Abstract
Parasitic nematodes cause serious diseases in humans, animals, and plants. They have limited lipid metabolism and are reliant on lipid-binding proteins to acquire these metabolites from their hosts. Several structurally novel families of lipid-binding proteins in nematodes have been described, including the fatty acid- and retinoid-binding protein family (FAR). In Caenorhabditis elegans, used as a model for studying parasitic nematodes, eight C. elegans FAR proteins have been described. The crystal structure of C. elegans FAR-7 is the first structure of a FAR protein, and it exhibits a novel fold. It differs radically from the mammalian fatty acid-binding proteins and has two ligand binding pockets joined by a surface groove. The first can accommodate the aliphatic chain of fatty acids, whereas the second can accommodate the bulkier retinoids. In addition to demonstrating lipid binding by fluorescence spectroscopy, we present evidence that retinol binding is positively regulated by casein kinase II phosphorylation at a conserved site near the bottom of the second pocket. far-7::GFP (green fluorescent protein) expression shows that it is localized in the head hypodermal syncytia and the excretory cell but that this localization changes under starvation conditions. In conclusion, our study provides the basic structural and functional information for investigation of inhibitors of lipid binding by FAR proteins.
Introduction
Hydrophobic lipophilic molecules such as fatty acids, eicosanoids, retinoids, and steroids have important functions both as energy sources and in metabolic signaling. They affect fundamental cellular processes such as gene transcription, cell development, inflammation, and immune response (1–3). The cellular cytosol is hydrophilic, and lipids need to be solubilized and protected from chemical damage. Their transport and availability are tightly regulated. Proteins that coordinate the lipid traffic include lipoproteins (such as the low density lipoprotein) and carrier proteins, known as lipid-binding proteins (LBPs).2 In vertebrates LBPs belong to the β-sheet calycin superfamily (lipocalins and fatty acid-binding proteins (FABPs)) or the α-helical serum albumin-like superfamily.
Nematodes are one of the most abundant groups of multicellular organisms. Parasitic nematodes cause serious and difficult to treat diseases in humans, animals, and plants affecting human health as well as having a negative impact on agricultural economics. It is estimated that more than one-sixth of the earth's population (mainly in developing countries), suffers from nematode infections, and at least 4 of the 15 neglected tropical diseases listed by the World Health Organization are caused by nematodes. Parasitic worms possess limited lipid metabolism and depend on import of essential lipids from their host (4), which makes the lipid transporters good targets for chemoprophylactic treatments. A 14-kDa FABP (Sm14) has been proposed as a vaccine candidate against Schistosoma mansoni in humans and Fasciola hepatica in cattle and sheep (5, 6). Work with parasitic species, which possess a complex life cycle often involving several hosts, is difficult, and therefore, Caenorhabditis elegans has been proposed as a suitable model organism for studying roundworm diseases and nematode metabolism (7, 8).
FABPs are found in vertebrates and invertebrates including parasitic worms and the free-living nematode C. elegans (Refs. 9 and 10 and the Wormbase database). They have gained medical importance as intracellular lipid chaperones (10), and they also play a role in metabolic diseases (2, 11, 12). It has even been suggested that inhibitors of FABPs could present a novel way of treating these metabolic diseases (11). Despite varying sequence identity (15–70%), different, tissue-specific, FABPs all have similar β-barrel structures that encase the bound fatty acid (Ref. 9 and references therein).
Nematodes have FABPs, but they also possess different and unique LBPs such as nematode polyprotein allergen/antigen proteins and fatty acid- and retinoid-binding proteins (FARs) (13). Both groups are allergens and are generally secreted from the parasite into the host tissues (13–15). There are no available three-dimensional structural data, but circular dichroism (CD) measurements and secondary structure predictions suggest these proteins are predominantly α-helical. Their importance for lipid metabolism, their antigenic properties, and the structural difference from their host FABP proteins makes them an interesting target for structural work. The first described FAR family member was Ov-FAR-1 from the filarial agent Onchocerca volvulus, which causes human river blindness (15). Ten more FAR proteins from filarial species, all causing serious sickness in humans and animals, have been studied (16). They belong to two major clusters and share high sequence similarity (79–100% as defined in Ref. 17). The first contains proteins from nodule species such as O. volvulus (Ov-FAR-1), and the second contains proteins from lymphatic species such as Brugia malayi (Bm-FAR-1), which causes elephantiasis (16). FAR proteins are classified as a pfam domain pfam05823:Gp-FAR-1 (17).
Parasitic nematodes possess one or two types of FAR proteins (16, 18) (see the Nematode Genome Sequencing Center website), but the free-living C. elegans produces eight FAR proteins (Ce-FAR-1–8) (19). They belong to three groups: group A (Ce-FAR-1, -2, and -6), group B (Ce-FAR-3, -4, and -5), and group C (Ce-FAR-7 and -8). Group A has the highest sequence identity to FARs from parasitic nematodes, such as Ov-FAR-1 (19). A majority of FAR proteins contain a signal peptide and are shown or are likely to be secreted. Some FARs are glycosylated (16, 19), and they apparently have a casein kinase II phosphorylation site (19).
There is a report of a NMR structure of a nematode polyprotein allergen protein (20), although coordinates are not available, but there is no structural information available on FAR proteins. Here we report the first high resolution x-ray crystallographic structure of a representative of the FAR family, Ce-FAR-7, from C. elegans, its affinity for some fatty acids, its phosphorylation effects, and its localization in C. elegans. The structure reveals a totally new α-helical fold, and although the sequence identity with other FAR proteins is low, structure-based sequence alignment suggests that this is the common FAR fold.
EXPERIMENTAL PROCEDURES
Protein Cloning, Expression, and Purification
Ce-FAR-7 was amplified from C. elegans cDNA and cloned into the pETM-11-LIC expression vector.3 The T26D mutant was produced by site-directed mutagenesis using the QuikChangeII® site-directed mutagenesis kit (Stratagene). All primers are given in supplemental Table S1. The recombinant full-length proteins contained an N-terminal His6 tag. Both were expressed in BL21 (DE3) pLysS Escherichia coli cells (Stratagene). Recombinant Ce-FAR-7 was produced using a Biostat B-DCU Quad benchtop fermenter system (B. Braun Biotech International) induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 20 °C overnight. Recombinant Ce-FAR-7 T26D was expressed in shaker cultures under the same conditions. Seleno-l-methionine was obtained from Sigma, and selenomethionine-labeled protein was expressed in B834 (DE3) pLysS E. coli cells using the standard protocol (21).
