Abstract
Accumulating evidence suggests that the mitochondrial molecular chaperone heat shock protein 60 (hsp60) also can localize in extramitochondrial sites. However, direct evidence that hsp60 functions as a chaperone outside of mitochondria is presently lacking. A 60-kDa protein that is present in the plasma membrane of a human leukemic CD4+ CEM-SS T cell line and is phosphorylated by protein kinase A (PKA) was identified as hsp60. An 18-kDa plasma membrane-associated protein coimmunoprecipitated with hsp60 and was identified as histone 2B (H2B). Hsp60 physically associated with H2B when both molecules were in their dephospho forms. By contrast, PKA-catalyzed phosphorylation of both hsp60 and H2B caused dissociation of H2B from hsp60 and loss of H2B from the plasma membrane of intact T cells. These results suggest that (i) hsp60 and H2B can localize in the T cell plasma membrane; (ii) hsp60 functions as a molecular chaperone for H2B; and (iii) PKA-catalyzed phosphorylation of both hsp60 and H2B appears to regulate the attachment of H2B to hsp60. We propose a model in which phosphorylation/dephosphorylation regulates chaperoning of H2B by hsp60 in the plasma membrane.
The molecular chaperone heat shock protein 60 (hsp60) is well characterized in eukaryotic cells with regard to its function in protein folding and assembly of oligomeric protein complexes (1–5). In yeast and mammalian cells, hsp60 is encoded by a single copy nuclear gene and contains a mitochondrial targeting presequence not present in the mature protein (6, 7). This mitochondrial targeting sequence, together with earlier biochemical and immunofluorescence studies demonstrating that this protein is present mainly in the matrix compartment (6–9), suggests that hsp60 functions as a molecular chaperone exclusively within mitochondria (1–5).
In recent years, however, accumulating evidence also has revealed the presence of hsp60 in extramitochondrial sites. Detailed immunoelectron microscopic analyses using several well characterized anti-hsp60 antibodies unequivocally have identified hsp60 in endoplasmic reticulum, peroxisomes, and insulin secretory granules (10–13). Moreover, using flow cytometry and immunogold labeling with backscattered electron imaging, hsp60 also has been identified on the cell surfaces of Daudi Burkitt’s lymphoma cells (14) and human leukemic T cells (13, 15) as well as stressed endothelial cells (16). Additionally, hsp60 has been observed to associate with p21ras and the A-system amino acid transporter, plasma membrane-associated proteins (17–19). Lastly, the presence of hsp60 in the plasma membrane has been confirmed by the specific labeling of this protein on radioiodination or biotinylation of intact cell surface proteins (13, 14).
Although the above observations are suggestive that hsp60 may subserve a functional role in extramitochondrial sites, direct evidence that hsp60 can act as a molecular chaperone outside of mitochondria presently is lacking. In our work on T cell plasma membrane-associated proteins that are phosphorylated on activation of type I protein kinase A (PKA-I), a 60-kDa protein was identified that was phosphorylated specifically on activation of either human primary T lymphocytes (20, 21) or a human CD4+ leukemic T cell line, CEM-SS. In this paper, we present evidence that this plasma membrane-associated protein, which undergoes phosphorylation by PKA-I, is hsp60. We also present evidence that membrane-associated hsp60 interacts with another protein, histone 2B (H2B), and that this membrane association with H2B is regulated by the phosphorylation status of hsp60 and H2B proteins. These results provide evidence that plasma membrane-associated hsp60 may be serving as a chaperone for H2B. A model is proposed in which phosphorylation/dephosphorylation regulates chaperoning of H2B by hsp60.
METHODS
Cell Line.
The human leukemic CD4+ T cell line CEM-SS (22, 23) was grown in RPMI medium 1640 supplemented with 10 mM Hepes, 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% (vol/vol) heat-inactivated fetal calf serum. Cell viability was ≥98.7%.
Reagents.
Monoclonal IgG anti-recombinant human lymphocyte mitochondrial hsp60 antibody (mAb II-13) was used (24). NS1 is an isotype-specific antibody control (14). Affinity-purified rabbit anti-human histone 2B peptide antibody (Ab) was kindly provided by S. Shaunak (Royal Postgraduate Medical School, London, U.K.) (25). Affinity-purified, monospecific rabbit anti-bovine mitochondrial NADH:ubiquinone oxidoreductase complex (51-kDa catalytic subunit of Complex I) was a gift from Y. Hatefi (Scripps Research Institute, La Jolla, CA) (26). Fluorescein isothiocyanate-goat F(Ab)2 anti-mouse IgG and rhodamine-goat F(Ab)2 anti-rabbit IgG (Cappel) were used at a final dilution of 1:50. Isoelectric point (pI) markers were from Bio-Rad. N6,O2′-dibutyryl cyclic adenosine 3′,5′-monophosphate (bt2-cAMP); protein A-Sepharose CL-4B; isobutylmethylxanthine (IBMX); and okadaic acid were obtained from Sigma. Ampholines were purchased from Pharmacia. [32Pi]-orthophosphate and γ-[32P]ATP (3,000 Ci/mmol; 5 mCi/ml) were from DuPont/NEN. Rainbow [14C] methylated protein markers and the enhanced chemiluminescence (ECL) kit were from Amersham. The PKA inhibitors PKI-(5–24) and Rp diastereomer of adenosine cyclic 3′,5′-phosphorothioate (Rp-cAMPS) were from Peninsula Laboratories and BioLog Chemical (San Diego), respectively.
Isolation of Plasma Membrane Fractions from CEM-SS T Cells.
After gentle disruption of cells with a Dounce homogenizer, membrane fragments were prepared as described (27). Immunoblotting of isolated membrane fragments with monospecific anti-51-kDa Complex I Ab to detect NADH:ubiquinone oxidoreductase demonstrated the absence of any contamination by mitochondrial constituents.
Protein Phosphorylation.
Plasma membrane protein (60 μg) was incubated in the absence or presence of 50 nM purified PKA C-subunit at 30°C for 0 and 10 min as described (21). Control samples were incubated with either C-subunit plus 10 μM PKI-(5–24) for 10 min or with enzyme for 0 min (21, 28). Protein phosphorylation in intact cells was performed in the absence or presence of 500 μM Rp-cAMPS as described (29).
