Abstract
Homocitrate synthase (HCS) catalyzes the first and committed step in lysine biosynthesis in many fungi and certain Archaea and is a potential target for antifungal drugs. Here we report the crystal structure of the HCS apoenzyme from Schizosaccharomyces pombe and two distinct structures of the enzyme in complex with the substrate 2-oxoglutarate (2-OG). The structures reveal that HCS forms an intertwined homodimer stabilized by domain-swapping between the N- and C-terminal domains of each monomer. The N-terminal catalytic domain is composed of a TIM barrel fold in which 2-OG binds via hydrogen bonds and coordination to the active site divalent metal ion, whereas the C-terminal domain is composed of mixed α/β topology. In the structures of the HCS apoenzyme and one of the 2-OG binary complexes, a lid motif from the C-terminal domain occludes the entrance to the active site of the neighboring monomer, whereas in the second 2-OG complex the lid is disordered, suggesting that it regulates substrate access to the active site through its apparent flexibility. Mutations of the active site residues involved in 2-OG binding or implicated in acid-base catalysis impair or abolish activity in vitro and in vivo. Together, these results yield new insights into the structure and catalytic mechanism of HCSs and furnish a platform for developing HCS-selective inhibitors.
Introduction
Many organisms synthesize lysine utilizing one of two distinct biosynthetic pathways. In plants and most bacteria, lysine is synthesized via the diaminopimelate pathway (1). Yeast and other fungi, including the human pathogens Cryptococcus neoformans, Candida albicans, and Aspergillus fumigatus, and certain archaebacteria, including Thermus thermophilus, utilize the α-aminoadipate (AAA)5 pathway to synthesize lysine (2–4). Because mammals are lysine auxotrophs, the enzymes composing the fungal AAA pathway represent excellent targets for small molecule inhibitors that may hold potential clinical value as broad-spectrum anti-fungal therapeutics, particularly for immunocompromised individuals, such as AIDS and cancer patients and transplant recipients, who are at a higher risk of developing invasive fungal infections (5).
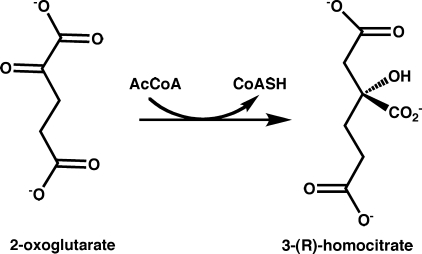
The AAA lysine biosynthetic pathway in fungi and yeast consists of eight enzymatic steps. Homocitrate synthase (HCS; EC 2.3.3.14) catalyzes the first and committed step in this pathway by transferring an acetyl group from acetyl-coenzyme A (AcCoA) to 2-oxoglutarate (2-OG) to form homocitrate and CoA (Fig. 1) (6). The kinetic mechanism of Saccharomyces cerevisiae HCS (ScHCS) Lys20 has been reported to obey an ordered Bi-Bi model in which 2-OG binding precedes AcCoA followed by the sequential release of the products CoA and homocitrate (7). The proposed reaction mechanism of HCS proceeds via a mixed aldol Claisen condensation that involves enzyme acid- and base-catalyzed steps (8), which is the same mechanism used by the citric acid cycle enzyme citrate synthase (CS) (9). Insights into this mechanism have been derived from analyses of the crystal structure of Mycobacterium tuberculosis α-isopropylmalate synthase (α-IPMS) encoded by the LeuA gene. α-IPMS shares homology to HCSs and catalyzes the chemically analogous first step in leucine biosynthesis by condensing 2-oxoisovalerate (2-OIV) and AcCoA to form isopropylmalate and CoA (10). Based on the structure of α-IPMS, mutagenesis and kinetic studies of ScHCS Lys20 have implicated a glutamate-histidine catalytic dyad in deprotonation of the acetyl group of AcCoA during the first step in catalysis (11). However, the absence of structural data for HCS have precluded definitive identification of the active residues involved in substrate binding and catalysis, limiting our understanding of the catalytic mechanism and regulation of these enzymes.
FIGURE 1.
Schematic of the reaction catalyzed by homocitrate synthase.
Here we describe the first crystal structure of a fungal HCS from Schizosaccharomyces pombe (SpHCS) as well as two distinct binary complexes of the enzyme bound to the substrate 2-OG. In the structure of one of the SpHCS·2-OG complexes, a lid motif obstructs the entrance to the active site within the TIM barrel, gating substrate binding to the enzyme. Steady state kinetic analysis and in vivo growth assays of wild type (WT) SpHCS and active site mutants reveal the contributions of these residues to substrate binding and catalysis. Taken together, our findings yield new insights into the mechanism of HCS and provide a basis for developing antifungal modulators of HCS.
EXPERIMENTAL PROCEDURES
Cloning
Full-length HCS gene encoded by the lys4+ gene was amplified from the S. pombe genomic clone SPBC1105.02c (Sanger Institute) and was subsequently subcloned into the parallel expression vector pHIS2, which contains an N-terminal His6 tag and a tobacco etch virus protease cleavage site, using BamHI and EcoRI (12). The lys4+ mutants were engineered using the QuikChange site-directed mutagenesis kit (Stratagene) and were confirmed by dideoxynucleotide sequencing.
Expression and Purification
SpHCS was overexpressed in Escherichia coli Rosetta2 (DE3) cells (EMD Biosciences), induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside, and grown at 18 °C overnight. Cells were lysed by adding 5 mg of lysozyme followed by sonication. The soluble enzyme was loaded onto either a Talon (Clontech) Co(II) immobilized metal affinity chromatography (IMAC) column (for crystallization) or a Zn(II)-charged IMAC-Sepharose (GE Healthcare) column (for kinetic studies) pre-equilibrated in 50 mm sodium phosphate, pH 7.0, 500 mm NaCl, and 5 mm β-mercaptoethanol and eluted with a linear gradient of 0–500 mm imidazole. The His6 tag was removed using tobacco etch virus protease during dialysis overnight against 50 mm sodium phosphate, pH 8.0, 150 mm NaCl, and 5 mm β-mercaptoethanol, and the tobacco etch virus protease was removed by batch binding to 5 ml of IMAC resin for 1 h. SpHCS was further purified by gel filtration chromatography using a Superdex 200 column (GE Healthcare) equilibrated in 25 mm Tris, pH 9.0, 50 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine. After purification, the protein was judged to be essentially pure by SDS-PAGE. In some instances, SpHCS was treated with 10 mm EDTA before gel filtration to remove the bound metal before crystallization trials. The SpHCS mutants were purified on the Zn(II)-charged IMAC-Sepharose column and gel-filtered as described for WT enzyme.
Inductively Coupled Plasma Mass Spectrometry
WT SpHCS from two different purifications on Zn(II)-charged IMAC-Sepharose was diluted to 20 μm in 50 mm HEPES, pH 7.5. Metal content was analyzed using a ThermoFisher Finnigan Element high resolution inductively coupled plasma mass spectrometer (University of Michigan, Dept. of Geology) with the protein buffer analyzed as a control. Metal standard calibration curves were performed at 0–100 ppb.
