA has to date been unclear. The detailed in vitro biochemical studies presented in this article show that EFdA inhibits HIV-1 reverse transcriptase mainly by blocking translocation after incorporation at the 3′-primer end and functioning as a TDRTI. The specificity of inhibition can vary depending on the type and sequence of the template (Fig. 2). Our studies also suggest that both the 3′-OH and the 4′-ethynyl groups of EFdA play important roles in the high antiviral potency exerted by this nucleoside analog.
The 4′-ethynyl group is essential for the mechanism of EFdA inhibition of RT-catalyzed DNA synthesis. The present work shows that EFdA-TP acts mainly as a potent terminator of RT-catalyzed DNA synthesis, despite having a 3′-OH group. It is possible that the presence of a 4′-ethynyl substitution on the ribose ring somehow affects the geometry and reactivity of its 3′-OH. However, in the presence of physiological concentrations of dNTPs (<50 μm) the chain terminating activity of EFdA appears to arise mainly from the difficulty of RT to translocate the 3′-EFdA-MP-terminated primer (T/PEFdA-MP) following incorporation of the inhibitor. Under these circumstances the dNTP-binding site is not accessible, and incorporation of the next complementary nucleotide is prevented (Fig. 7C). Hence, EFdA appears to act as a translocation-defective RT inhibitor. The 4′-ethynyl moiety appears to be the critical factor in the difficulty presented by translocation of DNA primers with a 3′-terminal EFdA-MP residue. Our molecular model of the RT·DNA·EFdA-TP ternary complex suggests that the 4′-ethynyl moiety fits nicely into a hydrophobic pocket in the RT active site defined by residues Ala-114, Tyr-115, Phe-160, and Met-184 and the aliphatic portion of Asp-185 (Fig. 7A), similar to the proposed interactions of 4′-Ed4T at the same site (32). Once EFdA-MP has been added to the primer end to form the pre-translocation (or N-site) reaction product, these same RT residues serve to stabilize the terminal EFdA-MP in the pre-translocation state (Fig. 7B). Hence, the stabilization of the primer 3′-terminal EFdA-MP in the pre-translocation state helps it to remain in a position antagonistic to further nucleotide addition and to inhibit DNA polymerization (Fig. 7C).
We have observed that in one instance RT stopped not only at the point of EFdA incorporation but also at the position following (Fig. 2A, positions 6 and 7, respectively). Interestingly, we found that RT has enhanced translocation efficiency at this site, both on T/PEFdA-MP and on T/PddAMP (data not shown). It appears that some translocated T/PEFdA-MP is also elongated by an additional nucleotide. Further polymerization may be inhibited by unfavorable interactions between the 4′-ethynyl group in the elongated template/primer (T/PEFdA-MP-dNMP) with protein residues upstream in the DNA-binding cleft. The effect of template on the inhibition mechanism by EFdA is the subject of an ongoing investigation.
RT-catalyzed phosphorolytic excision of chain-terminating NRTIs is a major mechanism of HIV-1 resistance to the nucleoside analog class of therapeutics (26–28, 33). Our previous studies have shown that the NRTI phosphorolytic excision reaction is favored when the primer 3′-terminal nucleotide is in the pre-translocation or N-site (19, 34). The preference of the primer 3′-terminal EFdA-MP to remain in this site suggests that terminal EFdA-MP should undergo facile phosphorolytic removal. EFdA-MP was subject to excision by pyrophosphorolysis somehow faster than ddAMP, which tends to localize in the post-translocation site when at the 3′ terminus of the primer (see Fig. 5A). Although EFdA-MP can undergo excision, this process is not overly efficient, apparently because once the nucleotide is excised through pyrophosphorolysis to form EFdA-TP, the latter is rapidly reincorporated. These findings suggest that phosphorolysis may not play a significant role in HIV-1 resistance to EFdA, consistent with the relatively small loss of antiviral potency of EFdA against excision-enhanced HIV-1-containing mutations associated with resistance to AZT (13).