Native or selenomethionine proteins were purified by nickel affinity chromatography on nickel-SepharoseTM 6 Fast Flow (GE Healthcare). The His6 tag was cleaved by incubation with tobacco etch virus protease, and the samples were then further purified by anion exchange chromatography on a 5/5 Mono Q column (GE Healthcare) and gel filtration on a 16/60 SuperdexTM 75 (GE Healthcare) column. Purified protein was treated with Lipidex-1000 (PerkinElmer Life Sciences) for two serial incubations of 1 h while shaking at 37 °C to remove residual lipids from the protein.
Crystallization
Initial crystallization conditions were determined at the high throughput crystallization facility at the EMBL Hamburg Outstation (22) using Ce-FAR-7 in 20 mm Tris, pH 8.5, 50 mm NaCl, and 5 mm 2-mercaptoethanol at concentrations ranging from 5 to 10 mg/ml. Subsequent optimization and additive screening resulted in crystals that diffracted to 1.8 Å. Clusters of fine plates were obtained from 2.1–2.9 m ammonium sulfate, 100 mm Tris, pH 7.8 to 8.5, or 100 mm MES, pH 6.2–6.5, at 20 °C with 3% of a carbohydrate such as d-(+)-glucose monohydrate, sucrose, or xylitol as an additive.
Data Collection and Structure Determination
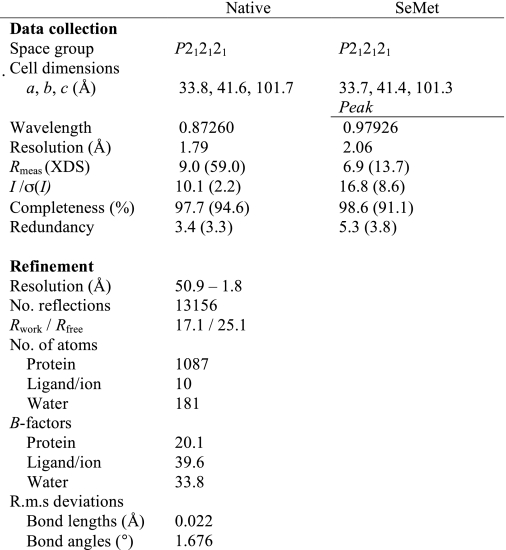
Data collection and refinement statistics are given in Table 2. The Ce-FAR-7 structure was solved at 2.5 Å using Se-SAD phasing with data collected on beamline ID29 at the European Synchrotron Radiation Facility, Grenoble. The model was refined against 1.8 Å data collected on ID23–2 at the European Synchrotron Radiation Facility. Data were processed with XDS (23) and scaled with SCALA (24, 25). Phases and initial maps were obtained by using the autoSHARP package (26). An initial model was built automatically using ARP/wARP (27) followed by cycles of manual rebuilding in Coot (28) and refinement with REFMAC5 (29). The final structure has good stereochemistry with 99.2% of the residues in core regions of the Ramachandran plot and only 0.8% outliers.
TABLE 2.
Data collection, phasing, and refinement statistics of Ce-FAR-7
One crystal was used for each of the datasets.
Phosphorylation
Ce-FAR-7 was phosphorylated in vitro with casein kinase II (CKII) (New England Biolabs) using 700 units of CKII/μg of protein in the presence of 1 mm ATP (New England Biolabs) and CKII buffer (New England Biolabs). The mixture was incubated for 3 h at 30 °C, and the phosphorylated sample and nonphosphorylated control were sent for mass spectral analysis at the Biomolecular Sciences Mass Spectrometry and Proteomics Unit, University of St. Andrews. The protein sample (20 μl, 10 μm) was desalted on-line through a MassPrep On-Line Desalting Cartridge 2.1 × 10 mm and delivered to an electrospray ionization mass spectrometer (LCT, Micromass, Manchester, UK) which had previously been calibrated using myoglobin. The envelope of multiply charged signals obtained was deconvoluted using MaxEnt1 software to give the molecular mass of the protein. The in-gel digestion was prepared according to Shevchenko et al. (30), and Lambda-protein phosphatase (New England Biolabs) was used for further dephosphorylation of the peptides. Experiments were performed using a Q-Star XL tandem mass spectrometer (Applied Biosystems, Foster City, CA) and a 4800 matrix-assisted laser desorption ionization time-of-flight (MALDI TOF/TOF) analyzer (Applied Biosystems) and analyzed with the Mascot 2.1 search engine (Matrix Science, London, UK) against the UniProt (Swiss-Prot and TREMBL combined) data base (April 2009).
Steady-state Fluorescence Binding Experiments
The fatty acids and retinol were purchased from Sigma. All ligands were dissolved in ethanol in concentrations of either 1, 0.1, 0.05, or 0.01 mm. The concentration of retinol was calculated from the absorption spectra using the molar extinction coefficient of 52.48 × 10−3 cm−1 m−1 at 325 nm. The protein concentration was calculated with molar extinction coefficients for protein with or without a His6 tag of 4.72 or 1.74 × 10−3 cm−1 m−1, respectively (31). Steady-state fluorescence was measured with a FluoroLog-3® (HORIBA Jobin Yvon) fluorimeter equipped with a thermostatically controlled cuvette holder.