Crystallization and Structure Determination
Selenomethionyl (SeMet) SpHCS was prepared following a modified protocol of Doublié (13) and was overexpressed and purified as described for WT SpHCS. SeMet apoenzyme crystals were obtained in 100 mm HEPES, pH 6.8–7.2, 200 mm magnesium acetate, 16–25% polyethylene glycol 400 at 24 °C using 24 mg/ml SeMet-SpHCS. A SeMet single wavelength anomalous diffraction experiment was conducted at the 23-ID-B beamline of General Medicine and Cancer Institutes Collaborative Access Team at the Advanced Photon Source Synchrotron, and a complete high resolution data set was also collected to 2.24 Å using a second SeMet-SpHCS crystal (Table 1). Data were processed and scaled using HKL2000 (14). Eight initial selenium sites were located in a P6222 space group with SOLVE/ RESOLVE (15), and AutoSHARP (16) was then used to refine the heavy atom sites, calculate and refine phases, perform density modification, and build a partial model. The remainder of the model was built using Coot (17) and refined using CNS (18). The resulting model was then used for molecular replacement with the high resolution SeMet-SpHCS data set in MOLREP (19), and the apoenzyme model was built and refined using Coot and REFMAC (20), respectively. During refinement it was discovered that the SpHCS structures are not perfect homodimers. The R-factors remained high during refinement in the higher symmetry space group P6222 because the crystallographic 2-fold axis is forced to coincide with the pseudo-symmetry of the molecular 2-fold axis of the dimer, masking subtle differences between the two monomers. This problem was resolved by refining in the lower space group P62 in which the entire homodimer was refined, allowing for minor variations in the conformation of each monomer. In the later stages of refinement, residues with alternative conformations were modeled, TLS refinement (21) was applied, and water molecules were added. Crystals of the SpHCS closed lid complex with 2-OG were obtained using 20 mg/ml EDTA-treated SpHCS, 1 mm Zn(II), and 50 mm 2-OG, whereas crystals of the SpHCS open lid 2-OG complex were grown using 24 mg/ml enzyme and 50 mm 2-OG. Crystals of both complexes were grown in 1.2–1.6 m sodium and potassium phosphate, pH 5.4–6.8, at 24 °C. The data sets were processed and scaled in space group P62, and the refined high resolution SeMet-SpHCS structure was used as a model for molecular replacement. The SpHCS complexes were built and refined as outlined for the SeMet-SpHCS apoenzyme structure. All structures were validated using MolProbity (22), and no residues were present in the disallowed regions of the Ramachandran plot (Table 1). Simulated annealing omits maps for 2-OG were calculated in CNS (18).
TABLE 1.
Data collection and refinement statistics
| SeMet SpHCS SAD (Co(II)) | SeMet SpHCS apoenzyme (Co(II)) | WT SpHCS/2OG closed lid (Zn(II)) | WT SpHCS/2OG open lid (Co(II)) | |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | 23-ID-B | 23-ID-B | 21-ID-G | 21-ID-G |
| Space group | P6222 | P62 | P62 | P62 |
| Cell dimensions a = b, c (Å) | 136.9, 120.9 | 136.0, 122.1 | 133.7, 125.9 | 133.8, 125.7 |
| Wavelength (Å) | 0.97913 | 0.97930 | 0.97856 | 0.97872 |
| Resolution (Å) | 40.0-2.70 (2.80-2.70)a | 50.0-2.24 (2.33-2.24) | 40.0-2.67 (2.78-2.67) | 40.0-2.72 (2.82-2.72) |
| Rmerge (%) | 7.9 (32.5) | 6.0 (54.0) | 6.0 (51.2) | 5.6 (49.5) |
| I/σI | 48.3 (15.4) | 28.1 (3.67) | 35.4 (4.54) | 22.2 (2.54) |
| Reflections | ||||
| Total | 657,489 | 446,827 | 343,456 | 125,123 |
| Unique | 18,936 | 60,860 | 36,145 | 33,176 |
| Redundancy | 34.7 (31.7) | 7.3 (7.0) | 9.5 (8.8) | 3.8 (3.7) |
| Completeness (%) | 100.0 (100.0) | 99.4 (99.7) | 100.0 (100.0) | 96.4 (98.5) |
| Phasing power | 2.789 | |||
| Sites found/total | 8/9 | |||
| FOMb | 0.4727/0.8464 | |||
| Refinement | ||||
| Resolution range (Å) | 29.45-2.24 | 39.4-2.67 | 39.41-2.72 | |
| No. of reflections | 60,728 | 36,061 | 31,491 | |
| No. of atoms | 6064 | 6324 | 5935 | |
| Protein atoms | 5788 | 6149 | 5794 | |
| Metals/ligand atoms | 4/– | 4/20 | 4/20 | |
| Water atoms | 272 | 151 | 117 | |
| Rwork/Rfreec | 19.1/22.1 | 16.5/20.7 | 17.9/21.9 | |
| Average B-factors (Å2) | ||||
| Overall | 54.5 | 54.5 | 63.3 | |
| Protein | 55.1 | 55.7 | 66.2 | |
| Metals/ligand | 54.9/– | 38.6/57.8 | 51.1/78.7 | |
| Water | 59.0 | 56.3 | 63.5 | |
| Root mean square deviation | ||||
| Bond length (Å) | 0.012 | 0.013 | 0.013 | |
| Bond angles (o) | 1.294 | 1.436 | 1.472 | |
| MolProbity score | 1.93d | 2.09e | 2.28f | |
| Ramachandran favored (%) | 96.69 | 96.20 | 96.43 | |
| Ramachandran allowed (%) | 3.31 | 3.80 | 3.57 | |
| Ramachandran outliers (%) | 0.00 | 0.00 | 0.00 | |
a Values in parentheses are for the highest resolution shell.
b Before and after density modification in SHARP.
c Rwork = Σ‖Fo| − | Fc‖/Σ|Fo|; Rfree = ΣT‖Fo| − |Fc‖/ΣT|Fo|, where T is a randomly chosen set of reflections consisting of 5% of the total reflections.
d 89th percentile for 1.99–2.49 Å structures.
e 97th percentile for 2.42–2.92 Å structures.
f 94th percentile for 2.47–2.97 Å structures.
Kinetics Studies
A fluorescent assay for AcCoA-dependent acetyltransferases (23) was used to determine the steady state kinetic parameters of WT SpHCS and its mutants. CoA production was measured using the sulfhydryl-sensitive fluorophore ThioGlo 1 (10-(2,5-dihydro-2,5-di-oxo-1H-pyrrol-1-yl)-9-methoxy-3-oxo-methyl ester 3H-naphthol(2,1-b)pyran-S-carboxylic acid) as previously described (24). HCS assays were performed in triplicate at room temperature in 100 mm HEPES, pH 7.5, using 10–100 nm SpHCS and were run for 3 min. To determine Km and kcat values for AcCoA, the 2-OG concentration was held at a saturating concentration, and the AcCoA concentration was varied from 0 to 400 μm. To determine Km and kcat values for 2-OG, the AcCoA concentration was held at a saturating concentration, and the 2-OG concentration was varied from 0 to 200 mm. To correct for the increased fluorescence of the ThioGlo 1 dye with increasing concentrations of 2-OG, CoA (ϵ260 nm = 15,400 m−1 cm−1) calibration curves were preformed at every 2-OG concentration used in each assay. Initial velocities were then plotted versus substrate concentration, and the Michaelis-Menten equation was fit to the data using SigmaPlot (Systat Software) or Prism (GraphPad Software) to derive the Km and kcat values. The folding of the inactive HCS mutants (Table 2) was verified by CD spectroscopy (supplemental Fig. S1).
TABLE 2.
Kinetic analysis of WT SpHCS and its active site mutants
NA indicates that no detectable activity was measured using 100 nm mutant assayed with 200 μm AcCoA and 50 mm 2-OG.