High resolution two-dimensional (2-D) SDS/PAGE was performed as detailed (21, 30, 31). After completion of the second dimension, proteins were electroblotted onto either an Immobilon-P or Immobilon-PSQ membrane. Autoradiograms and CBB-R-250-stained membrane were aligned to excise the relevant protein-containing spots from the membrane for sequence analysis.
Immunoprecipitation and Immunoblotting.
Immunoprecipitation of hsp60 was carried out by using mAb II-13 (1:20 dilution) as described (13). Bound proteins were eluted from the beads with Laemmli sample buffer by boiling for 5 min at 95°C if they were to be used for 1-D SDS/PAGE or with buffer A (9.5 M urea/2% [wt/vol] Triton X-100/1% [vol/vol] DTT/0.8% [vol/vol] of each ampholine) (final concentrations) at room temperature if they were to be used for 2-D SDS/PAGE. The eluted samples were loaded directly onto the gel.
Proteins fractionated by 1-D or 2-D SDS/PAGE were electroblotted onto an Immobilon membrane (Millipore). The membrane was blocked with Blotto (50 mM Tris⋅HCl, pH 7.4/100 mM NaCl/5% nonfat dry milk) for ECL-based detection for 1 hr. The membrane subsequently was incubated with 1:1,000 mAb II-13 or 1:250 anti-H2B Ab for 1 hr at room temperature and was washed with buffer B (50 mM Tris⋅HCl, pH 7.5/250 mM NaCl) (final concentrations). The membrane then was incubated for 1 hr with 1:5,000 peroxidase-conjugated anti-mouse IgG or with 1:4,500 peroxidase-conjugated anti-rabbit IgG diluted in Blotto. The blots were developed by ECL reagent.
To demonstrate that coimmunoprecipitation of H2B with hsp60 by mAb II-13 was specific, we performed two control experiments. First, immunoprecipitation of unphosphorylated plasma membrane fragments was performed in the absence or presence of mAb II-13 or with NS1. The immunoprecipitates were resolved on a 15% 1-D SDS/PAGE and were immunoblotted with anti-H2B antibody, and the blot was developed by ECL reagent. H2B was detected only in the lane in which mAb II-13 was used for immunoprecipitation (data not shown). This demonstrates the specificity of coimmunoprecipitation of H2B with hsp60. Second, we performed a competition experiment. Plasma membrane fragments were phosphorylated by PKA C-subunit as described above. Increasing concentrations of human recombinant hsp60 (rhsp60) (1–5 μg) were added simultaneously to the phosphorylation reaction mix, and the reaction mix was incubated for 10 min at 30°C. After incubation, the reaction mix was immunoprecipitated with mAb II-13. After resolving the immunoprecipitates on a gel and transferring to an Immobilon membrane, the membrane was immunoblotted with mAb II-13 and anti-H2B antibody and was developed by ECL reagent. Increasing concentrations of rhsp60 gradually competed away the association of dephosphoH2B with dephosphohsp60, demonstrating specificity of the interaction between these two molecules (data not shown).
Protein Sequencing.
Amino acid analyses and amino-terminal sequencing were performed by W. S. Lane (Harvard University, Cambridge, MA). Plasma membrane-associated p60 and rhsp60 were transferred to a poly(vinylidene difluoride) membrane for peptide mapping followed by matrix-assisted laser desorption ionization-time-of-flight (TOF) MS analysis (W. S. Lane). Sequence homologies of peptides derived from plasma membrane hsp60 were determined by using the blast server (http://www.ncbi.nlm.nih.gov) searching the Swiss Prot sequence databank (Release no. 35.0).
Protein Estimation.
Protein concentrations in samples were quantified by the method of Bradford (32).
Confocal Immunofluorescence Microscopy.
Samples were prepared by using standard procedures (33). The final dilution of anti-H2B antibody was 1:100 and of mAb II-13 was 1:4. Fluorescein isothiocyanate- and rhodamine-anti-antibodies were used to label mAb II-13 and anti-H2B antibody, respectively. Samples were examined with a confocal microscope (model TCS.NT, Leica, Deerfield, IL) equipped with an emission filter (590 nm for rhodamine and 515–565 nm for fluorescein isothiocyanate). Cells were analyzed in each field for both hsp60 and H2B.
RESULTS
Identification of Plasma Membrane-Associated Proteins Phosphorylated by the Purified PKA C-Subunit.
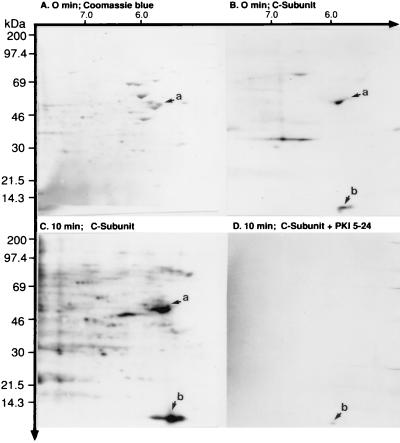
Primary human T cells and the CD4+ CEM-SS leukemic cell line constitutively express all of the R- and C-subunit isoform transcripts and have comparable PKA-I and PKA-II isozyme activities (data not shown). Previous studies have identified multiple plasma membrane-associated proteins from primary human T cells that are phosphorylated by PKA (20), including a 60-kDa protein (21). To determine whether a 60-kDa protein from the CEM-SS T cell plasma membrane is also a substrate of PKA, we used in vitro phosphorylation. Isolated plasma membrane fragments were incubated with 50 nM purified PKA C-subunit in the presence of γ-[32P]ATP for 0 and 10 min at 30°C. Using 2-D SDS/PAGE, we observed an array of CBB-stained proteins of differing Mrs and pI values (Fig. 1A). The Mrs varied between 10 and 200 kDa, and the pI values ranged between 3.5 and 10. Fig. 1 B and C shows autoradiograms at 0 and 10 min, respectively, after addition of purified C-subunit. At 0 min in the presence of C-subunit, basal phosphorylation of proteins was observed (Fig. 1B). By 10 min, an array of phosphoproteins were observed (Fig. 1C). These phosphoproteins were specific substrates of PKA, for their C-subunit-catalyzed phosphorylation could be competitively blocked by PKI-(5–24), a specific PKA peptide inhibitor (Fig. 1D) (21, 34). As anticipated, the Mrs and pI values were similar to CBB-stained proteins shown in Fig. 1A, suggesting that some of the T cell plasma membrane proteins may be substrates of PKA.