Ligand binding experiments for the fatty acids were performed with 1.3 μm Ce-FAR-7 (with or without His6 tag) and an initial sample volume of 1 ml. Binding affinities of Ce-FAR-7 for fatty acids were studied by changes in the intrinsic tyrosine/phenylalanine emission of the protein (excitation wavelength λexc of 275 nm and emission wavelength λem,max 307 nm). Ligand binding experiments for retinol were performed with either (i) a constant ligand concentration of 1 or 1.5 μm and titration with increasing protein concentrations or (ii) a constant protein concentration of 1.3 μm and following changes in specific emission intensity (λexc 350 nm and λem,max 420 nm) with increasing retinol concentration. Displacement experiments were performed after 5 min of incubation of 2 μm Ce-FAR-7 with 10 μm retinol (λexc 350 nm and λem,max 420 nm) followed by the addition of 10 μm fatty acid and a further 5–10 min of incubation time. The emission spectra were corrected for background fluorescence and inner filter effects where necessary. Samples were equilibrated until a steady emission reading was obtained (usually 3–5 min). Binding experiments were carried out in 1× phosphate-buffered saline, pH 7.4 (32), with 5 mm 2-mercaptoethanol at 20 °C. Binding of oleic acid to Ce-FAR-7 was also tested in 20 mm Tris, pH 8.0, 50 mm NaCl and 5 mm 2-mercaptoethanol, 1× phosphate-buffered saline, pH 7.4, and 5 mm 2-mercaptoethanol as well as in a non-reducing buffer (20 mm HEPES, pH 7.4, and 50 mm NaCl). The final concentration of the organic solvent did not exceed 3%. To minimize inner filter and self-absorption effects, absorbance of the samples at the excitation wavelength was always less than 0.05. All emission spectra were corrected for progressive dilution (≈3% maximum) (33).
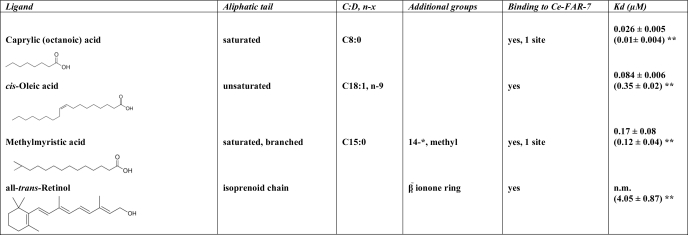
The dissociation constant (Kd) was calculated from the experimental data of three to six independent measurements. For all experiments the changes in the fluorescence emission were converted to percent values and analyzed with GraphPad Prism software package. The results are given in Table 1 including the standard error of the Kd.
TABLE 1.
Binding data of lipophilic ligands to Ce-FAR-7
C:D is the number of carbon atoms in the fatty acid, and D is the number of double bonds in the fatty acid. n-x is the double bond located on the xth carbon bond, counting from the terminal methyl carbon toward the carbonyl carbon. The single asterisk (*) indicates the position of the additional group, counting from the carbonyl group toward the terminal methyl group. The double asterisk (**) indicates the Kd calculated for the T26D mutant. n.m. indicates not measurable.
Tissue Localization of Ce-FAR-7
The C. elegans strain used in this study, pha-1(e2123) (34), was cultured at 15 °C on nematode growth media agar plates with the E. coli strain OP50 as food source using standard methods (35).
The entire far-7 gene along with 1000 bp upstream of the start codon was amplified from C. elegans genomic DNA by PCR (supplemental Table S1), and the insert was cloned into the pPD95.77 vector. The vector contains a promoterless green fluorescent protein (GFP) gene with the S65C mutation that improves fluorescence levels (1995 Fire Vector Kit). Germline transformation was performed using C. elegans pha-1(e2123) mutants by co-injecting the construct with the dominant marker gene pha-1 into the germline of L4 pha-1 mutants (a kind gift from R. Schnabel, Technisches Universität Braunschweig, Braunschweig, Germany). The selection of transgenic worms of the pha-1/pBX system is based on the temperature-sensitive embryonic lethal mutation pha-1. After microinjection, the animals were transferred to 25 °C, where only the transformed progeny survive. Worms were cultivated for 24 h on nematode growth media plates either seeded with OP50 (fed condition) or unseeded (fasted condition). Images were captured with a Zeiss axiovert 100 microscope equipped with fluorescein isothiocyanate/GFP filters.
RESULTS
Ce-FAR-7 Structure
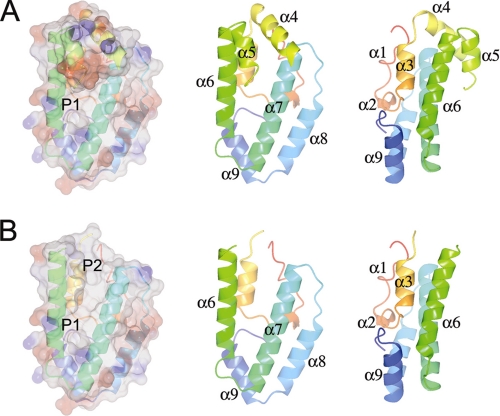
The structure (Fig. 1A and supplemental Figs. S3 and S4) is centered around two long amphipathic helices, α6 and α7, which do not directly contact each other but are inclined to each other by about 20 degrees. This angle is constrained at one end by the turn and at the other by the amphipathic helices α4 and α5. α6 and α7 together with α8 are roughly coplanar and are covered on one side by the helices α1, α2, α3, and α9. α4 and α5 cover the wider part of the other side.
FIGURE 1.
The structure of Ce-FAR-7. A, shown is a transparent electrostatic surface rendering (produced with ccp4mg (50)) with the underlying schematic colored from the N to the C terminus from red to blue to the right. The helices are labeled in two schematic representations rotated relative to each other by 90° about the vertical axis. B, the same images are shown with the helices α4 and α5 removed to better illustrate the two pockets (P1 and P2) and the surface groove between them that is partly covered by α4, α5, and the connecting, partially missing, loop (L4–5).
This structural organization (Fig. 1B) results in two deep hydrophobic pockets (P1 and P2) joined by a cleft, which would allow Ce-FAR-7 to accommodate a variety of ligands with different lengths of aliphatic chain. This hypothesis is confirmed by the ligand binding experiments described below. The cleft is capped by the helices α4 and α5 and the linker (L4–5) which joins them. This “lid” is flexible, at least when no ligand is bound, and the region (Ser-41 to Cys-46) is ill-defined in the electron density map and is not modeled in the structure. P1 is relatively narrow and would appear able only to accommodate an aliphatic chain with a maximum length of seven to eight carbon atoms. P2 could accommodate the bulkier isoprenoid chain of retinol. The DALI server (36) yields no structural homologs for Ce-FAR-7, demonstrating that this structure has a new fold and suggesting that the entire FAR family is structurally unique.