| Km AcCoA | kcat varying AcCoA | kcat/Km | Km 2-OG | kcat varying 2-OG | Kcat/Km | |
|---|---|---|---|---|---|---|
| μm | min−1 | min−1 μm−1 | mm | min−1 | min−1 mm−1 | |
| WT | 10.7 ± 0.6a | 299 ± 7 | 28.0 ± 1.7 | 0.159 ± 0.015 | 308 ± 4 | 1940 ± 190 |
| R43A | NA | NA | NA | NA | NA | NA |
| R43K | NA | NA | NA | NA | NA | NA |
| R43Q | NA | NA | NA | NA | NA | NA |
| Q47A | 57.6 ± 12.6 | 35.0 ± 0.1 | 0.608 ± 0.133 | 4.94 ± 1.28 | 33.5 ± 0.08 | 6.78 ± 1.76 |
| E74A | NA | NA | NA | NA | NA | NA |
| E74Q | 30.7 ± 3.0 | 44.5 ± 1.4 | 1.45 ± 0.15 | 0.791 ± 0.081 | 46.7 ± 1.7 | 59.0 ± 6.3 |
| H103A | 35.2 ± 5.7 | 20.0 ± 0.9 | 0.568 ± 0.096 | 3.56 ± 0.61 | 18.2 ± 0.7 | 5.11 ± 0.35 |
| R163A | NA | NA | NA | NA | NA | NA |
| R163K | 146 ± 28b | 33.9 ± 2.8b | 0.232 ± 0.048 | 24.4 ± 4.3 | 26.1 ± 1.4c | 1.07 ± 0.20c |
| R163Q | NA | NA | NA | NA | NA | NA |
| S165A | 18.6 ± 2.3 | 158 ± 5.8 | 8.5 ± 1.0 | 0.536 ± 0.070 | 160 ± 6 | 298 ± 40 |
| E167A | NA | NA | NA | NA | NA | NA |
| E167Q | NA | NA | NA | NA | NA | NA |
| T197A | 7.44 ± 0.35 | 10.7 ± 0.1 | 1.44 ± 0.07 | 0.341 ± 0.083 | 11.9 ± 0.7 | 34.9 ± 8.7 |
| T197S | 6.00 ± 0.59 | 124 ± 3 | 20.6 ± 2.1 | 0.229 ± 0.034 | 137 ± 4 | 597 ± 91 |
| T197V | NA | NA | NA | NA | NA | NA |
| Y332A | NA | NA | NA | NA | NA | NA |
| Y332F | 40.0 ± 4.6 | 23.5 ± 0.8 | 0.59 ± 0.07 | 0.461 ± 0.070 | 20.7 ± 0.79 | 44.9 ± 6.9 |
a Errors are reported as the error in curve fitting.
b Values are approximate because the mutant is inhibited at AcCoA concentrations above 400 μm so saturation could not be achieved.
c Values were underestimated because the mutant is inhibited at AcCoA concentrations above 400 μm.
In Vivo Cell Growth Assay for HCS Activity
Yeast strain LPY 11411 (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1 ura3-1 rDNA::ADE2CAN1 lys20Δ::KanMX lys21Δ::NatMX4) was transformed using the lithium acetate method with empty 2 μm URA3 vector pRS202 (pLP 1402), pRS202 containing LYS20 (pLP1412), pRS426 containing lys4+ under the control of the S. cerevisiae LYS20 promoter (pLP2211), or pLP2211 containing point mutations of lys4+. Yeast cells were grown on synthetic complete medium lacking uracil or uracil and lysine according to standard procedures (25). Western blot analysis demonstrated that the steady state levels of the SpHCS mutant proteins were comparable to that of the WT SpHCS.
RESULTS
Structure Determination of SpHCS
The crystal structure of full-length (residues 1–418) SpHCS was determined by selenomethionine single wavelength anomalous diffraction phasing to 2.7 Å resolution (Table 1). The partially refined model of the SpHCS dimer was then used in molecular replacement to solve the 2.24 Å resolution apoenzyme structure and the structures of the 2-OG bound complexes. In the course of screening crystals of the SpHCS·2-OG complex, we identified two different conformations of the lid motif (residues 320–333) that encloses the entrance to the active site, which we term the closed and open conformations. In the closed lid complex (2.67 Å resolution), the lid motif is clearly visible in the electron density map and obstructs the entrance to the active site, whereas in the open complex (2.72 Å resolution), density for the lid is absent, resulting in a solvent-exposed active site. Additionally, two divalent metal-bound forms of the enzyme are reported here. Synchrotron radiation x-ray fluorescence scans of the SeMet-derivatized crystals detected the presence of Co(II) that was presumably introduced during the Co(II) IMAC step in the enzyme purification. The structure of Zn(II)-bound SpHCS closed lid complex with 2-OG was solved by chelating the Co(II) using EDTA followed by the addition of 1–2 mm Zn(II) during crystallization. For kinetic analyses, SpHCS and its mutants were purified on a Zn(II) IMAC column, and the WT enzyme had a Zn(II) content of 87 ± 5% as confirmed by inductively coupled plasma high resolution mass spectrometry (data not shown).
Crystal Structure of SpHCS
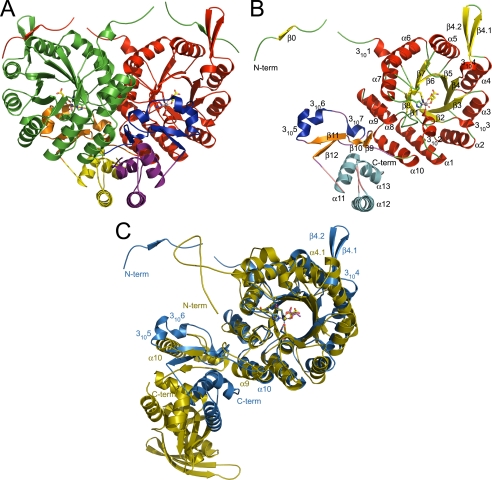
The crystal structure of SpHCS reveals that the enzyme forms a tightly associated homodimer within the asymmetric unit (Fig. 2A). The 47-kDa SpHCS monomer is composed of an N-terminal catalytic domain and a C-terminal domain of mixed α-helical and β-sheet topology (Fig. 2B). The N-terminal domain (residues 1–300) is composed of a canonical (α/β)8 TIM barrel (residues 35–300) with additional secondary structural elements flanking and inserted within the barrel. In the SpHCS·2-OG complexes, an N-terminal extension (residues 1–34) consisting of a short β-strand (β0) followed by a 310 helix (3101) precedes the β1 strand of the TIM barrel in the SpHCS·2-OG complexes, whereas the residues preceding the 3101 helix are disordered in the apoenzyme structure. Within the TIM barrel, several short 310 helices are interspersed throughout the (α/β)8 fold, and a region of mixed topology is inserted between β4 and α4. In the SpHCS·2-OG complexes, this region is fully ordered in the electron density maps and consists of the 3104 helix followed by a β-hairpin motif formed by β4.1-β4.2, whereas these structural elements are disordered in the apoenzyme structure (see below). After the TIM barrel, α-helices α9 and α10 buttress the barrel C terminus, packing against the α1 and α8 helices.
FIGURE 2.
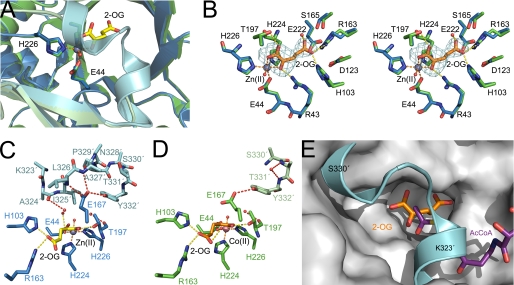
Crystal structure of S. pombe SpHCS and its overlay with M. tuberculosis α-IPMS. A, shown is a ribbon diagram of the SpHCS homodimer bound to 2-OG (closed lid motif) with the N-terminal domain, C-terminal subdomain I, and C-terminal subdomain II of monomer A depicted in red, orange, and yellow, respectively, and of monomer B depicted in green, blue, and violet, respectively. The Zn(II) atom is modeled as a dark gray sphere, and the bound 2-OG is rendered as sticks with yellow carbon atoms. B, shown is a ribbon diagram of the secondary structure of a monomer of SpHCS in complex with 2-OG with the secondary structure assigned following standard nomenclature for TIM barrel enzymes. The secondary structure is colored to distinguish between the N-terminal domain and C-terminal subdomains I and II. C, superimposition of a monomer of SpHCS (blue) in complex with 2-OG and a monomer of α-IPMS (gold) bound to 2-OIV. Coordination of the Zn(II) metal site in SpHCS and in α-IPMS is depicted as dashed orange and blue lines, respectively, whereas 2-OG and 2-OIV are depicted with yellow and magenta carbon atoms, respectively.