Figure 1.
In vitro PKA C-subunit-catalyzed phosphorylation of CEM-SS T cell plasma membrane-associated proteins. Phosphorylation was performed as described in the Methods section. The phosphorylated plasma membrane fragments were analyzed by 10% 2-D SDS/PAGE, the proteins were transblotted to Immobilon-PSQ membrane, the membranes were stained with CBB, and the blot was exposed to x-ray film. (A) CBB stain showing the distribution of plasma membrane-associated proteins. The arrow marked a points to a 60-kDa protein with a pI of 5.5. (B) Autoradiogram of the same blot at 0 min revealing constitutively phosphorylated substrates. The arrow marked a corresponds to the CBB-stained protein shown in A and reveals that the 60-kDa protein has a low level of constitutive phosphorylation. Arrow b points to a protein with an apparent Mr of 14 kDa and a pI of 5.5, is not stained by CBB in A, and is constitutively phosphorylated. (C) In vitro phosphorylation of plasma membrane-associated proteins by 50 nM purified PKA C-subunit over 10 min at 30°C. Arrows a and b reveal enhanced phosphorylation of the 60- and 14-kDa proteins compared with B. (D) Specific inhibition of C-subunit-catalyzed phosphorylation by PKI-(5–24).
Identification of Plasma Membrane-Associated Phosphoprotein 60 (pp60) as hsp60.
Coomassie blue staining of the plasma membrane revealed a discrete 60-kDa protein having a pI of 5.5 (Fig. 1A) that was phosphorylated specifically by the PKA C-subunit in vitro (Fig. 1C). This protein was termed phosphoprotein 60 (pp60). To determine whether pp60 was hsp60, we performed amino-terminal amino acid sequencing of the protein after its excision from the gel. The amino-terminal sequence was A K D V K F G A D A R A L M L Q G V D L. A search of the Swiss-Prot database demonstrated that this 60-kDa protein (termed p60 in its dephosphorylated state) shares 100% identity, in the first 20 amino-terminal amino acids that were analyzed, with hsp60 (7, 35).
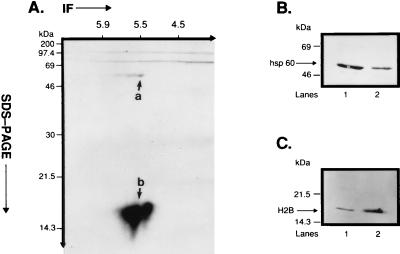
To confirm that p60 is hsp60, we immunoblotted enriched plasma membrane fragments derived from CEM-SS T cells with mAb II-13, a monoclonal antibody directed against human rhsp60 (24). Fig. 2B, lane 1 shows that mAb II-13 immunoblotted a 60-kDa protein. The estimated content of hsp60 protein is 4.85% of total CEM-SS T cell plasma membrane protein. This estimate was derived from multiple, individual samples immunoblotted with mAb II-13 and quantified by laser densitometry. As a positive control, mAb II-13 also immunoblotted the 60-kDa rhsp60 (Fig. 2B, lane 2). Taken together with the sequence analysis, these data confirm that p60 is hsp60. Moreover, these data verify our recent identification of hsp60 in the plasma membrane of CEM-SS T cells (13, 15).
Figure 2.
Immunoprecipitation of plasma membrane-associated phosphoproteins by mAb II-13 and immunoblotting of p60 and p18. (A) Isolated CEM-SS T cell plasma membrane fragments (60 μg protein) were phosphorylated in vitro by 50 nM PKA C-subunit in the presence of γ-[32P]ATP. In vitro phosphorylated fragments were immunoprecipitated with mAb II-13, and the immunoprecipitate was resolved on a 15% 2-D SDS/PAGE. Coimmunoprecipitation of phsp60 and pp18 was observed on the autoradiogram. Also notable was that both phsp60 and pp18 were comprised of isoforms whose pI values varied between ≈5.5 and 5.8. Differences in the relative intensity of phosphorylation of phsp60 and pp18 are addressed in Discussion. (B) Isolated CEM-SS T cell plasma membrane fragments (lane 1, 60 μg protein) and 1 μg of rhsp60 (lane 2) were run on a 15% 1-D SDS/PAGE and were immunoblotted with mAb II-13. (C) Isolated CEM-SS T cell plasma membrane fragments (lane 1, 60 μg protein) and 2 μg of purified human H2B (lane 2) were run on a 15% 1-D SDS/PAGE and were immunoblotted with affinity-purified polyclonal anti-H2B Ab.
Immunoprecipitation of Phosphorylated hsp60 (phsp60) by mAb II-13 Yields both phsp60 and Phosphorylated H2B (pH2B).
To probe the function of phsp60, we initially immunoprecipitated hsp60 phosphorylated in vitro by the PKA C-subunit from plasma membrane fragments with mAb II-13 and analyzed the immunoprecipitate by 10% 1-D SDS/PAGE. Phsp60 ran in an identical position as hsp60 immunoblotted with mAb II-13 (data not shown). Surprisingly, the autoradiogram also revealed a more heavily phosphorylated band that coimmunoprecipitated with phsp60, had a relative Mr < 14.3, and was not detected by CBB staining (data not shown).