The importance of the 3′-OH in antiviral activity of EFdA is perhaps best highlighted by the observation that EFdA is 10,000-fold more potent at inhibiting HIV-1 replication in PBMCs than is the identical nucleoside lacking a 3′-OH, namely EFddA (Table 2). The 3′-OH on EFdA appears to play a number of roles in contributing to the exceptional antiviral potency of the compound. The 3′-OH on natural dNTP substrates contributes to the efficiency with which RT uses these substrates, and in general NRTIs that lack a 3′-OH are used less efficiently by RT than the base-analogous dNTP (35) (Table 3). Our in vitro biochemical data demonstrate that EFdA-TP is approximately 1 and 2 orders of magnitude more potent an inhibitor of RT-catalyzed DNA synthesis in vitro than are the adenine-based NRTIs ddATP and TFV-DP, respectively (Fig. 1D), neither of which has a 3′-OH. Indeed, it appears that under identical conditions, RT is at least twice as likely to use EFdA-TP as a substrate over the natural nucleotide dATP (Table 3). This suggests that during HIV-1 reverse transcription, EFdA-TP might be preferentially incorporated, thereby leading to early and profound chain termination and contributing to the observed potent antiviral activity of this nucleoside analog.
NRTIs are administered therapeutically as prodrugs; these must undergo phosphorylation in target cells in order to exert their antiviral activity. The lack of a 3′-OH on current clinically used NRTIs can reduce recognition by cellular nucleoside/nucleotide kinases that have evolved to interact with the 3′-OH present in their natural nucleoside substrates (9). Preliminary studies suggest that EFdA appears to undergo rapid and facile intracellular conversion to the active antiviral EFdA-TP (36), showing that the 4′-ethynyl group does not interfere with recognition by cellular nucleoside/nucleotide kinases. Furthermore, the presence of fluorine at position 2 of the adenine base of EFdA helps to stabilize intracellular levels of EFdA and its phosphorylated products by hindering adenosine deaminase-catalyzed degradation of the molecule (13). Thus, both the 2-fluoro and the 3′-OH of EFdA may contribute to the intracellular accumulation of the antiviral EFdA-TP, thereby leading to its pronounced antiviral activity. We are presently carrying out detailed studies of the intracellular pharmacokinetics of EFdA in comparison with other NRTIs to better understand the dynamics of EFdA phosphorylation and turnover as contributors to the exceptional potency and persistence of its antiviral activity.
Other nucleoside analog inhibitors of HIV-1 RT that possess a 3′-OH have been described (21, 37–42), although these have mechanisms of action quite distinct from that of EFdA. North-methanocarba-2′-deoxyadenosine triphosphate and North-methanocarba-2′-thymidine triphosphate inhibit HIV-1 RT in vitro by a mechanism of delayed chain termination, where RT-catalyzed DNA synthesis pauses after the addition of several nucleotides following incorporation of the inhibitor (38, 39). Neither of these compounds has antiviral activity, presumably because of poor intracellular phosphorylation. Entecavir is a nucleoside analog with a 3′-OH that is approved for treatment of hepatitis B infection. Entecavir-TP also has been shown to inhibit HIV-1 RT-catalyzed DNA synthesis by a mechanism of delayed chain termination (40). Entecavir has only weak antiviral activity against HIV-1.
Additionally, nucleoside analogs substituted at the 4′-position have also been described previously (43–46). For example, 4′-azidothymidine and 4′-azidoadenosine both inhibit HIV-1 replication (46) although with potencies 200–2000-fold less than that of EFdA. Both 4′-azido nucleosides also have poor in vitro selectivity indices because of significant cytotoxicity. Azidothymidine-TP was shown to inhibit RT-catalyzed DNA synthesis by a type of delayed chain termination; incorporation of two sequential azidothymidine-MP molecules blocked DNA synthesis (44, 45). 4′-Methyl thymidine and 4′-ethyl thymidine both seem to cause pauses and stops in DNA synthesis at the point of incorporation (39). However, neither of these compounds has antiviral activity, because they cannot be phosphorylated by cellular nucleoside kinases. An analog of d4T that has a 4′-ethynyl substitution (Ed4T) is ∼10 times more active than the parent compound (47, 48). Because Ed4T lacks a 3′-OH, it inhibits RT as a conventional chain terminator. Interestingly, Ed4T is a better substrate than d4T for phosphorylation by human thymidine kinase 1 (47–50), a property that leads to its increased antiviral potency compared with d4T. The antiviral activity of Ed4T is ∼50-fold lower than that of EFdA.