The C-terminal domain of SpHCS (residues 301–405) is composed of two distinct subdomains (Fig. 2B). Subdomain I (residues 301–350) has mixed topology consisting of a two-stranded anti-parallel β-sheet (β9- β10) and a two-stranded parallel β-sheet (β11-β12) with three consecutive 310 helices (3105–3107) interleaved between the latter two strands. In the structures of the apoenzyme and 2-OG closed lid complex, the 3105 and 3106 helices form the lid motif that occludes access to the active site within the TIM barrel. Subdomain II (residues 351–405) consists of a three-helix bundle composed of α11-α13. The residues after the α13 helix are disordered in SpHCS structures and are not reported here.
The quaternary structure of SpHCS forms an intricately intertwined homodimer typified by domain-swapping (Fig. 2A) (26). This oligomerization state is consistent with gel filtration analysis of SpHCS, which elutes as a ∼110-kDa species during purification (data not shown). Dimerization buries a large surface area of ∼5900 Å2 per monomer, representing ∼41% of the total monomer surface. The dimer interface includes extensive contacts between the N-terminal and C-terminal domains of each SpHCS monomer. A large portion of the dimer interface is formed by the lateral association of the TIM barrel domains through interactions between the α6-α10 helices in each monomer (Fig. 2, A and B). The remaining interactions are mediated predominantly by domain swapping of the N-terminal extension preceding the TIM barrel and the C-terminal domain. In the SpHCS·2-OG complexes, the β0 strand in the N- terminal extension forms an anti-parallel β-sheet with the β4.1-β4.2 hairpin, whereas this interaction is absent in the apoenzyme structure due to the disorder of these β-strands.
The C-terminal domain also contributes to domain swapping in the SpHCS dimer. Deletion of residues 301–405 in SpHCS yields insoluble recombinant protein demonstrating the importance of the C-terminal domain in maintaining its quaternary structure (data not shown). Both subdomains I and II contribute to the dimerization interface. In subdomain I, the β9-β10 and β11-β12 sheets and the 3105 and 3106 helices that compose the lid motif contact the TIM barrel of the neighboring monomer (Fig. 2, A and B), whereas the interactions with the 310 helices are absent in the open 2-OG complex due to the lid disorder. The three-helix bundle in subdomain II also contributes to the dimer interface with the α12 and α13 packing anti-parallel to their counterpart helices in the adjacent monomer.
Structural Comparison of SpHCS and α-IPMS
After determining the structure of SpHCS, we sought to identify structural homologs of the enzyme by searching the Protein Data Bank using the three-dimensional structure comparison server Dali (27). Using a SpHCS monomer (residues 4–405), the Dali search identified structural homology with multiple TIM barrel enzymes, including 3-hydroxy-3-methylglutaryl-CoA lyase (28), the aldolase dehydrogenase DmpG/DmpF (29), and M. tuberculosis α-IPMS (10). The latter enzyme was identified as the top hit (Z-score 31.1), consistent with the predicted structural homology between SpHCS and α-IPMS and the analogous reactions catalyzed by these enzymes. The TIM barrel domains of α-IPMS (residues 73–357) and SpHCS (residues 36–298) align closely with a root mean square deviation in the aligned main chain atoms of 1.38 Å (Fig. 2C). The active sites of α-IPMS and SpHCS also harbor structural homology in the residues which coordinate the divalent metal ion and that are involved in substrate binding and catalysis (see below). Aside from these similarities, there are minor structural differences in the TIM barrel domains that distinguish the enzymes; notably, the 310 helices and the β-hairpin of SpHCS are absent in α-IPMS.
In contrast to their conserved catalytic domains, the C-terminal domains of SpHCS and α-IPMS share limited structural homology. Both enzymes have structural similarity in subdomain I, with the exception that the 3105 and 3106 helices in SpHCS which are substituted by the α10-helix in α-IPMS (Fig. 2C). Subdomain II of α-IPMS and SpHCS are each composed of three-helix bundles that exhibit different spatial arrangements in relation to the TIM barrel. The greatest divergence between the two structures is the βββα-fold regulatory module at the C terminus of α-IPMS that is absent in SpHCS. In α-IPMS, the regulatory modules bind free leucine to mediate allosteric feedback inhibition, whereas lysine inhibition of HCS has been reported to be competitive versus 2-OG, suggesting that the amino acid binds within the active site (30–32). Thus, the absence of this domain in SpHCS reflects distinct mechanisms for feedback inhibition for HCSs and α-IPMSs.
Active Site and 2-OG Binding
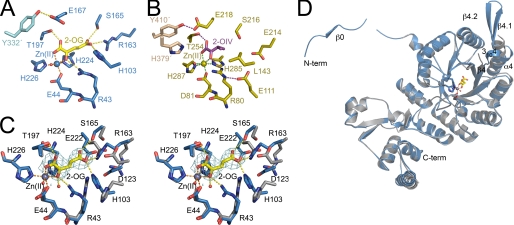
The active site of SpHCS is located within the C-terminal end of the β1-β8 strands that forms the interior of the TIM barrel (Fig. 2B). The divalent metal cation in the active site, which is either Co(II) or Zn(II) in the structures reported here (Table 1), is coordinated in an octahedral geometry by the side chains of Glu-44, His-224, and His-226 in SpHCS, by the C1 carboxylate and the 2-oxo carbonyl group of 2-OG, and by a water molecule (Fig. 3A). 2-OG is bound within the center of the β-barrel through its coordination to the divalent metal ion and through a network of hydrogen bonds and salt bridge interactions. The C1 carboxylate and 2-oxo groups of 2-OG hydrogen-bond to the hydroxyl and guanidinium groups of Thr-197 and Arg-43, respectively, whereas the C5 carboxylate group engages in hydrogen bonds with the side chains of His-103, Arg-163, and Ser-165 in SpHCS. The substrate binding mode for 2-OG is analogous to the orientation and interactions of 2-OIV within the active site of α-IPMS, with the exception that the methyl groups of 2-OIV are recognized through hydrophobic contacts with Leu-143 in the TIM barrel (Figs. 3, A and B).
FIGURE 3.
SpHCS active site and 2-OG binding. A, shown is the active site of SpHCS (closed lid conformation; blue carbon atoms) in complex with 2-OG (yellow carbons) and Zn(II) (blue). Orange and yellow dashes represent coordination of the Zn(II) ion and hydrogen bonding to 2-OG, respectively. B, shown is the active site of α-IPMS (gold carbons) in complex with 2-OIV (magenta carbons) and Zn(II) (gold). The coordination of the Zn(II) ion and hydrogen bonds to 2-OIV are colored green and magenta, respectively. C, shown is the stereoview of the active site of the SpHCS 2-OG closed lid complex (rendered as in panel A) overlaid with the SpHCS apoenzyme (gray carbons). The Zn(II) ion and water molecules are represented as gray and red spheres, respectively. Electron density of the Fo − Fc simulated annealing omit map (contoured at 2.5 σ) corresponding to the 2-OG in the closed loop complex is depicted in cyan. D, shown is a ribbon diagram of a monomer of the SpHCS 2-OG closed lid complex (blue) superimposed with the corresponding monomer in the SpHCS apoenzyme (gray). The secondary structural elements that undergo conformational changes between the two structures are denoted.
Conformational changes within the catalytic domain of SpHCS accompany 2-OG binding. A superimposition of the active sites of the apoenzyme and 2-OG closed complex reveals that the carboxylate groups of Asp-123 and Glu-222 rotate away from the center of the β-barrel to accommodate the binding of the C5 carboxylate, whereas side chains of Arg-43, His-103, Arg-163, and Ser-165 reorient toward the barrel center to form hydrogen bonds or salt bridge interactions with the substrate (Fig. 3C). This reorganization appears to be coupled to conformational changes in the TIM barrel. Although the overall alterations between the catalytic domains of the apoenzyme and 2-OG complex are relatively modest (root mean square deviation value of 1.25 Å for all aligned atoms), pronounced structural changes occur in the linker between β4 and α4 that is disordered in the apoenzyme. Binding of 2-OG induces ordering of the linker, resulting in the formation of the 3104 helix and the β4.1-β4.2 hairpin. In addition, the α4 helix is splayed outward, and the first turn of the helix unwinds to accommodate the formation of the β-hairpin. These conformational changes contribute to the ordering of the domain-swapped N-terminal extension through the interaction of β0 with the β-hairpin (Fig. 3D).