To investigate the low molecular weight phosphoprotein further, phosphorylated plasma membrane fragments were immunoprecipitated with mAb II-13 and were resolved by 15% 2-D SDS/PAGE electrophoresis. Autoradiograms of gels showed both phsp60 and a low molecular weight phosphoprotein (Fig. 2A). The latter phosphoprotein had a Mr of 18 kDa on 15% 2-D SDS/PAGE and was, therefore, termed pp18. Of interest, phsp60 was comprised of a series of phosphoproteins migrating at 60 kDa with pI values varying between 5.5 and 5.8, suggesting the presence of isoforms (Fig. 2A, arrow a). Also notable was that pp18 was composed of at least two heavily phosphorylated proteins with pI values varying between 5.5 and 5.8 (Fig. 2A, arrow b). These two phosphoproteins also may be isoforms. Both phsp60 and pp18 share a predominant pI of ≈5.5. Thus, these experiments revealed the presence of an 18-kDa phosphoprotein that coimmunoprecipitates with phsp60.
To identify this 18-kDa protein, amino-terminal amino acid sequencing was performed. The amino-terminal sequence was P E P A K S A P A P K K. The Swiss Prot database search demonstrated 100% identity, in the first 12 N-terminal amino acids that were analyzed, with H2B. To verify that p18 is H2B, we immunoblotted plasma membrane fragments from CEM-SS T cells isolated on a 15% 1-D gel with an affinity-purified polyclonal rabbit anti-human H2B Ab (25). Fig. 2C, lane 1 demonstrates that anti-H2B immunoblotted an 18-kDa protein that ran at an identical position as purified human H2B (Fig. 2C, lane 2). In contrast to hsp60, it is estimated that H2B makes up only 0.95% of the total CEM-SS T cell plasma membrane. Taken together with the sequencing data, these experiments revealed that p18 is H2B.
Stoichiometry of Recombinant hsp60 Phosphorylation by PKA C-Subunit.
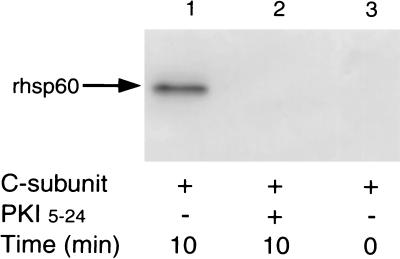
To verify PKA-catalyzed phosphorylation of hsp60 and to quantify the stoichiometry of phosphorylation, we used rhsp60 (24). In two independent experiments, phosphorylation of rhsp60 by PKA C-subunit peaked at 10 min and revealed a stoichiometry of 0.5 mole of radiolabeled phosphate per mole of rhsp60 at 10 min (Fig. 3). PKA-catalyzed phosphorylation could be inhibited competitively by 10 μM PKI-(5–24) (Fig. 3), demonstrating the specificity of PKA for rhsp60. Thus, using rhsp60, these data verify our findings in Fig. 2B and suggest that PKA-I effectively can phosphorylate hsp60.
Figure 3.
In vitro phosphorylation of rhsp60. rhsp60 (1 μg) was phosphorylated by 50 nM of purified PKA C-subunit, and phosphorylated rhsp60 was resolved by 10% 1-D SDS/PAGE. Resolved prhsp60 then was transferred to poly(vinylidene difluoride) membrane, and autoradiography was performed. Lanes: 1, 10 min without PKI-(5–24); lane 2, 10 min with 10 μM PKI-(5–24); lane 3, 0 min without PKI-(5–24).
cAMP-Dependent, PKA-I-Catalyzed Phosphorylation of Intact CEM-SS T Cells Fails to Identify Coimmunoprecipitation of pp60 and pH2B.
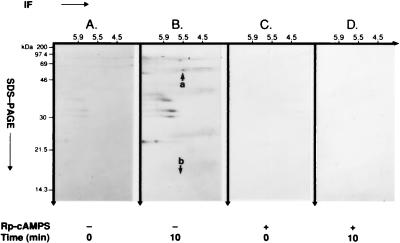
Previous experiments used plasma membrane fragments phosphorylated in vitro by purified PKA C-subunit to identify hsp60 and H2B as substrates of PKA. In complementary experiments, we asked whether phsp60 and pH2B are present and can be coimmunoprecipitated from isolated plasma membrane fragments after bt2-cAMP-stimulated, PKA-I isozyme-catalyzed phosphorylation in the intact CEM-SS T cell. Fig. 4A and B reveals the absence and presence of phsp60, respectively, on 2-D autoradiograms at 0 and 10 min after the addition of bt2-cAMP to intact cells. Although hsp60 makes up almost 5% of the total plasma membrane protein, the intensity of phosphorylation is limited by its stoichiometry. Fig. 4 C and D shows that Rp-cAMPS, a cell-permeant diastereomer inhibitor of cAMP (36), completely blocked phosphorylation of hsp60, demonstrating specificity of PKA-I-catalyzed phosphorylation of hsp60.
Figure 4.
cAMP-dependent, PKA-catalyzed phosphorylation of hsp60 in intact CEM-SS T cell isolated plasma membrane fragments. After loading with [32Pi]orthophosphate for 3 hr, cells were exposed to 2.5 mM bt2-cAMP in the presence of 200 μM isobutylmethylxanthine and 10 μM okadaic acid for 0 min and 10 min. Plasma membrane fragments then were isolated, and phsp60 was immunoprecipitated with mAb II-13. Immunoprecipitated pp60 was analyzed by 15% 2-D SDS/PAGE. (A) At 0 min, shows the absence of basal phosphorylation of hsp60. Note that other, unidentified phosphoproteins are coimmunoprecipitated with hsp60, but pH2B is not observed. (B) At 10 min, shows immunoprecipitated phsp60 (arrow a). Although other, unidentified phosphoproteins are coimmunoprecipitated with phsp60, pH2B is not observed (arrow b). (C) At 0 min with 500 μM Rp-cAMPS. (D) At 10 min with 500 μM Rp-cAMPS blocks cAMP-dependent, PKA-catalyzed phosphorylation of hsp60.