In summary, EFdA is a TDRTI with two functionalities lacking in current therapeutic NRTIs, namely a 4′-ethynyl group and a 3′-OH. These additional properties impart superior antiviral activity to the compound and contribute to its mechanism of action, namely inhibition of primer translocation following EFdA-MP incorporation. This mechanism allows EFdA to act mainly as a de facto chain terminator of RT-catalyzed DNA synthesis, despite the presence of a 3′-OH. The present study validates RT nucleic acid translocation as a potential therapeutic target.
This work was supported, in whole or in part, by National Institutes of Health Grants AI076119, AI074389, AI076119-S1, and AI076119-02S1 (to S. G. S.) and AI079801 (to M. A. P.).
K. A. Kirby, K. Singh, E. Michailidis, B. Marchand, E. N. Kodama, E. Nagy, N. Ashida, H. Mitsuya, M. A. Parniak, and S. G. Sarafianos, unpublished data.
- NRTI
- nucleoside reverse transcriptase inhibitor
- TDRTI
- translocation-defective RT inhibitor
- RT
- reverse transcriptase
- HIV
- human immunodeficiency virus
- EFdA
- 4′-ethynyl-2-fluoro-2′-deoxyadenosine
- MP
- monophosphate
- TP
- triphosphate
- AZT
- azidothymidine
- EdA
- 4′-ethynyl-2′-deoxyadenosine
- EFddA
- 4′-ethynyl-2-fluoro-2′,3′-dideoxyadenosine
- EFd4A
- 4′-ethynyl-2-fluoro-2′,3′-dihydro-2′,3′-dideoxyadenosine
- Ed4T
- 4′-ethynyl-2′,3′-dihydro-3′-deoxythymidine
- TFV
- tenofovir
- PBMC
- peripheral blood mononuclear cell
- T/P
- template/primer
- T/PEFdA-MP or T/PddA-MP
- template/primer possessing either EFdA-MP or ddAMP at the 3′-primer terminus (or T/P chain terminated by EFdA or ddA)
- N-site
- nucleotide-binding site
- P-site
- primer site
- PDB
- Protein Data Bank
- d4T
- stavudine.
REFERENCES
- 1.Hammer S. M., Saag M. S., Schechter M., Montaner J. S., Schooley R. T., Jacobsen D. M., Thompson M. A., Carpenter C. C., Fischl M. A., Gazzard B. G., Gatell J. M., Hirsch M. S., Katzenstein D. A., Richman D. D., Vella S., Yeni P. G., Volberding P. A. (2006) Top. HIV Med. 14, 827–843 [PubMed] [Google Scholar]
- 2.Schinazi R. F., Hernandez-Santiago B. I., Hurwitz S. J. (2006) Antiviral Res. 71, 322–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parniak M. A., Sluis-Cremer N. (2000) Adv. Pharmacol. 49, 67–109 [DOI] [PubMed] [Google Scholar]
- 4.De Clercq E. (2007) Verh. K. Acad. Geneeskd. Belg. 69, 81–104 [PubMed] [Google Scholar]
- 5.Sluis-Cremer N., Tachedjian G. (2008) Virus Res. 134, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deval J. (2009) Drugs 69, 151–166 [DOI] [PubMed] [Google Scholar]
- 7.Sharma P. L., Nurpeisov V., Hernandez-Santiago B., Beltran T., Schinazi R. F. (2004) Curr. Top. Med. Chem. 4, 895–919 [DOI] [PubMed] [Google Scholar]
- 8.Sarafianos S. G., Marchand B., Das K., Himmel D. M., Parniak M. A., Hughes S. H., Arnold E. (2009) J. Mol. Biol. 385, 693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallois-Montbrun S., Schneider B., Chen Y., Giacomoni-Fernandes V., Mulard L., Morera S., Janin J., Deville-Bonne D., Veron M. (2002) J. Biol. Chem. 277, 39953–39959 [DOI] [PubMed] [Google Scholar]