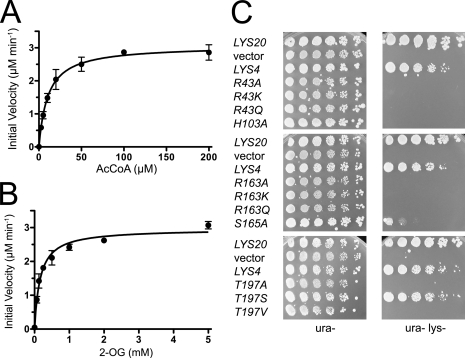
To understand the structural basis for these conformational changes, we analyzed the active site residues that are involved in 2-OG binding. Steady state kinetic analysis of the Zn(II)-bound form of WT SpHCS yielded kcat and Km values for AcCoA and 2-OG (Fig. 4, A and B, and Table 2) that are comparable with those reported for other HCSs, including ScHCS Lys20 and T. thermophilus HCS (kcat = 63–92 min−1, Km(2-OG) = 0.044–0.140 mm, and Km(AcCoA) = 32–42 μm) (11, 32). We then measured the kinetic parameters for conservative and alanine substitutions of the active site residues that interact with 2-OG (Fig. 3A). Mutation of Thr-197 to a serine resulted in a modest decrease in catalytic efficiency (as defined by kcat /Km) due to a minor decrease in its turnover number in comparison to WT SpHCS. In contrast, a T197A mutant of SpHCS exhibited a 25-fold decrease in the kcat value of SpHCS, whereas a T197V substitution abolished activity (Table 2), illustrating that the hydrogen bond between the Thr-197 hydroxyl group and C1-carboxylate of 2-OG group contributes to catalysis. Interactions between SpHCS and the 2-oxo carbonyl group of 2-OG are also important to homocitrate synthesis. Alanine, glutamine, and lysine substitutions of Arg-43 abolished activity in SpHCS. Together, these results demonstrated that the hydrogen bond between the 2-oxo atom of 2-OG and the Arg-43 guanidinium group is critical for catalysis.
FIGURE 4.
In vitro and in vivo activity of SpHCS. A and B, shown are Michaelis-Menten plots of the initial velocity versus AcCoA concentration (A) and 2-OG concentration (B) for the formation of homocitrate catalyzed by WT SpHCS. Data points are the average of triplicate measurements, and the error bars represent 1 S.D. C, in vivo yeast growth assays of LYS4 and LYS4 2-OG binding mutants.
Residues that interact with the C5 carboxylate group of 2-OG are also essential for substrate binding (Fig. 3A). Alanine and glutamine substitutions of Arg-163 in SpHCS abolished activity, whereas H103A and R163K substitutions substantially impaired catalytic efficiency compared with the WT enzyme (Table 2). Notably, the R163K mutation resulted in a >150-fold increase in the Km for 2-OG, consistent with the observed salt bridge interaction between its guanidinium group and the substrate C5 carboxylate moiety. Unexpectedly, this mutant also exhibited an elevated Km for AcCoA, which may be a consequence of the severely diminished affinity for 2-OG, whose binding precedes that of the AcCoA in the ordered Bi-Bi reaction mechanism (7). In contrast, an S165A substitution resulted in a moderate decrease in catalytic efficiency compared with WT SpHCS. Collectively, these results illustrate that the residues in SpHCS that interact with the C5 carboxylate group of 2-OG are pivotal to its binding within the active site.
To ascertain the effects of these SpHCS mutations in vivo, we generated an S. cerevisiae strain in which the genes encoding the two ScHCS paralogs, LYS20 and LYS21, were deleted to generate a lys20Δ lys21Δ double mutant strain. This strain displayed robust growth on lysine-supplemented media but failed to grow on lysine-deficient media, illustrating its auxotrophy for this amino acid (Fig. 4C). We then tested whether expression of SpHCS encoded by lys4+, whose protein sequence is ∼80% identical to ScHCS Lys20 and Lys21 (Supplementary Fig. S2), could complement lysine auxotrophy in the lys20Δ lys21Δ deletion strain by cloning the lys4+ gene under the control of the LYS20 promoter. Transformants expressing LYS20 or lys4+ were able to grow on medium that lacks lysine, whereas those transformed with vector alone were inviable on Lys− media, demonstrating that lys4+ complemented the loss of HCS activity in S. cerevisiae (Fig. 4C). Next, we investigated whether the previously described 2-OG binding mutants in SpHCS could support growth in lysine-deficient media. The 2-OG binding mutants expressed at steady state levels comparable with WT SpHCS as confirmed by protein immunoblotting and behaved in a recessive manner when assayed for genetic dominance (data not shown). The 2-OG interacting mutants failed to support growth on lysine deficient media, with the exception of the SpHCS T197S mutant and, to a lesser extent, the S165A mutant. These results correlate with our kinetic data wherein the T197S and S165A mutants exhibited only 3- and 7-fold reductions in catalytic efficiency, respectively, compared with WT SpHCS (Table 2) and, thus, were able to partially complement lysine auxotrophy in the lys20Δ lys21Δ deletion strain. In contrast, the other 2-OG binding mutants of SpHCS manifested more severe decreases in catalytic efficiency and fail to function in vivo.
Functions of the Lid Motif
An unexpected finding that emerged from the SpHCS structures is the presence of the lid motif that encloses the active site of SpHCS. This motif is absent in the structurally related α-IPMS, revealing that it is a unique feature conserved in fungal HCSs (supplemental Fig. S2). The lid motif, composed of the 3105–3106 helices in the C-terminal domain, participates in a domain-swapped interaction with the catalytic domain of the neighboring subunit in the SpHCS homodimer by straddling the entrance to the TIM barrel. The presence of the lid accounts for the complete burial of 2-OG in the structure of the SpHCS·2-OG closed complex, whereas its absence in the SpHCS·2-OG open complex results in a solvent-accessible active site, permitting the exchange of 2-OG and water molecules (Fig. 5A). Superimposition of the active sites of the open and closed SpHCS structures reveals that the lid movement does not result in overt changes in 2-OG coordination (Fig. 5B). This observation is consistent with the fact that the lid motif does not directly contact 2-OG. Instead, several water molecules are sandwiched between 2-OG and the lid that are presumably trapped during its closure. The lid closed state is maintained through a network of hydrogen bonds to the carboxylate group of Glu-167, which includes the amide nitrogens of Leu-326′ and Ala-327′ and the hydroxyl group of Tyr-332′ in the lid motif (the prime (′) denotes residues in the lid motif from the neighboring monomer) (Fig. 5C). In the open complex, residues 323′-329′ in the lid motif are disordered, resulting in a loss of the hydrogen bonds to Leu-226′ and Ala-227′. Furthermore, the phenol side chain of Tyr-332′ is partially disordered in the open lid complex and is modeled with a partial occupancy of 0.5. The hydroxyl group of the modeled Tyr-332′ appears to maintain its hydrogen bond with the side chain of Glu-167, which adopts an alternative conformation over the 2-OG binding site that presumably orients its carboxylate group for base catalysis (Fig. 5D) (see below).
FIGURE 5.
The lid motif of SpHCS. A, structural superimposition of the SpHCS·2-OG complexes with the open lid motif (monomer A is green, and monomer B is pale green) and the closed lid motif (monomer A is blue, and monomer B is cyan). The 2-OG and Zn(II) ion of the closed lid structure are depicted in yellow carbons and gray, respectively. B, shown is a stereoview of the active site of SpHCS·2-OG open lid complex (green carbon atoms with 2-OG rendered with orange carbons) overlaid with the SpHCS·2-OG closed lid complex (blue carbons). Electron density for the Fo − Fc-simulated annealing omit map (contoured at 2.0 σ) corresponding to the 2-OG in the open lid complex is shown in cyan. The orange and yellow dashes represent coordination of the Zn(II) ion and hydrogen bonding to 2-OG, respectively. C and D, shown are comparisons of the active site and lid motif of the SpHCS·2-OG open and closed lid complexes. Monomer A (blue carbons) and B (cyan carbons) of the closed lid complex are depicted with the Zn(II) ion (gray) and 2-OG (yellow carbons) (C), whereas monomer A (green carbons) and B (light green carbons) of the open lid complex are illustrated with the Co(II) ion (pink) and 2-OG (orange carbons) (D). Dashed lines denote the coordination of the metal ion (orange), hydrogen bonding to 2-OG (yellow), and hydrogen bonding within the lid motif (red). Residues in the lid motif that lack interpretable side-chain electron density are modeled as alanines, whereas the side chain of Tyr-332 in the open lid complex (D) is modeled with an occupancy of 0.5. E, shown is a model of a ternary complex of SpHCS bound to 2-OG (orange carbons) and AcCoA (violet carbons). The lid motif of the SpHCS·2-OG closed lid complex (cyan) is superimposed on the gray surface of the SpHCS·2-OG open lid complex.