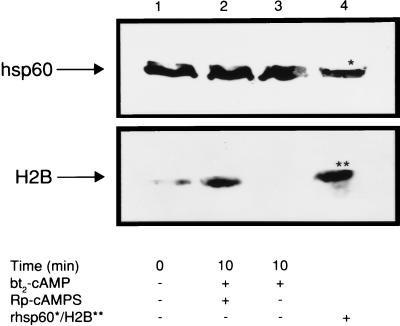
Based on our in vitro phosphorylation data demonstrating that pH2B coimmunoprecipitated with phsp60 (Fig. 2A), we conjectured that pH2B may be associated physically with phsp60 and that a similar finding would be observed after phosphorylation in intact cells. However, we did not observe coimmunoprecipitation of pH2B with phsp60 in plasma membrane after cAMP-dependent, PKA-I-catalyzed phosphorylation in intact T cells (Fig. 4B, arrow b). To determine the relationship between hsp60 and H2B before and after cAMP-dependent phosphorylation in intact T cells, we immunoblotted isolated plasma membrane fragments with mAb II-13 and anti-H2B Ab. Fig. 5, lane 1 reveals that, before cAMP-dependent, PKA-I-catalyzed phosphorylation of intact T cells, H2B is present in the plasma membrane; 10 min after phosphorylation, H2B has been lost from the plasma membrane (Fig. 5, lane 3). Thus, this loss of H2B is a rapid event. If cAMP-dependent, PKA-I-catalyzed phosphorylation is inhibited in the intact cell by Rp-cAMPS, H2B is not only present in the plasma membrane but accumulates 3.5-fold within 10 min over baseline (Fig. 5, compare lane 2 to lane 1). By contrast, there is no change in the amount of hsp60 in the plasma membrane (Fig. 5). That phosphorylation of both hsp60 and H2B by the PKA-I isozyme results in loss of H2B, but not hsp60, from the membrane suggests that phosphorylation may regulate chaperoning of H2B by hsp60.
Figure 5.
Effect of cAMP-dependent, PKA-catalyzed phosphorylation on the presence of H2B and hsp60 in the T cell plasma membrane. T cells were either pretreated or not pretreated with 500 μM Rp-cAMPS and subsequently were exposed to 2.5 mM bt2-cAMP for 0 or 10 min at 30°C in the absence or presence of Rp-cAMPS, and plasma membrane fragments were isolated. The membrane fragments were run on a 10% 1-D SDS/PAGE, were transferred, and were immunoblotted with polyclonal anti-H2B Ab or mAb II-13. Lanes: 1, constitutive H2B and hsp60; 2, immunoblots of pH2B and phsp60 at 10 min after initiation of phosphorylation in the presence of Rp-cAMPS; 3, immunoblots of pH2B and phsp60 at 10 min after initiation of phosphorylation in the absence of Rp-cAMPS; 4, rhsp60 and H2B controls.
Confocal Immunofluorescence Microscopy of Plasma Membrane hsp60 and H2B.
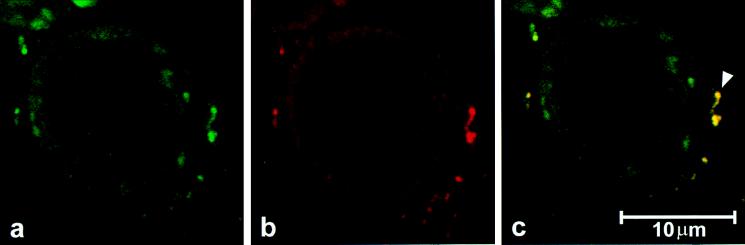
The data suggest that dephosphoH2B is associated with dephosphohsp60 in the plasma membrane. To confirm this association, we used confocal immunofluorescence microscopy. Fig. 6a and b demonstrate that both hsp60 and H2B are present in the plasma membrane, respectively. In Fig. 6c, computer-assisted superimposition of the images revealed colocalization of the two molecules. This observation supports the concept that H2B is associated with hsp60 in the plasma membrane.
Figure 6.
Confocal laser photomicrograph shows the distribution of (a) hsp60 (green) and (b) H2B (red) in the CEM-SS T cell. (c) An image generated by computer-assisted superimposing a and b. Colocalization of hsp60 and H2B is recognized as yellow (▹) in the representative cell.
Peptide Mapping and Mass Spectrometric Analysis of Plasma Membrane-Associated hsp60.
To determine whether plasma membrane-associated hsp60 is homologous to mitochondrial hsp60, we carried out peptide mapping of plasma membrane-associated hsp60. One-hundred and nineteen fractions were resolved by reverse-phase HPLC after Lys-C digestion of plasma membrane hsp60. By using rhsp60 as a control, candidate HPLC peptides were selected by matrix-assisted laser desorption ionization-TOF MS analysis on the basis of mass variation. Amino acid analysis of peptides 66, 72, and 92 was performed. By using the Swiss Prot database, peptides 66 (amino acids 555 to 569), 72 (amino acids 251 to 269), and 92 (amino acids 270 to 291) revealed 100% sequence homology with mitochondrial hsp60. Thus, the sequence of plasma membrane-associated hsp60 did not differ from mitochondrial hsp60.
DISCUSSION
Using several cell types, it has been demonstrated that hsp60 localizes in widely divergent subcellular compartments, including mitochondria, endoplasmic reticulum, peroxisomes, granules, and plasma membrane (2, 10, 13, 15, 16, 37–39). Among cell types in which hsp60 has been identified in the plasma membrane are T lymphocytes (13, 15). Although the role of hsp60 in the mitochondrion as a molecular chaperone responsible for membrane transport and protein folding is now well accepted (37, 38), its biological function in extramitochondrial sites, such as the plasma membrane, remains uncertain (13). The data presented herein verify that hsp60 is present in the T cell plasma membrane and demonstrate for the first time that its function as a molecular chaperone of H2B is regulated by PKA-I-catalyzed phosphorylation.
Our current experiments confirmed the presence of hsp60 in the CEM-SS T cell plasma membrane by four essential criteria. First, we demonstrated that hsp60 has an identical electrophoretic mobility on 2-D SDS/PAGE as rhsp60. Second, we immunoprecipitated hsp60 with mAb II-13, a monoclonal antibody specific for hsp60 (24). Third, we immunoblotted hsp60 with mAb II-13, demonstrating specificity. Finally, we performed peptide mapping and amino acid sequencing by matrix-assisted laser desorption ionization-TOF MS analysis, which revealed that there was 100% sequence identity between rhsp60 and T cell plasma membrane hsp60. Taken together with our previous identification of hsp60 by flow cytometry and immunogold labeling on backscattered electron imaging (13, 15), there is now incontrovertible evidence for the presence of hsp60 in the T cell plasma membrane.