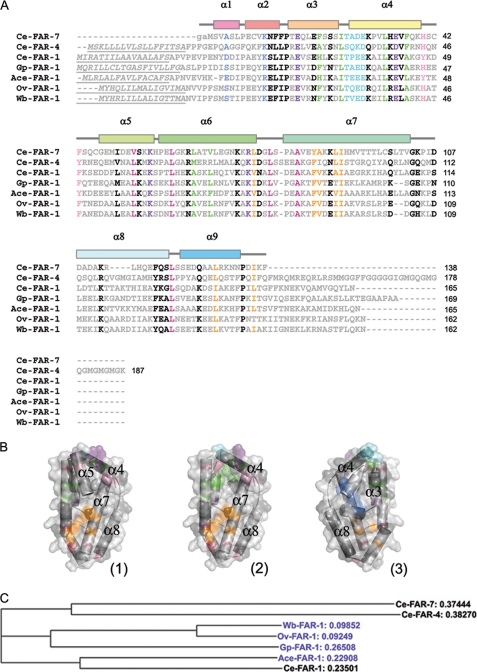
The C. elegans genome codes for eight FAR proteins that belong to three distinct groups (19), and FAR proteins are found in many parasitic nematodes, including filarial nematodes (16). Fig. 2 shows a sequence alignment between Ce-FAR-7 (group C), one representative of each of the other C. elegans FAR groups (Ce-FAR-4 from group B and Ce-FAR-1 from group A), one of each of the main clusters from the highly similar filarial FARs (Ov-FAR-1 from O. volvulus and Wb-FAR-1 from Wuchereria bancrofti), one from the plant parasite Globodera pallida (Gp-FAR-1), and one from the hookworm Ancylostoma ceylanicum. (Ace-FAR-1). An extended alignment is provided in supplemental Fig. S1. The residues Leu-78, Ala-82, Leu-121, and Leu-129 are well conserved and help determine the angle between α6 and α7. At the other end of the groove formed by these helices the well conserved residues Leu-33, Val-53, and Leu-60 are involved in determining the orientation between α6 and both α4 and α5. A salt bridge between a conserved acidic (Glu-35) and basic (Lys-56) residue helps to maintain the orientation between α4 and α5. The residues lining the pocket P1 (Leu-75, Tyr-85, Ala-86, Leu-89, Ile-90, Leu-129, and Ile-136) are all hydrophobic/ aromatic residues, conserved to a high degree within the family members. The aliphatic chain of Arg-74 also forms part of the cavity wall, and this residue forms a conserved salt bridge with Glu-17, although acidic and basic residues are exchanged in a few FARs. Ce-FAR-7 also differs from most other family members in that the C terminus is also stabilized by the interaction between Asp-135 and Arg-74. The residues lining the pocket P2 (Phe-21, Ile-25, Leu-33, Phe-37, Leu-64, and Val-67) are also always hydrophobic or aromatic except at the N terminus (Met-1) and the end of α7 (Thr-101). However, the residues lining P2 vary more than for P1. Toward the bottom of this cavity is also a conserved CKII phosphorylation site (Thr-26, Ala-27, Asp-28, Glu-29) that is followed in Ce-FAR-7 by a proline (Pro-31) residue in the middle of helix α4, suggesting that the cavity might be altered in shape or extent upon phosphorylation. The walls of the groove are formed primarily from the hydrophobic residues of α6 and α7. The orientation between helices α1 and α2, which forms the base of P2, is maintained by a hydrogen bond between the conserved Lys-11 and the carbonyl oxygen of Ala-4. The helix initiating prolines of α2 (Pro-7), α3 (Pro-15), and α7 (Pro-80) are especially well conserved, as is Pro-134, and it is possible that in other, longer, FAR family members this residue might initiate an additional C-terminal helix (α10).
FIGURE 2.
Sequence-related information. A, shown is multiple sequence alignment between representatives of the FAR proteins from C. elegans groups A, B, and C as well as from parasitic nematodes. The figure was prepared with Clustal W 1.83 (51). Ce-FAR-7 is Q9TZ51_CAEEL, Ce-FAR-4 is Q19477_CEAEL, and Ce-FAR-1 is FAR1_CAEEL from C. elegans, Gp-FAR-1 is Q94569 from G. pallida, Ace-FAR-1 is B3U0R8_9BILA from A. ceylanicu, Ov-FAR-1 is FAR1_ONCVO from O. volvulus, and Wb-FAR-1 is FAR1_WUCBA from W. bancrofti (protein IDs are from UniProtKB/TrEMBL). Conserved residues are colored as follows; residues determining helix orientation are in raspberry, residues on the surface of pocket P1 are in orange, residues on the surface of pocket P2 are in green, conserved salt bridges are in purple, hydrogen-bonded residues are in blue, the conserved CKII phosphorylation site is colored cyan, residues from the flexible loop L4–5 are pink, and all others are black. Underlined italics signify the predicted signal peptide regions. Lowercase characters are residues appended to the gene sequence after tobacco etch virus cleavage of the His6 tag. Helices represent the secondary structure of Ce-FAR-7 and are colored as in Fig. 1. B, shown are a surface representation of Ce-FAR-7 with conserved residue regions colored as in A (1) and the surface representation without α5 to show more clearly the P2 pocket (2) and 180° rotation relative to 2 (3). The figures were produced with Pymol (52). C, shown is a non-rooted phylogenic tree prepared with Clustal W (51) for sequences in A. Relative distances are indicated, and proteins from parasitic nematodes are colored blue.
Ligand Binding to Ce-FAR-7
Ligand binding affinities of Ce-FAR-7 were investigated by steady-state fluorescence spectroscopy titration experiments. Four chemically and structurally different ligands were used (Table 1). All lipids were bound by Ce-FAR-7 although with quite different affinities (Table 1). Caprylic acid was bound with the highest affinity (Fig. 3), with lower affinities for oleic acid and 13-methyl myristic acid (see supplemental Fig. S2). The binding of the fatty acids was monitored by measuring the changes in the Tyr/Phe fluorescence of Ce-FAR-7.