In addition to gating access to 2-OG, the lid motif may also regulate AcCoA binding to SpHCS. Because exhaustive attempts to co-crystallize SpHCS with CoA were unsuccessful, we modeled an SpHCS Michaelis complex with 2-OG and AcCoA based on the superimposition of the coordinates of the 2-OG open complex and a previously reported model of the α-IPMS·2-OIV·AcCoA ternary complex (10). The model reveals a groove in the enzyme surface leading into the active site that may accommodate the binding of the pantetheine moiety of AcCoA (Fig. 5E). Superimposition of the AcCoA-bound model with the closed SpHCS·2-OG complex reveals that the lid motif occupies the putative pantetheine binding groove, resulting in a steric clash between the lid residues and AcCoA. Taken together, these findings imply that the lid motif is inherently flexible and serves as a gatekeeper to regulate substrate binding to SpHCS.
Catalytic Mechanism
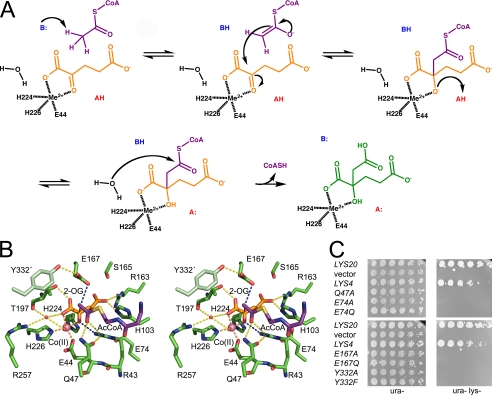
The catalytic mechanism of HCS is believed to proceed via a mixed aldol Claisen condensation reaction (8) (Fig. 6A). The first step in this reaction is the ordered binding of 2-OG and AcCoA to the enzyme, in agreement with kinetic studies of ScHCS Lys20 (7) and the spatial arrangement of the substrates in the modeled SpHCS ternary complex (Fig. 5E). The acetyl group of AcCoA is then deprotonated by a catalytic base to form either an enol or enolate (Fig. 6A). The enol/enolate nucleophile subsequently attacks the C2 carbonyl carbon of 2-OG, yielding an alkoxide intermediate that is protonated by a catalytic acid to generate homocitryl-CoA. The water molecule coordinated to the active site metal has been proposed to hydrolyze the thioester bond of homocitryl-CoA, yielding CoA and homocitrate. An analogous reaction mechanism has been postulated for α-IPMS (10).
FIGURE 6.
Catalytic mechanism and the SpHCS ternary complex model. A, shown is the proposed catalytic mechanism of HCS with 2-OG and AcCoA depicted as in Fig. 5E and homocitrate illustrated in green. B, shown is a stereoview of the active site of the modeled SpHCS ternary complex with 2-OG and the pantetheine arm of AcCoA rendered as in Fig. 5E. Dashed lines represent coordination of the Zn(II) ion (orange), hydrogen bonding to 2-OG (yellow), hydrogen bonding to the putative catalytic residues (blue), and the direction of the nucleophilic attack from the enolate tautomer of AcCoA to the C2 atom of 2-OG (red). C, in vivo yeast growth assays of the LYS4 mutants implicated in catalysis are shown.
The model of the SpHCS·AcCoA·2-OG ternary complex provides insights into the residues involved in homocitrate synthesis (Fig. 6B). The acetyl group of AcCoA is positioned near the C2 carbon of 2-OG where nucleophilic attack by the enolate occurs. Several residues that are invariant in HCS (supplemental Fig. S2) and that are structurally conserved in α-IPMS cluster around the AcCoA acetyl group, implicating them in catalysis. In the ternary complex model, the carbonyl oxygen of the coenzyme acetyl group is within hydrogen-bond distance to the amide side chain of Gln-47 and the guanidinium group of Arg-43 that also hydrogen-bonds to 2-OG. A similar model has been proposed for the corresponding Arg-80 in α-IPMS whereby its guanidinium group bridges interactions between AcCoA and 2-OIV (10). The hydrogen bonds to Gln-47 and Arg-43 in SpHCS stabilize the enol tautomer of the AcCoA, promoting nucleophilic attack on the C2 atom of 2-OG. In the modeled substrate complex, the methyl group of AcCoA is located ∼3 Å from the carboxylate of Glu-167, which is poised to function as a catalytic base to deprotonate the coenzyme acetyl group, yielding the enolate tautomer. The conformation of the Glu-167 side chain is stabilized via hydrogen bonding to the hydroxyl group of Tyr-332′ in the neighboring monomer. This glutamate-tyrosine pair is conserved in HCSs (supplemental Fig. S2) and in α-IPMS (Glu-218–Tyr410′, Fig. 3B) and has been proposed to orient the glutamate carboxylate group for acetyl group deprotonation in α-IPMS (10) and in ScHCS Lys20 (11). After enolization of AcCoA, nucleophilic attack occurs at the carbonyl C2 atom of 2-OG, yielding homocitryl-CoA through the formation of a carbon-carbon bond (Fig. 6, A and B). The C2 center is rendered electrophilic due to the polarization of the 2-oxo moiety of 2-OG through its metal ion coordination and its hydrogen bond to the guanidinium cation of Arg-43. The conformation of Arg-43 side chain is maintained via a hydrogen bond to Gln-47 and a salt-bridge interaction with Glu-74 in the TIM barrel. Arg-43 and Glu-74 are invariant in the HCS family (supplemental Fig. S2) and are structurally conserved in α-IPMS, suggesting that they have conserved catalytic roles.
To investigate the functions of the residues highlighted above, we generated alanine and conservative point mutations and assayed their activities in vitro and in vivo (Table 2 and Fig. 6C). We first examined the effects of mutations of the putative catalytic base, Glu-167, and the adjacent Tyr-332′ in SpHCS (Fig. 6B). Alanine and glutamine substitutions of Glu-167 abolished activity in vitro, consistent with the role of this residue in AcCoA enolization. These data also concur with the >1000-fold decrease in kcat reported for the alanine and glutamine mutations of the homologous Glu-155 in ScHCS Lys20 (11). Mutations of Tyr-332′, which hydrogen-bonds to the Glu-167 carboxylate group (Fig. 6B), also disrupted activity in SpHCS. A phenylalanine substitution of Tyr-332 decreased kcat ∼13-fold and resulted in modest increases in the Km values for each substrate compared with WT SpHCS, whereas a Y332A mutant exhibited no activity. Finally, plate growth assays using the S. cerevisiae lys20Δ lys21Δ deletion strain demonstrated that the SpHCS Glu-167 and Tyr-332′ mutants were unable to complement growth on lysine-deficient media (Fig. 6C), correlating with the catalytic defects of these mutants in vitro (Table 2).