- 10.Kodama E. I., Kohgo S., Kitano K., Machida H., Gatanaga H., Shigeta S., Matsuoka M., Ohrui H., Mitsuya H. (2001) Antimicrob. Agents Chemother. 45, 1539–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohrui H., Mitsuya H. (2001) Curr. Drug Targets Infect. Disord. 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 12.Ohrui H., Kohgo S., Hayakawa H., Kodama E., Matsuoka M., Nakata T., Mitsuya H. (2006) Nucleic Acids Symp. Ser. (Oxf.) 2006, 1–2 [DOI] [PubMed] [Google Scholar]
- 13.Kawamoto A., Kodama E., Sarafianos S. G., Sakagami Y., Kohgo S., Kitano K., Ashida N., Iwai Y., Hayakawa H., Nakata H., Mitsuya H., Arnold E., Matsuoka M. (2008) Int. J. Biochem. Cell Biol. 40, 2410–2420 [DOI] [PubMed] [Google Scholar]
- 14.White K. L., Chen J. M., Feng J. Y., Margot N. A., Ly J. K., Ray A. S., Macarthur H. L., McDermott M. J., Swaminathan S., Miller M. D. (2006) Antivir. Ther. 11, 155–163 [DOI] [PubMed] [Google Scholar]
- 15.Mascolini M., Larder B. A., Boucher C. A., Richman D. D., Mellors J. W. (2008) Antivir. Ther. 13, 1097–1113 [PubMed] [Google Scholar]
- 16.Le Grice S. F., Grüninger-Leitch F. (1990) Eur. J. Biochem. 187, 307–314 [DOI] [PubMed] [Google Scholar]
- 17.Meyer P. R., Matsuura S. E., So A. G., Scott W. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13471–13476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bøyum A., Løvhaug D., Tresland L., Nordlie E. M. (1991) Scand. J. Immunol. 34, 697–712 [DOI] [PubMed] [Google Scholar]
- 19.Marchand B., Götte M. (2003) J. Biol. Chem. 278, 35362–35372 [DOI] [PubMed] [Google Scholar]
- 20.Biaglow J. E., Kachur A. V. (1997) Radiat. Res. 148, 181–187 [PubMed] [Google Scholar]
- 21.Siddiqui M. A., Hughes S. H., Boyer P. L., Mitsuya H., Van Q. N., George C., Sarafinanos S. G., Marquez V. E. (2004) J. Med. Chem. 47, 5041–5048 [DOI] [PubMed] [Google Scholar]
- 22.Stewart J. J. P. (1989) J. Comput. Chem. 10, 209–220 [Google Scholar]
- 23.Hattori S., Ide K., Nakata H., Harada H., Suzu S., Ashida N., Kohgo S., Hayakawa H., Mitsuya H., Okada S. (2009) Antimicrob. Agents Chemother. 53, 3887–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quan Y., Liang C., Inouye P., Wainberg M. A. (1998) Nucleic Acids Res. 26, 5692–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong W., Lu C. D., Sharma S. K., Matsuura S., So A. G., Scott W. A. (1997) Biochemistry 36, 5749–5757 [DOI] [PubMed] [Google Scholar]
- 26.Sluis-Cremer N., Arion D., Parniak M. A. (2000) Cell. Mol. Life Sci. 57, 1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arion D., Kaushik N., McCormick S., Borkow G., Parniak M. A. (1998) Biochemistry 37, 15908–15917 [DOI] [PubMed] [Google Scholar]
- 28.Meyer P. R., Matsuura S. E., Mian A. M., So A. G., Scott W. A. (1999) Mol. Cell 4, 35–43 [DOI] [PubMed] [Google Scholar]
- 29.Sarafianos S. G., Clark A. D., Jr., Das K., Tuske S., Birktoft J. J., Ilankumaran P., Ramesha A. R., Sayer J. M., Jerina D. M., Boyer P. L., Hughes S. H., Arnold E. (2002) EMBO J. 21, 6614–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarafianos S. G., Clark A. D., Jr., Tuske S., Squire C. J., Das K., Sheng D., Ilankumaran P., Ramesha A. R., Kroth H., Sayer J. M., Jerina D. M., Boyer P. L., Hughes S. H., Arnold E. (2003) J. Biol. Chem. 278, 16280–16288 [DOI] [PubMed] [Google Scholar]