FIGURE 3.
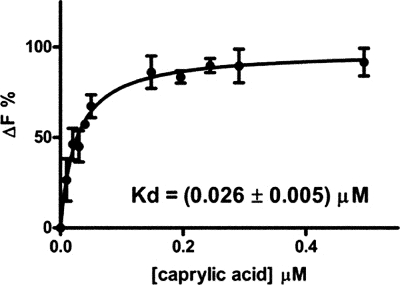
Binding of caprylic acid to Ce-FAR-7. Binding was followed by the changes of the Tyr/Phe emission of the protein at 307 nm after excitation at 275 nm. The derived curve is fitted by a nonlinear regression using GraphPad Prism software package. The saturation isotherm corresponds to ligand biding to one site, and the calculated Kd is 0.026 ± 0.005 μm. Other quantitative binding data are given in Table 1 and the supplemental Fig. S2. ΔF is the % change in fluorescence.
Retinol binding was monitored by changes in its own fluorescence, namely, a blue shift and increased intensity upon the addition of protein to the ligand (14, 37). It was bound with a low affinity, and saturation was not reached even at protein concentrations above 13 μm. The lower affinity observed for retinol could be an indirect result of the bulky ionone ring. The changes observed in the self-fluorescence of retinol were similar to those previously reported (19) (supplemental Fig. S2). Bound retinol was not displaced by caprylic or methyl myristic acids but was by oleic acid. Because the Hill coefficient for oleic acid was greater than unity (supplemental Fig. S2), it is possible that this ligand binds to more than one site and is, therefore, able the displace retinol bound in P2.
The dissociation constant and binding mode for oleic acid to Ce-FAR-7 was not affected by varying the buffer (HEPES, Tris, or phosphate-buffered saline), by the presence of reducing agent (5 mm 2-mercaptoethanol), by pH within the range from 7.4–8.0, or by ionic strength as probed by salt concentrations of 50 mm NaCl or phosphate-buffered saline (137 mm NaCl and 2.7 mm KCl) (32). The affinity did not depend upon the presence of the N-terminal His6 tag. The lack of in vitro dependence of ligand binding on reducing agent is an important observation, because Cys-42 in the flexible loop is probably close enough to Cys-98 to form a disulfide bridge. Neither cysteine is conserved in other FAR proteins, but such a disulfide link might constrain the flexibility of the loop L4–5, thus making the ligand binding affinity or even specificity of Ce-FAR-7 dependent upon redox potential.
Potential Regulation by Casein Kinase II
Using casein kinase II, Ce-FAR-7 was phosphorylated in vitro. Intact mass analysis showed an 81-Da (expected 80 Da) difference between phosphorylated and non-phosphorylated samples, confirming that Ce-FAR-7 has been phosphorylated. To map the phosphorylation, site-phosphorylated and non-phosphorylated Ce-FAR-7 samples were subjected to in-gel digestion and mass spectrometry. The peptide coverage for the non-phosphorylated control was 94%, whereas the coverage for the phosphorylated sample was 74%, as the peptide (NFFPTEQLEFSSSITADEKPVLHEVFQ) signal was missing. Specifically, neither this signal at 1090 Da (triply charged) nor that of the phosphopeptide at 1116 Da (also triply charged) was present. Upon treatment of the phosphorylated peptide with phosphatase, the signal at 1090 Da was again detected.
The phosphorylated product was unstable, and so to mimic phosphorylation, Thr-26 was mutated to Asp (Ce-FAR-7 T26D mutant) and used in comparative ligand binding experiments. Binding of retinol and fatty acids was studied by the addition of increasing concentrations of ligand to the T26D mutant. T26D bound retinol with higher affinity, and unlike for the native protein, saturation was reached (Kd 4.05 ± 0.87 μm). In contrast the affinity for fatty acids was not affected. As for the native protein, caprylic acid was bound to Ce-FAR-7 with the highest affinity followed by 13-methyl myristic and oleic acids (Table 1 and supplemental Fig. S2). These results would suggest that the retinol binding site (P2) is probably regulated by casein kinase II, whereas the fatty acid binding pocket (P1) is not affected by phosphorylation of the protein. However, the affinity for retinol of T26D is an order of magnitude lower than that for fatty acids, implying that, in contrast to the other family members, transport of retinoids is not the major function of Ce-FAR-7. Nevertheless we suggest that FAR protein affinity for signaling lipids, such as retinoids, is probably regulated in all members, as the CKII phosphorylation site is conserved within the family (cyan in Fig. 2).
GFP Localization in C. elegans
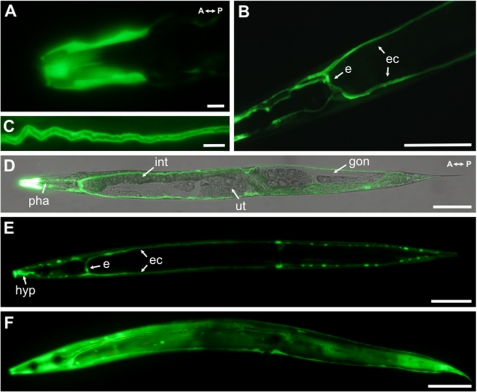
Ce-FAR-7 contains no classical secretion signal, and consequently we analyzed the expression pattern of far-7. Transgenic worms were created that expressed far-7::GFP under the control of the far-7 promoter (Pfar-7far-7::GFP) (Fig. 4). GFP signals were detected during all stages of C. elegans development (larvae L1-L4 and adult hermaphrodites). Strong far-7::GFP expression was observed in parts of the hypodermis, in particular the syncytia covering the lips and parts of the head region (Fig. 4, A, D, and E). The C. elegans hypodermis acts in nutrient storage, secretes the cuticle, and takes up apoptotic cell bodies by phagocytosis (Ref. 38; see also the Wormbase database). Because Ce-FAR-7 becomes intensively localized in the hypodermis of the head and the lips, this could point to an indirect role of the protein in the interaction of the worm with the external environment.
FIGURE 4.