In addition to probing the functions of Glu-167 and Tyr-332, we also examined the effects of mutations of Arg-43, Gln-47, and Glu-74 in SpHCS. As previously discussed, substitutions of Arg-43 to alanine, glutamine, and lysine abolished activity of SpHCS in vitro and in vivo (Table 2 and Fig. 4C), demonstrating that the Arg-43 guanidinium group is required for catalysis. Substitutions of Gln-47 and Glu-74 that hydrogen-bond to Arg-43 also impaired enzymatic activity (Fig. 6B). An alanine substitution of Glu-74 in SpHCS abrogated activity in vitro, whereas an E74Q mutation resulted in a moderate decrease in the turnover number and a slight increase in the Km value for each substrate in comparison to the WT enzyme (Table 2). Analogous effects were observed for the Q47A mutant, although the elevation in the substrates' Km values was more pronounced. This may be due to the ablation of the Gln-47 side chain that would weaken hydrogen bonding to the acetyl group of AcCoA in the ternary complex model and perturb the orientation of the Arg-43 side chain that hydrogen-bonds to 2-OG. As with the SpHCS Glu-167 and Tyr-332 mutants, the Q47A, Arg-43, and Glu-74 mutants were inviable on lysine-deficient media when transformed into the lys20Δ lys21Δ deletion strain (Figs. 4C and 6C), illustrating the importance of these residues in lysine biosynthesis in vivo.
DISCUSSION
The structures of the SpHCS presented here furnish molecular insights into the basis of substrate binding and catalysis in the HCS family. Our results also highlight the similarities in the HCS and α-IPMS enzymes, including the structural conservation of their TIM barrel catalytic domains, divalent metal ion coordination, substrate binding and catalytic residues, and 2-oxo acid substrate binding modes. Recent studies have proposed that a glutamate-histidine dyad in α-IPMS (Glu-218—His-379′) and ScHCS Lys20 (Glu-155-His-309′) participates in base catalysis to mediate the enolization of AcCoA (10, 11). In α-IPMS, the side chains of Glu-218 and His-379′ are within an appropriate distance to deprotonate the acetyl group of AcCoA as suggested by the modeled α-IPMS·2-OIV·AcCoA ternary complex (10). Based on this model, a similar arrangement has been proposed for Glu-155 and His-309′ in the active site of ScHCS Lys20 (11). However, in SpHCS, His-321′ (corresponding to His-309′ in ScHCS Lys20) is located in the 3105 within the lid motif (supplemental Fig. S2) and is disordered in one of the two subunits in the 2-OG open lid complex that was used to generate the ternary complex model. In monomer B, which exhibits clear density for His-321′, the histidine imidazole ring is located ∼13 Å from the Glu-167 carboxylate group. Comparable distances are observed for the Glu-167 and His-321′ side chains in the structures of the apoenzyme and the SpHCS·2-OG closed lid complex. Although these distances are too great to form a functional glutamate-histidine catalytic dyad, it is conceivable that AcCoA binding to SpHCS could induce a conformational change in the lid motif that would bring the Glu-167 and His-321′ side chains into the appropriate proximity and geometry for base catalysis. Consistent with this model, a H321A mutant decreased the activity compared with WT SpHCS by more than 200-fold (data not shown), in agreement with His-309 mutations that abrogate activity in ScHCS Lys20 (11). Alternatively, mutations of residues within the lid motif, such as His-321′ and Tyr-332′, may perturb substrate binding by altering lid conformational dynamics without directly affecting the chemical steps in catalysis. Additional studies are needed to address whether a histidine-glutamate dyad participates in catalysis in HCSs.
Although there is a general consensus concerning the involvement of the structurally conserved glutamate in base catalysis in HCS (10, 11), the identity of the catalytic acid in the reaction mechanisms of these enzymes remains an open question (Fig. 6A). Our structural and biochemical studies of SpHCS suggest that the Arg-43 may fulfill this role by protonating the alkoxide of homocitryl-CoA that is formed during condensation of AcCoA and 2-OG. Although it is unusual for an arginine guanidinium cation to function as a proton donor due to its high pKA value, several lines of evidence support a role for Arg-43 in acid catalysis. First and most importantly, there are no other ionizable residues or solvent molecules in the active site of the ternary complex model that are located at an appropriate distance to the 2-oxo atom of 2-OG to function as a catalytic acid barring a substantial conformational change within the TIM barrel (Fig. 6B). Second, Arg-43 is invariant in the HCS family (supplemental Fig. S2) and mutation of this residue abolishes activity (Table 2), illustrating its importance to catalysis. Third, the pKA value of an alkoxide ion is greater than 12.5, favoring protonation by the Arg-43 guanidinium group (aqueous pKA = 12.5). Fourth, a recent survey of enzyme reaction mechanisms has highlighted a role for arginine in acid-base chemistry (33), such as in soluble fumarate reductase, in which Arg-402 has been reported to protonate fumarate to form succinate during flavin-dependent reduction (34). Fifth, the close proximity of the divalent metal ion to the Arg-43 guanidinium group (∼5 Å) may contribute to depressing its pKA value, facilitating acid catalysis. Although these observations collectively point toward a role for Arg-43 as the proton donor, it is possible that an acid-catalyzed step is not required in the reaction mechanisms of HCSs and α-IPMSs. Evidence for this alternative model is based on structural and functional studies of the TIM barrel enzyme malate synthase that catalyzes the Claisen condensation of AcCoA and glyoxylate to yield malate, analogous to the reactions catalyzed by HCSs and α-IPMSs. The active site of E. coli malate synthase isoform G shares certain structural similarities with α-IPMS and SpHCS, including an octahedrally coordinated Mg(II) ion that binds glyoxylate and an arginine residue (Arg-338) whose guanidinium cation hydrogen bonds to the 2-oxo atom of the substrate (35, 36). Site-directed mutagenesis of Arg-338 in malate synthase suggests that it does not participate in the proton transfer during malate synthesis and that the alkoxide intermediate remains deprotonated during the subsequent steps in the catalytic cycle through its coordination to Mg(II) cation (35). Future studies are needed to ascertain whether the reaction mechanisms of HCSs and α-IPMSs proceed via an acid-catalyzed step and, if so, whether the structurally conserved arginine protonates the alkoxide intermediate.
Despite the structural and catalytic similarities shared by the HCSs and α-IPMSs, the mechanisms by which their activities are regulated are distinct. Among the most notable differences are the relative accessibilities of their active sites. In α-IPMS, the entrance of its TIM barrel is solvent-exposed, indicating that the enzyme can readily accommodate the binding of 2-OIV and AcCoA within its active site (Fig. 2C). In contrast, the entrance to the TIM barrel of SpHCS is effectively plugged by the flexible lid motif that gates access of 2-OG to the active site and competes with AcCoA binding to the enzyme (Figs. 5, A and E). The closed conformation of the lid motif is stabilized through hydrogen bonding to the carboxylate group of Glu-167, illustrating its dual roles in catalysis and in regulating substrate access to the active site. The intrinsic flexibility of the lid motif may serve a regulatory function by interacting with factors that modulate its open or closed states, thereby governing the rate of homocitrate synthesis and the metabolic flux through the AAA pathway.
In contrast to the structural and mechanistic similarities shared by HCS and α-IPMS, these enzymes have little in common with CS. Although CS catalyzes an analogous mixed aldol Claisen condensation of oxaloacetate and AcCoA to yield citrate and CoA, the details of its catalytic mechanism as well as its sequence and structure differ substantially from HCS. CS functions as a homodimer with each subunit composed of two domains that adopt a predominantly α-helical fold which is unrelated to the TIM barrel architecture of HCS (37, 38). The active site of CS is located in the cleft between the two domains and is dominated by histidine and arginine residues that form hydrogen bonds or salt bridge interactions with oxaloacetate. This substrate binding mode is distinct from that of HCS in which 2-OG is bound within the TIM barrel through its coordination to the divalent metal ion and through hydrogen bonds to basic and polar amino acids (Fig. 3A). Furthermore, there are notable differences in the residues involved in acid-base catalysis in the Claisen condensation reactions catalyzed by each enzyme. Based on the structures of CS binary and ternary complexes, it has been proposed that an aspartate functions as the catalytic base in deprotonating the methyl group of AcCoA to initiate the condensation reaction, whereas a histidine acts as the catalytic acid to protonate the alkoxide form of citryl-CoA (9, 38). In contrast, our structural and functional studies of SpHCS suggest that Glu-167 functions as the catalytic base in AcCoA enolization, whereas Arg-43 may participate in acid catalysis by protonating the alkoxide intermediate. Finally, there are significant differences in the conformational changes that accompany substrate binding in CS and HCS. Crystallographic analyses of CS have revealed that the enzyme can exist in open and closed forms due to large-scale rotation of its two domains, the focal point of which coincides with the enzyme active site (37, 39). The binding of oxaloacetate to CS has been shown to induce the closure of the active site over the substrate, stimulating AcCoA binding and the subsequent steps in catalysis. Conversely, the lid motif in HCS appears to function as a flexible gate that regulates substrate binding (Fig. 5, C–E), and the conformational changes in the TIM barrel associated with 2-OG binding are localized to the β4-α4 region of the TIM barrel (Fig. 3D) as opposed to the large domain shift observed in CS. In summary, the differences in the structures and catalytic mechanisms of CS and HCS provide remarkable insights into the molecular evolution of two distinct enzymatic mechanisms that catalyze the Claisen condensations of closely related 2-oxo acid substrates.