We have observed that primary human T cells possess multiple plasma membrane-associated proteins that are substrates of PKA-I, including a 60-kDa protein (p60) (20, 21, 42). Because p60 proved to be hsp60, we reviewed its amino acid sequence (7) for the presence of serine and threonine residues. The presence of such residues predict that hsp60 should be phosphorylatable by PKA-I so long as these serine/threonine residues are in the general consensus sequence Arg-Arg-Xaa-Ser/Thr or Lys-Arg/Xaa-Ser/Thr (43). In fact, the amino acid sequence of hsp60 (7) contains only six potential phosphorylation sites: Lys72-Val73-Thr74 (KVT), Lys130-Ile131-Ser132 (KIS), Lys157-Gln158-Ser159 (KQS), Lys250-Ile251-Ser252 (KIS), Lys396-Leu397-Ser398 (KLS), and Lys469-Arg470-Thr471 (KRT). This relatively limited number of phosphorylatable motifs in hsp60 probably accounts for the lower intensity of phosphorylation compared with H2B on 2-D autoradiograms as well as the stoichiometry of 0.5 mole of radiolabeled phosphate per mole of rhsp60 at 10 min. Thus, this is the first demonstration that hsp60 can exist in both dephospho and phospho forms, and that it is phosphorylated by PKA-I.
Immunoprecipitation of phsp60 from plasma membrane fragments consistently coprecipitated two unidentified, heavily phosphorylated 18-kDa molecules with pI values between 5.5 and 5.8. We initially termed these phosphorylated molecules pp18. Coimmunoprecipitation of phsp60 and pp18 suggested that these structures may be associated physically in the plasma membrane. Amino-terminal amino acid sequencing unexpectedly revealed that p18 is H2B. The presence of H2B in the plasma membrane was verified by (i) its identical electrophoretic mobility on 1-D SDS/PAGE as purified human H2B; (ii) immunoprecipitation by an affinity-purified, polyclonal anti-H2B Ab (25); (iii) recognition of p18 by anti-H2B Ab on immunoblot; and (iv) amino acid sequencing. Although histone has a Mr of ≈14 and a pI of 11, phosphorylation shifts its charge and apparent Mr and may, therefore, explain both its apparent Mr of 18 and its pI of 5.5. Our identification of H2B in CEM-SS T cell plasma membrane fragments verified the recent recognition of this molecule in the plasma membrane of human primary T cells (25). Our results suggest, therefore, that hsp60 and H2B are associated physically in the T cell plasma membrane.
We observed an apparent dichotomy between in vitro and in vivo PKA-I-catalyzed phosphorylation of hsp60 and H2B. Immunoprecipitation of in vitro phosphorylated hsp60 in isolated plasma membrane fragments yielded a complex with pH2B. By contrast, pH2B did not coimmunoprecipitate with phsp60 after cAMP-dependent, PKA-I-catalyzed phosphorylation in intact T cells. Nevertheless, both dephosphohsp60 and dephosphoH2B are present in plasma membrane before PKA-I-catalyzed phosphorylation in intact cells, as demonstrated by immunoblotting. This apparent disparity may be accounted for if one proposes that dephosphoH2B is docked to dephosphohsp60 and that phosphorylation of both H2B and hsp60 by PKA-I in the intact cell plasma membrane results in separation of pH2B from phsp60 and rapid loss of the former from the plasma membrane. Within 10 min after phosphorylation, pH2B, but not phsp60, disappeared from the plasma membrane. By contrast, inhibition of PKA-I-catalyzed phosphorylation of hsp60 and H2B in the intact T cell by Rp-cAMPS actually resulted in a 3.5-fold increase of H2B, but not hsp60, protein in the plasma membrane. Compared with intact cells, in vitro phosphorylation of both hsp60 and H2B in isolated plasma membrane fragments does not appear to permit dissociation of pH2B from phsp60. Thus, both phosphoproteins are retained in the membrane fragments and can coimmunoprecipitate. We propose, therefore, that failure to observe coimmunoprecipitation of phsp60 and pH2B after phosphorylation in the intact cell may be caused by the transient nature of their association in the plasma membrane. It is conceivable that phosphorylation of both hsp60 and H2B in the intact cell rapidly severs their physical association, resulting in immediate expulsion of H2B from the plasma membrane so that the phosphorylated forms cannot be coimmunoprecipitated.
Our data are consistent with the concept that PKA-I-catalyzed phosphorylation regulates the association of hsp60 with H2B. Concomitant phosphorylation of hsp60 and H2B may alter the conformation of these molecules. By altering their conformation, phosphorylation of both hsp60 and H2B may be a mechanism to effect release of H2B from hsp60 and, potentially, to expel H2B from the plasma membrane. As we discussed above, this may account for why both phsp60 and pH2B are present in isolated plasma membrane fragments but not in plasma membrane fragments from intact cells.
Our proposal that hsp60 docks H2B is supported by three independent experiments. First, immunoprecipitation of dephosphohsp60 from isolated plasma membrane fragments by mAb II-13 and subsequent immunoblotting with anti-H2B antibody revealed the presence of H2B at the expected relative molecular mass. This finding suggests that hsp60 is associated with H2B and that this is a specific interaction. Second, competition of H2B by increasing concentrations of rhsp60 blocked immunoprecipitation of H2B by mAb II-13, further demonstrating specificity of the interaction between H2B and hsp60. Finally, confocal microscopy revealed colocalization of hsp60 and H2B in the plasma membrane. We interpret these data, therefore, to support the concept that dephosphohsp60 is associated with dephosphoH2B in the intact T cell plasma membrane.