Analysis of the expression pattern of Ce-FAR-7 in C. elegans. GFP images of adult C. elegans carrying Pfar-7far-7::GFP are shown. A, strong GFP signals detected in hypodermal syncytia (hyp) covering the lips and head portion are shown. B, the large H-shaped excretory cell (e) that extends bilateral canals (ec) anteriorly and posteriorly nearly the whole length of the animal is shown. C, GFP signals are located in the excretory cell cytoplasm, not within the lumen of the excretory duct. D, shown are combined confocal differential interference contrast and fluorescence micrographs; pha, pharynx; int; intestine; ut, uterus; and gon, gonads. E, GFP expression profile over the whole animal is shown. F, GFP expression profile of a nematode starved for 24 h is shown. Scale bar for A and C, 10 μm; scale bar for B, D, E, and F, 100 μm.
The second major localization of far-7::GFP expression was in the H-shaped excretory cell (Fig. 4, B, D, and E). The excretory cell, the largest cell in C. elegans, forms two canals running the entire length of the nematode. The canals are connected to the hypodermis (via extensive gap junctions), and their basal surface remains in contact with the body cavity (pseudocoelom) (39). The proposed functions of the excretory system are osmoregulation, excretion of metabolic waste, and secretion of molting fluid and/or secretion/export of hormones to target tissues (Ref. 38; see also the Wormbase database). All this suggests metabolites accumulate in this cell, necessitating a good intracellular transport system for hydrophobic ligands as well as other metabolites. far-7::GFP expression is observed in the cytoplasm of the excretory cell and not in the lumen of the excretory duct (Fig. 4C), in agreement with the lack of classical signal peptide in the Ce-FAR-7. The fact that no expression was observed in other interfacial or gland cells that are associated with the excretory system (Ref. 38; see also the Wormbase database) also supports a potential intracellular function for Ce-FAR-7. Additional experiments by feeding double-stranded Ce-FAR-7 RNA to the far-7::GFP worms strongly inhibited GFP fluorescence, clearly indicating that the GFP signal observed is not an artifact (data not shown). Expression in the head hypodermis and the excretory cell remains upon fasting (Fig. 4F), but additional strong expression of far-7::GFP is observed in the hypodermal syncytium (hyp) covering the body of the animal (Fig. 4F).
DISCUSSION
Structure
The sequence alignment (Fig. 2 and supplemental Fig. S1) and the structural analysis given above would suggest that the structure (Fig. 1A) is representative of the complete family of FAR proteins. Previous structural analysis of this family has been limited to a small angle x-ray scattering study (40), and although the overall molecular dimensions and shape are consistent with our work, the assumption of a structural similarity of the FAR proteins with the ligand binding domain of the retinoic acid receptor (RXRα (41)) and the nematode polyprotein allergens (20, 42) is incorrect. The fundamental differences between Ce-FAR-7 and other FAR proteins are the absence of an N-terminal secretion signal (see below) and the possible existence of an additional C-terminal helix. The latter possibility has been previously predicted (19). The loop L7–8 may also be longer in other FARs, or alternatively, α8 might have one more helical turn. Ce-FAR-7 is monomeric in both liganded and unliganded forms (data not shown). Ce-FAR-4 has been reported to be monomeric in the unliganded state, whereas Ov-FAR-1 is reported to be dimeric in both liganded and unliganded states (40). The structure reported here indicates that the dimerization interface would not include the ligand binding face involving helices α4 through α7 (Fig. 1A) but could well be related to the existence of an additional C-terminal helix.
Ligand Binding
The structure shows a complex potential ligand binding site comprised of two pockets with a surface groove joining them (Fig. 1B). This groove is partially covered by helices α4, α5, and the flexible loop (L4–5) between them. This loop shows strong conservation of residues with planar ring systems in two places (His-40 and Phe-43). If the loop was closed, it would cover most of the binding groove but would not occlude pocket P1. These residues are unlikely to stabilize binding of an aliphatic chain but may well help stabilize the binding of a more complex head group such as that found in retinol. Circular dichroism experiments show that there is no detectable change in secondary structure (data not shown) upon binding of fatty acids.
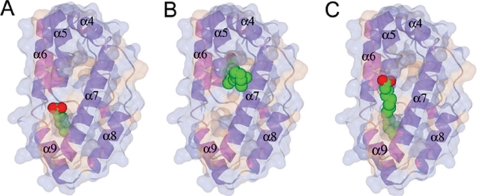
Of the ligands we have examined, the highest affinity observed is for caprylic (octanoic) acid. It is probably no coincidence that the binding pocket P1 is sufficiently long and narrow to accommodate almost perfectly the aliphatic chain of caprylic acid (Fig. 5A and supplemental Movie S1), with the head group just extending out of the cavity. The bottom of P1 is hydrophobic in nature and possibly too small to be able to bind the carboxylate head group of a fatty acid. Indeed, modeling suggests that P1 can barely accommodate the branched aliphatic tail of methyl myristic acid (see Table 2). Methyl myristic acid would not be expected to bind well in P2 because the pocket is too large. We believe that longer saturated or unsaturated fatty acids bind using P1, and the groove as shown for methyl myristic acid in Fig. 5C and supplemental Movie S3. For Ce-FAR-7, retinol cannot use cavity P1, because of the more bulky and rigid isoprenoid chain. Nevertheless other FARs do bind retinol with an affinity similar to that for fatty acids (15, 16, 18, 19), which means that P2 in other FARs might be better matched to these ligands. That the binding pockets for fatty acids and retinol are distinct (Fig. 5B and supplemental Movie S2) is consistent with the observation that caprylic and 13-methyl myristic acids do not displace retinol from the wild type protein despite having a higher affinity. However, oleic acid does displace retinol, which suggests that it could bind in P2 as well as in P1. Its binding to more than one site is consistent with the higher Hill coefficient (supplemental Fig. S2).
FIGURE 5.
Docking of fatty acids to Ce-FAR-7. A, caprylic acid bound to P1 is shown. B, retinol docked in P2 is shown. C, methyl myristic acid bound in P1 is shown. Ligands were docked manually and the geometry was idealized using REFMAC5 (29). 360° rotation movies are given in the supplemental Movies S1–S3.