- 31.Tuske S., Sarafianos S. G., Clark A. D., Jr., Ding J., Naeger L. K., White K. L., Miller M. D., Gibbs C. S., Boyer P. L., Clark P., Wang G., Gaffney B. L., Jones R. A., Jerina D. M., Hughes S. H., Arnold E. (2004) Nat. Struct. Mol. Biol. 11, 469–474 [DOI] [PubMed] [Google Scholar]
- 32.Yang G., Wang J., Cheng Y., Dutschman G. E., Tanaka H., Baba M., Cheng Y. C. (2008) Antimicrob. Agents Chemother. 52, 2035–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sluis-Cremer N., Arion D., Parikh U., Koontz D., Schinazi R. F., Mellors J. W., Parniak M. A. (2005) J. Biol. Chem. 280, 29047–29052 [DOI] [PubMed] [Google Scholar]
- 34.Marchand B., White K. L., Ly J. K., Margot N. A., Wang R., McDermott M., Miller M. D., Götte M. (2007) Antimicrob. Agents Chemother. 51, 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng J. Y., Murakami E., Zorca S. M., Johnson A. A., Johnson K. A., Schinazi R. F., Furman P. A., Anderson K. S. (2004) Antimicrob. Agents Chemother. 48, 1300–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakata H., Amano M., Koh Y., Kodama E., Yang G., Bailey C. M., Kohgo S., Hayakawa H., Matsuoka M., Anderson K. S., Cheng Y. C., Mitsuya H. (2007) Antimicrob. Agents Chemother. 51, 2701–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strerath M., Cramer J., Restle T., Marx A. (2002) J. Am. Chem. Soc. 124, 11230–11231 [DOI] [PubMed] [Google Scholar]
- 38.Boyer P. L., Julias J. G., Marquez V. E., Hughes S. H. (2005) J. Mol. Biol. 345, 441–450 [DOI] [PubMed] [Google Scholar]
- 39.Boyer P. L., Julias J. G., Ambrose Z., Siddiqui M. A., Marquez V. E., Hughes S. H. (2007) J. Mol. Biol. 371, 873–882 [DOI] [PubMed] [Google Scholar]
- 40.Tchesnokov E. P., Obikhod A., Schinazi R. F., Götte M. (2008) J. Biol. Chem. 283, 34218–34228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summerer D., Marx A. (2005) Bioorg. Med. Chem. Lett. 15, 869–871 [DOI] [PubMed] [Google Scholar]
- 42.Di Pasquale F., Fischer D., Grohmann D., Restle T., Geyer A., Marx A. (2008) J. Am. Chem. Soc. 130, 10748–10757 [DOI] [PubMed] [Google Scholar]
- 43.Maag H., Rydzewski R. M., McRoberts M. J., Crawford-Ruth D., Verheyden J. P., Prisbe E. J. (1992) J. Med. Chem. 35, 1440–1451 [DOI] [PubMed] [Google Scholar]
- 44.Chen M. S., Suttmann R. T., Papp E., Cannon P. D., McRoberts M. J., Bach C., Copeland W. C., Wang T. S. (1993) Biochemistry 32, 6002–6010 [DOI] [PubMed] [Google Scholar]
- 45.Chen M. S., Suttmann R. T., Wu J. C., Prisbe E. J. (1992) J. Biol. Chem. 267, 257–260 [PubMed] [Google Scholar]
- 46.Maag H., Nelson J. T., Steiner J. L., Prisbe E. J. (1994) J. Med. Chem. 37, 431–438 [DOI] [PubMed] [Google Scholar]
- 47.Nitanda T., Wang X., Kumamoto H., Haraguchi K., Tanaka H., Cheng Y. C., Baba M. (2005) Antimicrob. Agents Chemother. 49, 3355–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka H., Haraguchi K., Kumamoto H., Baba M., Cheng Y. C. (2005) Antivir. Chem. Chemother. 16, 217–221 [DOI] [PubMed] [Google Scholar]
- 49.Hsu C. H., Hu R., Dutschman G. E., Yang G., Krishnan P., Tanaka H., Baba M., Cheng Y. C. (2007) Antimicrob. Agents Chemother. 51, 1687–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang G., Dutschman G. E., Wang C. J., Tanaka H., Baba M., Anderson K. S., Cheng Y. C. (2007) Antiviral Res. 73, 185–191 [DOI] [PubMed] [Google Scholar]