Because hsp60 is a chaperonin (7, 44), we propose that it chaperones H2B within the plane of the plasma membrane. There is precedent for this proposal. In intact 70z cells, p21ras was found to complex transiently with hsp60 in the plasma membrane, where it appeared to act as a chaperone and to be integral to the function of p21ras as a GTP-binding signaling molecule (17). Second, hsp60 has been identified to associate with the A system of amino acid transporters, which are plasma membrane-associated proteins (18, 19). Thus, evidence supports the concept that hsp60 may be involved with protein handling in the plasma membrane as well as in mitochondria.
Our recognition that both hsp60 and H2B are concomitantly phosphorylated by PKA-I and that this post-translational modification appears to regulate the capacity of hsp60 to chaperone H2B in the T cell plasma membrane prompts a tentative model of the process. We propose that H2B is imported into the plasma membrane from the cytosol. However, the mechanism by which H2B is exported from the nucleus and traverses the cytosol remains to be elucidated. Once in the plasma membrane, we envisage that dephosphohsp60 docks dephosphoH2B and that hsp60 then chaperones H2B within the plane of the plasma membrane. PKA-I-catalyzed phosphorylation of both hsp60 and H2B then would result in the release of H2B from hsp60 and its expulsion from the plasma membrane, possibly into the extracellular milieu. After dephosphorylation of hsp60 by a phosphatase(s), hsp60 then would be able to repeat the cycle. Whether phosphorylation of H2B by PKA-I can occur before its entry into the plasma membrane and prevent its import is being investigated. If so, it is conceivable that the loss of pH2B from the plasma membrane after its phosphorylation is a product of both expulsion from the membrane as well as inhibition of its import into the membrane. Thus, this phosphorylation/dephosphorylation cycle is a newly identified cycle whose biological significance may be disposal of H2B from the plasma membrane.
We have documented the presence of both hsp60 and H2B in a seemingly unusual site: the plasma membrane. Hsp60 generally is regarded as a mitochondrial chaperonin whereas H2B is a component of nuclear chromatin. In addition to our localization of hsp60 to extramitochondrial sites (10, 13, 15), others also have observed hsp60 in similar subcellular compartments (14, 45). Our recent work also has revealed that heat shock as well as other cellular stresses significantly enhance the epitope density of hsp60 on T cells (unpublished data). That hsp60 also has been detected on the surfaces of live Chinese hamster ovaries, Daudi lymphoma, and aortic endothelial cells (13, 14, 16) by various methods raises the possibility that this chaperonin also may serve as a receptor. Although the presence of both hsp60 and H2B in plasma membrane initially may seem unusual and even surprising, other mitochondrial and cytoplasmic proteins, including aspartate aminotransferase (46), early pregnancy factor (47), transforming growth factor-β1 (48), actin, tubulin, glycolytic enzymes, glycosyltransferases, and kinases (49) also have been identified in this unconventional site. It is anticipated that novel, physiologic functions for these proteins will be identified as organizational, structural, and regulatory systems are elucidated.
Acknowledgments
We thank Drs. Greg Shelness and Mark Lively for their invaluable discussions during the course of this work and Danielle Mathis for dedicated technical assistance. This work was supported by a grant from the National Institutes of Health (AR-39501) (to G.M.K.); a Wake Forest University School of Medicine Research Support Grant (2127-741-2096); a pilot grant from the National Institutes of Health Wake Forest University Comprehensive Cancer Center Grant (CA-12197-22); a grant from the National Cancer Institute (5 P30 CA12197-21, 21S1); and a grant from the North Carolina Biotechnology Center (9510-IDG-1006).
ABBREVIATIONS
- PKA-I
type I protein kinase A
- bt2-cAMP
N6,O2′-dibutyryl cyclic adenosine 3′,5′-monophosphate
- Rp-cAMPS
Rp diastereomer of adenosine cyclic 3′,5′-phosphorothioate
- hsp60
60-kDa heat shock protein
- H2B
histone 2B
- phsp60
phosphorylated hsp60
- pH2B
phosphorylated H2B
- CBB
Coomassie brilliant blue
- rhsp60
human recombinant hsp60
- TOF
time-of-flight
- pp60
phosphoprotein 60
- Ab
antibody
- 2-D
two-dimensional
References
- 1.Rothman J E. Cell. 1989;59:591–601. doi: 10.1016/0092-8674(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 2.Gething M-J, Sambrook J. Nature (London) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 3.Ellis R J, van der Vies S M. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- 4.Hartl F-U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 5.Neupert W. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 6.Reading D S, Hallberg R L, Myers A M. Nature (London) 1988;337:655–659. doi: 10.1038/337655a0. [DOI] [PubMed] [Google Scholar]
- 7.Jindal S, Dudani A K, Singh B, Harley C B, Gupta R S. Mol Cell Biol. 1989;9:2279–2283. doi: 10.1128/mcb.9.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMullin T W, Hallberg R L. Mol Cell Biol. 1988;8:371–380. doi: 10.1128/mcb.8.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta R S. Biochem Cell Biol. 1990;68:1352–1363. doi: 10.1139/o90-198. [DOI] [PubMed] [Google Scholar]
- 10.Brudzynski K, Martinez V, Gupta R S. Diabetologia. 1992;35:316–324. doi: 10.1007/BF00401198. [DOI] [PubMed] [Google Scholar]