In FABPs the ligand is bound within a large hydrophobic cavity (Ref. 10 and references therein), and the fatty acids are generally observed in a twisted horseshoe shape (Ref. 9 and references therein). It appears that nematodes have developed a different mechanism for binding a variety of different, predominantly hydrophobic ligands, which in turn means that inhibitors of FABPs are unlikely to be very effective at inhibiting FARs. Conversely, inhibitors specifically designed for FARs are not likely to interact with host FABPs.
All FAR proteins have a conserved casein kinase II phosphorylation site (cyan in Fig. 2A), and here we show that Ce-FAR-7 is indeed phosphorylated by this kinase in vitro and that mimicking phosphorylation results in increased affinity for retinol but not for simple fatty acids. This would suggest that protein function and/or localization are regulated by phosphorylation in vivo. Two CKII genes exist in C. elegans, corresponding to the two subunits of CKII (see the Wormbase database). Phosphorylation of a Tyr residue is also observed in mammalian adipocyte (43) and heart (or muscle) FABPs, although the reports on the latter are contradictory (44, 45).
A second isoform of Ce-FAR-7, Ce-FAR-7b (Q86s19_ CAEEL), has been postulated on the basis of cDNA (see the Wormbase database). This isoform is truncated at the N terminus and misses the first 4 helices. As mentioned above, α1 and α2 form the bottom of pocket P2, which could in principle produce a protein capable of binding an aliphatic chain of almost any length in a nonspecific manner with low affinity. Further work on this isoform is required to confirm its existence and to establish its specific function.
Localization and Function
The result of chaperoning poorly soluble organic molecules, such as lipids and fatty acids, can have many outcomes. If transport is desired, the affinity for the ligand should not be too great, as high affinity binding could act to negatively regulate signaling molecules of this type. Ce-FAR-7 possesses two binding sites, P1 and P2, for two types of ligands, fatty acids and retinoids, with only the latter regulated by CKII. The phosphorylation results in increased affinity for retinol, and this would imply that FARs play a role in the signaling processes in the nematode even if the lower affinity for retinol relative to that for fatty acids would suggest that this is not the major function of Ce-FAR-7. As already mentioned, Ce-FAR-7 does not contain a classical secretion signal peptide. However, the SecretomeP Version 2.0 algorithm (46) does predict that it could be secreted via an alternative pathway (data not shown).
Complete information on C. elegans lipid-binding protein expression is not available. From localization and expression patterns of FAR proteins and a comparison with other lipid transporters, we can conclude that there is indeed a tissue and stage-specific expression as well as different subcellular localizations. C. elegans LBPs are expressed in muscles, pseudocoelom, intestine, hypodermis, pharynx, and reproductive system (see the Wormbase database); however, Ce-FAR-7 is the first to be localized in the excretory cell and the lips.
far-7
GFP expression is observed in two functionally different parts of C. elegans; that is, the hypodermis, involved in nutrient storage and external environment contact, and the excretory cell, which functions in osmotic regulation and metabolite excretion. We propose that Ce-FAR-7 plays an important role in the intracellular lipid trafficking of both tissues and is, therefore, important for more than one process in C. elegans. Fasting stimulates the expression of various genes that are involved in converting fat stores into energy, and fasted C. elegans display decreased fat deposits and considerable changes in fatty acid composition (47). Interestingly, Taubert et al. (48) found that far-7 expression is up-regulated in response to fasting, and our experiments support this observation (Fig. 4F). Their work demonstrated that up-regulation of far-7 does not depend on the nuclear hormone receptor NHR-49 or require the mediator subunit MDT-15 (48), and thus, unidentified regulatory complexes modulate far-7 expression in response to fasting. Expression of Ce-FAR-7 in the whole hypodermis during starvation suggests an up-regulation to mobilize to the greatest extent the fatty acids necessary for nematode survival. This implies a function in nutritional rather than signaling processes, in agreement with the higher affinity for nutritional molecules, such as fatty acids, over retinol.
Although we describe the general structure-function relationships for this family of proteins, there are many differences in detail. Parasitic nematodes, as opposed to the free-living C. elegans, appear to have only one or two FAR proteins despite their limited lipid metabolism and their strong dependence on lipid transport proteins (see the Nematode Genome Sequencing Center website). Immunolocalization studies of Gp-FAR-1 from the potato cyst nematode G. pallida show that, like Ce-FAR-7, it localizes to the hypodermis as well as to material shed from the surface of the worm, making host-parasite interaction indirectly feasible (18).
FAR domains do not necessarily occur alone; for example, another unique nematode LBP, Ag-lbp55 from Ascaridia galli, has also been described and contains a C-terminal FAR domain, although its N-terminal part has no known homologues (49). In conclusion we provide the basic structural information for understanding the mode of action of FAR proteins and investigation of inhibitors of lipid binding.
Acknowledgments
We thank the staff at the European Synchrotron Radiation Facility for support in data collection and acknowledge the use of the X12, X13, and BW7A EMBL beamlines at DESY (Deutsches Elektronen Synchrotron). We also thank Dr. Catherine Botting from the University of St. Andrews for mass spectral analysis. We thank Fatma Abdala for help with protein purification and Santosh Panjikar for advice during structure solution.
This work was supported by Marie Curie Intra-European Fellowship Contract MEIF-CT-2006-025860 (to R. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Table S1, and Movies S1–S3.
The atomic coordinates and structure factors (code 2W9Y) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
A. Geerlof, unpublished information.
- LBP
- lipid-binding protein
- FABP
- fatty acid-binding protein
- CKII
- casein kinase II
- FAR
- fatty acid- and retinoid-binding protein
- MES
- 2-(N-morpholino)ethanesulfonic acid
- GFP
- green fluorescent protein
- Gp-
- G. pallida
- Ce-
- C. elegans
- Ov-
- O. volvulus
- Ace
- A. ceylanicu
- Wb
- W. bancrofti.