The enzymes composing the AAA pathway have been proposed as potential targets for antifungal inhibitors. In particular, HCS is an attractive target for inhibition because it catalyzes the first committed step in the pathway and is a nexus for regulation via feedback inhibition by lysine (6). The loss of function exhibited by the majority of the lys4+ mutants in vivo further underscores the importance of HCS in maintaining fungal viability in nutrient-poor conditions (Figs. 4C and 6C). Our structural and functional characterization of SpHCS provides a framework for developing selective inhibitors of HCSs. The enzyme active site possesses a conserved divalent metal center and numerous polar and charged residues through which small molecule compounds could interact. Additionally, bisubstrate analog inhibitors that mimic homocitryl-CoA could be synthesized that maximize contacts with the active site and surrounding surface. Efforts to rationally design inhibitors must account for the intrinsic plasticity of the active site residues that accommodate 2-OG binding as well as the flexibility of the lid motif that regulates access to the active site. Small molecules that are rationally designed or identified via high-throughput screening may offer opportunities to exploit this conformational flexibility to yield potent inhibitors that may serve as lead compounds for the development of new antifungal drugs.
Acknowledgments
We thank J. Woodward at the Sanger Institute for providing the genomic clone of lys4+ and J. Aris for supplying the yeast HCS antibody. We also thank E. Townsend and C. Burbank for assistance in protein purification and preparing site-directed mutants, respectively. In addition, we acknowledge S. Anderson and R. Sanishvili for assistance in x-ray data collection, T. Huston for performing the inductively coupled plasma mass spectrometry analysis, and A. Gafni and E. Lee for use of the CD spectropolarimeter. We kindly thank T. Baker and N. Koon for furnishing the coordinates of the α-IPMS·AcCoA·2-OIV model and P. O'Brien and P. Del Rizzo for reading the manuscript and providing useful comments. Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science under Contract DE-AC02-06CH11357. General Medicine and Cancer Institutes Collaborative Access Team was funded in whole or in part with National Institutes of Health Grants Y1-CO-1020 (NCI) and Y1-GM-1104 (NIGMS). Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corp. and the Michigan Technology Tri-Corridor for support of this research program (Grant 085P1000817).
This work was supported, in whole or in part, by National Institutes of Health Grants GM056469 (to L. P.) and GM073839 (R. C. T.). This work was also supported by a University of Michigan Biomedical Research Council pilot grant (to R. C. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3IVS, 3IVT, and 3IVU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AAA
- α-aminoadipate
- HCS
- homocitrate synthase
- 2-OG
- 2-oxoglutarate
- AcCoA
- acetyl coenzyme A
- CoA
- coenzyme A
- CS
- citrate synthase
- α-IPMS
- α-isopropylmalate synthase
- 2-OIV
- 2-oxoisovalerate
- WT
- wild type
- IMAC
- immobilized metal affinity chromatography
- SeMet
- selenomethionyl.
REFERENCES
- 1.Vogel H. J. (1960) Biochim. Biophys. Acta 41, 172–174 [DOI] [PubMed] [Google Scholar]
- 2.Garrad R. C., Bhattacharjee J. K. (1992) J. Bacteriol. 174, 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosuge T., Hoshino T. (1998) FEMS Microbiol. Lett. 169, 361–367 [DOI] [PubMed] [Google Scholar]
- 4.Tucci A. F., Ceci L. N. (1972) Arch. Biochem. Biophys. 153, 742–750 [DOI] [PubMed] [Google Scholar]
- 5.Walsh T. J., Groll A. H. (1999) Transpl. Infect. Dis. 1, 247–261 [DOI] [PubMed] [Google Scholar]
- 6.Xu H., Andi B., Qian J., West A. H., Cook P. F. (2006) Cell Biochem. Biophys. 46, 43–64 [DOI] [PubMed] [Google Scholar]
- 7.Andi B., West A. H., Cook P. F. (2004) Biochemistry 43, 11790–11795 [DOI] [PubMed] [Google Scholar]
- 8.Qian J., West A. H., Cook P. F. (2006) Biochemistry 45, 12136–12143 [DOI] [PubMed] [Google Scholar]
- 9.Karpusas M., Branchaud B., Remington S. J. (1990) Biochemistry 29, 2213–2219 [PubMed] [Google Scholar]
- 10.Koon N., Squire C. J., Baker E. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian J., Khandogin J., West A. H., Cook P. F. (2008) Biochemistry 47, 6851–6858 [DOI] [PubMed] [Google Scholar]
- 12.Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expression Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 13.Doublié S. (1997) Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 14.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 17.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 19.Vagin A. A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 20.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21.Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 22.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trievel R. C., Li F. Y., Marmorstein R. (2000) Anal. Biochem. 287, 319–328 [DOI] [PubMed] [Google Scholar]
- 24.Collazo E., Couture J. F., Bulfer S., Trievel R. C. (2005) Anal. Biochem. 342, 86–92 [DOI] [PubMed] [Google Scholar]
- 25.Amberg D. C., Burke D. J., Strathern J. N. (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Liu Y., Gotte G., Libonati M., Eisenberg D. (2002) Protein Sci. 11, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Z., Runquist J. A., Forouhar F., Hussain M., Hunt J. F., Miziorko H. M., Kim J. J. (2006) J. Biol. Chem. 281, 7526–7532 [DOI] [PubMed] [Google Scholar]
- 29.Manjasetty B. A., Powlowski J., Vrielink A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andi B., West A. H., Cook P. F. (2005) J. Biol. Chem. 280, 31624–31632 [DOI] [PubMed] [Google Scholar]
- 31.Jaklitsch W. M., Kubicek C. P. (1990) Biochem. J. 269, 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wulandari A. P., Miyazaki J., Kobashi N., Nishiyama M., Hoshino T., Yamane H. (2002) FEBS Lett. 522, 35–40 [DOI] [PubMed] [Google Scholar]
- 33.Guillén Schlippe Y. V., Hedstrom L. (2005) Arch. Biochem. Biophys. 433, 266–278 [DOI] [PubMed] [Google Scholar]
- 34.Mowat C. G., Moysey R., Miles C. S., Leys D., Doherty M. K., Taylor P., Walkinshaw M. D., Reid G. A., Chapman S. K. (2001) Biochemistry 40, 12292–12298 [DOI] [PubMed] [Google Scholar]
- 35.Anstrom D. M., Kallio K., Remington S. J. (2003) Protein Sci. 12, 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard B. R., Endrizzi J. A., Remington S. J. (2000) Biochemistry 39, 3156–3168 [DOI] [PubMed] [Google Scholar]
- 37.Remington S., Wiegand G., Huber R. (1982) J. Mol. Biol. 158, 111–152 [DOI] [PubMed] [Google Scholar]
- 38.Wiegand G., Remington S. J. (1986) Annu. Rev. Biophys. Biophys. Chem. 15, 97–117 [DOI] [PubMed] [Google Scholar]
- 39.Wiegand G., Remington S., Deisenhofer J., Huber R. (1984) J. Mol. Biol. 174, 205–219 [DOI] [PubMed] [Google Scholar]