- 11.Velez-Granell C S, Arias A E, Torres-Ruiz J A, Bendayan M. J Cell Sci. 1994;107:539–549. doi: 10.1242/jcs.107.3.539. [DOI] [PubMed] [Google Scholar]
- 12.Velez-Granell C S, Arias A E, Torres-Ruiz J A, Bendayan M. Biol Cell. 1995;85:67–75. doi: 10.1111/j.1768-322x.1995.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 13.Soltys B J, Gupta R S. Exp Cell Res. 1996;222:16–27. doi: 10.1006/excr.1996.0003. [DOI] [PubMed] [Google Scholar]
- 14.Kaur I, Voss S D, Gupta R S, Schell K, Fisch P, Sondel P M. J Immunol. 1993;150:2046–2055. [PubMed] [Google Scholar]
- 15.Soltys B J, Gupta R S. Cell Biol Int. 1997;21:315–320. doi: 10.1006/cbir.1997.0144. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Schett G, Seitz C S, Hu Y, Gupta R S, Wick G. Circ Res. 1994;75:1078–1085. doi: 10.1161/01.res.75.6.1078. [DOI] [PubMed] [Google Scholar]
- 17.Ikawa S, Weinberg R A. Proc Natl Acad Sci USA. 1992;89:2012–2016. doi: 10.1073/pnas.89.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones M, Gupta R S, Englesberg E. Proc Natl Acad Sci USA. 1994;91:858–862. doi: 10.1073/pnas.91.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodlock T J, Chen X, Young D A, Bethlendy G, Lichtman M A, Segel G B. Arch Biochem Biophys. 1997;338:50–56. doi: 10.1006/abbi.1996.9798. [DOI] [PubMed] [Google Scholar]
- 20.Laxminarayana D, Berrada A, Kammer G M. J Clin Invest. 1993;92:2207–2214. doi: 10.1172/JCI116823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kammer G M, Khan I U, Malemud C J. J Clin Invest. 1994;94:422–430. doi: 10.1172/JCI117340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foley G E, Lazarus H, Farber S, Uzman B G, Boone B A, McCarthy R E. Cancer. 1965;18:522–529. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 23.Nara P L, Hatch W C, Dunlop N M, Robey W G, Arthur L O, Gonda M A, Fischunger P J. AIDS Res Hum Retroviruses. 1987;3:287–302. doi: 10.1089/aid.1987.3.283. [DOI] [PubMed] [Google Scholar]
- 24.Singh B, Gupta R S. DNA Cell Biol. 1992;11:489–496. doi: 10.1089/dna.1992.11.489. [DOI] [PubMed] [Google Scholar]
- 25.Watson K, Edwards R J, Shaunak S, Parmelee D C, Sarraf C, Gooderham N J, Davies D S. Biochem Pharmacol. 1995;50:299–309. doi: 10.1016/0006-2952(95)00142-m. [DOI] [PubMed] [Google Scholar]
- 26.Han A-L, Yagi T, Hatefi Y. Arch Biochem Biophys. 1988;267:490–496. doi: 10.1016/0003-9861(88)90055-0. [DOI] [PubMed] [Google Scholar]
- 27.Hasler P, Moore J J, Kammer G M. FASEB J. 1992;6:2735–2741. doi: 10.1096/fasebj.6.9.1319361. [DOI] [PubMed] [Google Scholar]
- 28.Kemp B E, Pearson R B, House C M. Methods Enzymol. 1991;201:287–304. doi: 10.1016/0076-6879(91)01026-x. [DOI] [PubMed] [Google Scholar]
- 29.Hasler P, Schultz L A, Kammer G M. Proc Natl Acad Sci USA. 1990;87:1978–1982. doi: 10.1073/pnas.87.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Farrell P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 31.Wallin R, Culp E N, Coleman D B, Goodman S R. Proc Natl Acad Sci USA. 1984;81:4095–4099. doi: 10.1073/pnas.81.13.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Ojcius D M, Niedergang F, Subtil A, Hellio R, Dautry-Varsat A. Res Immunol. 1996;147:175–188. doi: 10.1016/0923-2494(96)83169-5. [DOI] [PubMed] [Google Scholar]
- 34.Kemp B E, Parker M W, Hu S, Tiganis T, House C. Trends Biochem Sci. 1994;19:440–444. doi: 10.1016/0968-0004(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 35.Waldinger D, Eckerskorn C, Lottspeich F, Cleve H. Biol Chem Hoppe-Seyler. 1988;369:1185–1189. doi: 10.1515/bchm3.1988.369.2.1185. [DOI] [PubMed] [Google Scholar]
- 36.Rothermel J D, Perillo N L, Marks J S, Botelho L H P. J Biol Chem. 1984;259:15294–15300. [PubMed] [Google Scholar]
- 37.Abdel-Ghany M, El-Gendy K, Zhang S, Racker E. Proc Natl Acad Sci USA. 1990;87:7061–7065. doi: 10.1073/pnas.87.18.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen M Y, Hartl F-U, Horwich A L. Nature (London) 1990;348:455–458. doi: 10.1038/348455a0. [DOI] [PubMed] [Google Scholar]
- 39.Fisch P, Malkovsky M, Kovats S, Sturm E, Braakman E, Klein B S, Voss S D, Morrisey L W, DeMars R, Welch W J, et al. Science. 1990;250:1269–1273. doi: 10.1126/science.1978758. [DOI] [PubMed] [Google Scholar]
- 40.Cheng M, Hartl F-U, Martin J, Pollock R, Kabousek F, Neupert W, Hallberg E, Hallberg R L, Horwich A. Nature (London) 1989;337:620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- 41.Langer T, Neupert W. Curr Top Microbiol Immunol. 1991;167:3–30. doi: 10.1007/978-3-642-75875-1_1. [DOI] [PubMed] [Google Scholar]
- 42.Laxminarayana D, Kammer G M. J Immunol. 1996;156:497–506. [PubMed] [Google Scholar]
- 43.Edelman A M, Blumenthal D K, Krebs E G. Annu Rev Biochem. 1987;56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- 44.Gupta R S. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 45.Le Gall I M, Bendayan M. J Histochem Cytochem. 1996;44:743–749. doi: 10.1177/44.7.8675995. [DOI] [PubMed] [Google Scholar]
- 46.Betting J, Seufert W. J Biol Chem. 1996;271:25790–25796. doi: 10.1074/jbc.271.42.25790. [DOI] [PubMed] [Google Scholar]
- 47.Elder M E, Lin D, Clever J, Chan A C, Hope T J, Weiss A, Parslow T G. Science. 1994;264:1595–1599. doi: 10.1126/science.8202712. [DOI] [PubMed] [Google Scholar]
- 48.Maicas E, Pluthero F G, Freissen F G. Mol Cell Biol. 1988;8:169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smalheiser N R. Mol Biol Cell. 1996;7:1003–1014. doi: 10.1091/mbc.7.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]