REFERENCES
- 1.Hotamisligil G. S. (2006) Nature 444, 860–867 [DOI] [PubMed] [Google Scholar]
- 2.Cao H., Gerhold K., Mayers J. R., Wiest M. M., Watkins S. M., Hotamisligil G. S. (2008) Cell 134, 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funk C. D. (2001) Science 294, 1871–1875 [DOI] [PubMed] [Google Scholar]
- 4.Barrett J. (2001) in Perspectives on Helminthology, pp. 309–334, Science Publishers Inc., Enfield, NH [Google Scholar]
- 5.Tendler M., Simpson A. J. (2008) Acta Trop. 108, 263–266 [DOI] [PubMed] [Google Scholar]
- 6.Tendler M., Brito C. A., Vilar M. M., Serra-Freire N., Diogo C. M., Almeida M. S., Delbem A. C., Da Silva J. F., Savino W., Garratt R. C., Katz N., Simpson A. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garofalo A., Kennedy M. W., Bradley J. E. (2003) Med. Microbiol. Immunol. 192, 47–52 [DOI] [PubMed] [Google Scholar]
- 8.Plenefisch J., Xiao H., Mei B., Geng J., Komuniecki P. R., Komuniecki R. (2000) Mol. Biochem. Parasitol. 105, 223–236 [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman A. W., Veerkamp J. H. (2002) Cell. Mol. Life Sci. 59, 1096–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuhashi M., Hotamisligil G. S. (2008) Nat. Rev. Drug Discov. 7, 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuhashi M., Tuncman G., Görgün C. Z., Makowski L., Atsumi G., Vaillancourt E., Kono K., Babaev V. R., Fazio S., Linton M. F., Sulsky R., Robl J. A., Parker R. A., Hotamisligil G. S. (2007) Nature 447, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuncman G., Erbay E., Hom X., De Vivo I., Campos H., Rimm E. B., Hotamisligil G. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6970–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDermott L., Cooper A., Kennedy M. W. (1999) Mol. Cell. Biochem. 192, 69–75 [PubMed] [Google Scholar]
- 14.Kennedy M. W., Brass A., McCruden A. B., Price N. C., Kelly S. M., Cooper A. (1995) Biochemistry 34, 6700–6710 [DOI] [PubMed] [Google Scholar]
- 15.Kennedy M. W., Garside L. H., Goodrick L. E., McDermott L., Brass A., Price N. C., Kelly S. M., Cooper A., Bradley J. E. (1997) J. Biol. Chem. 272, 29442–29448 [DOI] [PubMed] [Google Scholar]
- 16.Garofalo A., Kläger S. L., Rowlinson M. C., Nirmalan N., Klion A., Allen J. E., Kennedy M. W., Bradley J. E. (2002) Mol. Biochem. Parasitol 122, 161–170 [DOI] [PubMed] [Google Scholar]
- 17.Marchler-Bauer A., Anderson J. B., Cherukuri P. F., DeWeese-Scott C., Geer L. Y., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Marchler G. H., Mullokandov M., Shoemaker B. A., Simonyan V., Song J. S., Thiessen P. A., Yamashita R. A., Yin J. J., Zhang D., Bryant S. H. (2005) Nucleic Acids Res. 33, D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prior A., Jones J. T., Blok V. C., Beauchamp J., McDermott L., Cooper A., Kennedy M. W. (2001) Biochem. J. 356, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garofalo A., Rowlinson M. C., Amambua N. A., Hughes J. M., Kelly S. M., Price N. C., Cooper A., Watson D. G., Kennedy M. W., Bradley J. E. (2003) J. Biol. Chem. 278, 8065–8074 [DOI] [PubMed] [Google Scholar]
- 20.Meenan N. A., Cooper A., Kennedy M. W., Smith B. O. (2005) J. Biomol. NMR 32, 176. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) EMBO J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller-Dieckmann J. (2006) Acta Crystallogr D. Biol. Crystallogr 62, 1446–1452 [DOI] [PubMed] [Google Scholar]
- 23.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 24.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr 50, 760–76315299374 [Google Scholar]
- 25.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26.Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 27.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29.Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 30.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 31.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) The Proteomics Protocols Handbook, pp. 571–607, Humana Press Inc., Totowa, NJ [Google Scholar]
- 32.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., pp. B20–B26, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 33.Lehrer S. S. (1971) Biochemistry 10, 3254–3263 [DOI] [PubMed] [Google Scholar]
- 34.Granato M., Schnabel H., Schnabel R. (1994) Nucleic Acids Res. 22, 1762–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S. (1974) Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson T. C., Wilton D. C. (1986) Biochem. J. 238, 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altun Z. F., Hall D. H. (eds) (2006) WormAtlas, http://www.wormattas.org/
- 39.Buechner M. (2002) Trends Cell Biol. 12, 479–484 [DOI] [PubMed] [Google Scholar]
- 40.Solovyova A. S., Meenan N., McDermott L., Garofalo A., Bradley J. E., Kennedy M. W., Byron O. (2003) Eur. Biophys. J. 32, 465–476 [DOI] [PubMed] [Google Scholar]
- 41.Bourguet W., Ruff M., Chambon P., Gronemeyer H., Moras D. (1995) Nature 375, 377–382 [DOI] [PubMed] [Google Scholar]
- 42.Meenan N. (2004) Structure, Folding, and Function of a Nematode Polyprotein Allergen, Ph.D. thesis, Department of Chemistry, University of Glasgow, Glasgow [Google Scholar]
- 43.Buelt M. K., Shekels L. L., Jarvis B. W., Bernlohr D. A. (1991) J. Biol. Chem. 266, 12266–12271 [PubMed] [Google Scholar]
- 44.Prinsen C. F., Werten P. J., Maassen J. A., Veerkamp J. H. (1994) Biochim. Biophys. Acta 1215, 103–108 [DOI] [PubMed] [Google Scholar]
- 45.Nielsen S. U., Spener F. (1993) J. Lipid Res. 34, 1355–1366 [PubMed] [Google Scholar]
- 46.Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S. (2004) Protein Eng. Des. Sel. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 47.Van Gilst M. R., Hadjivassiliou H., Yamamoto K. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13496–13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taubert S., Van Gilst M. R., Hansen M., Yamamoto K. R. (2006) Genes Dev. 20, 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordanova R., Radoslavov G., Fischer P., Torda A., Lottspeich F., Boteva R., Walter R. D., Bankov I., Liebau E. (2005) J. Biol. Chem. 280, 41429–41438 [DOI] [PubMed] [Google Scholar]
- 50.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 51.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]