Abstract
Mitophagy is an autophagic process that degrades mitochondria by an intracellular engulfment that leads to their delivery into the lumen of the cell's hydrolytic compartment, such as the lysosome in animal cells or the vacuole in yeast. It is hypothesized that such processes serve a quality control function to prevent or slow the accumulation of malfunctioning mitochondria, which are thought in turn to underlie central aspects of the aging process in eukaryotic organisms. We recently identified a conserved mitochondrial protein phosphatase homolog, Aup1, which is required for efficient stationary phase mitophagy in yeast. In the present report, we demonstrate that the retrograde signaling pathway (RTG) is defective in aup1Δ mutants. In agreement with a role for Aup1 in the regulation of the RTG pathway, we find that deletion of RTG3, a transcription factor that mediates the RTG response, causes a defect in stationary phase mitophagy and that deletion of AUP1 leads to changes in Rtg3 phosphorylation patterns under these conditions. In addition, we find that mitophagic conditions lead to induction of RTG pathway target genes in an Aup1-dependent fashion. Thus, our results suggest that the function of Aup1 in mitophagy could be explained through its regulation of Rtg3-dependent transcription.
Introduction
Mitochondria play an essential role in the physiology of eukaryotic cells. They are the sites of aerobic energy production and biosynthesis of amino acid precursors as well as nucleotide and lipid production. At the same time, mitochondria also present a threat to the cell, through production of reactive oxygen species and through the potential leakage of toxic factors, such as cytochrome c.
It is generally hypothesized that quality control mechanisms scavenge malfunctioning mitochondrial compartments, safely neutralizing them before they damage cellular physiology (1, 2). One such quality control mechanism is autophagy, a catabolic process used by cells to transport cytoplasmic material for degradation in the lysosome (in animal cells) or the vacuole (in yeast, filamentous fungi, and plants) (3–5). Autophagic pathways can be classified either according to their molecular mechanism or according to their specificity. Thus, macroautophagy, the most well studied form of autophagy, is thought to be a relatively nonspecific scavenging mechanism that is induced in response to starvation cues (6–8). Mechanistically, macroautophagy involves the generation of a cytosolic intermediate, an autophagosome, that consequently fuses with the lytic compartment or with elements of the endomembrane system that flow into the lytic compartment. On the other hand, microautophagy is a form of autophagy that involves direct involution of the vacuolar membrane, thus allowing import of material into the vacuolar lumen for degradation. There are several well documented types of selective autophagy. When Pichia pastoris is grown on methanol as a carbon source, it massively induces peroxisomes. When these cells are shifted to a better carbon source, such as glucose or ethanol, one observes a selective and quantitative degradation of peroxisomes. Interestingly, shifting to glucose medium induces micropexophagy, whereas shifting to ethanol induces macropexophagy (reviewed in Refs. 9–11). A second form of selective autophagy is piecemeal microautophagy of the nucleus, a process in which parts of the nucleus are engulfed and digested by the vacuole (12). Ribophagy, or selective autophagy of ribosomes, has also been documented (13), and ER-phagy has been suggested to occur in cells that are aggressively inducing the unfolded protein response (14, 15).
Several recent reports indicate the existence of a selective process that targets mitochondria. In mammalian cells, mitochondrial autophagy has been linked to permeability transition pore opening (16, 17), and a sophisticated statistical analysis has indicated that defectively functioning mitochondria are selectively degraded by autophagic mechanisms (18).
A number of laboratories have reported the existence of mitophagic mechanisms in Saccharomyces cerevisiae. It was suggested by Kissova et al. (19) that rapamycin or starvation treatments lead to a mitophagic response in yeast and that the mitochondrial outer membrane protein Uth1 is required for this response. Several additional groups have demonstrated that, under certain pathological conditions in which mitochondrial function is impaired by mutation or toxins, mitochondrial degradation occurs via an autophagic pathway (20, 21).
We recently reported that yeast cells naturally undergo nearly quantitative mitophagy in very long term cultures (incubation times of 3 days and longer) that are grown on lactate as carbon source (22). This is detected as import of mitochondrial material into the vacuolar lumen by observation of mitochondrially targeted GFP2 fluorescence in the vacuolar lumen as well as by degradation of the mitochondrial matrix enzyme aconitase in a Pep4-dependent fashion (22). In contrast with other reports of mitophagic processes in S. cerevisiae, our system appears to reflect a cytoprotective response, in line with the hypothesized role of mitophagy as a quality control mechanism. This implies that our assay may be useful for studying potential implications of mitophagy on the aging process in eukaryotic systems. Furthermore, we found that a mitochondrially localized protein phosphatase homolog, Aup1, is required for efficient mitophagy under these conditions (22). A previous large scale screening effort for rapamycin-sensitive mutants identified aup1Δ mutants as showing rapamycin sensitivity (23), whereas other reports showed these cells to be caffeine-sensitive (24). Because both of these reagents have been shown to inhibit the Tor (target of rapamycin) protein kinase (25, 26), it is possible that Aup1 is part of a signaling mechanism that functionally overlaps with Tor.
In this study, we have tried to identify cellular pathways by which Aup1 regulates mitophagy. We find that aup1Δ mutants are defective in certain aspects of the retrograde signaling pathway (27–29), a mechanism that induces nuclear responses to mitochondrial stress cues. In this pathway, various signals related to mitochondrial function and nutrient availability are channeled to a cytosolic regulator, Rtg2, that inactivates the sequestering factor Mks1, in turn releasing the Rtg3/Rtg1 heterodimer to enter the nucleus and activate a specific transcription program. We find that the function of the RTG pathway transcription factor, Rtg3, is required in stationary phase mitophagy. The dependence of RTG signaling on Aup1 is correlated with changes in the phosphorylation states of Rtg3 during mitophagy, suggesting that Aup1 regulates the retrograde pathway. Finally, we show that mutations in a motif conserved between Aup1 and its closest mammalian homologs, the PP1K phosphatase homolog subgroup within the PP2C protein superfamily, lead to loss of function, suggesting that the Aup1 mechanism of action is conserved between yeast and mammals.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Growth Conditions
Strains used in this study are listed in Table 1. Cells were grown in SD (0.67% yeast nitrogen base without amino acids (Difco), 2% glucose, and auxotrophic amino acids and vitamins as needed) or SL (0.67% yeast nitrogen base, 2% sodium lactate, pH 6.0, and auxotrophic amino acids and vitamins as needed). All culture growth and manipulations were at 26 °C unless indicated otherwise. Deletion mutants and epitope-tagged strains were constructed by the cassette method (30), using plasmids pHAB102 (31), ME3 (36), or pFA6a-KanMX6 as template, unless otherwise specified. To make plasmid pHAB162, the Aup1 reading frame, together with 500 bases of 5′ and 500 bases of 3′ sequences was amplified using a PCR and primers containing SacI and HindIII linkers. The PCR product was verified by sequencing. To clone the C-terminal PP2C motif of Aup1 into the pET14b expression vector, the reading frame was amplified using the oligonucleotide primers ATACTACATATGGGAGGTCCGTATGATGATCCGAATG (5′) and ATACTAGGATCCCTAAACCACCACGCAAGTGGC (3′). The PCR product (spanning nucleotides 636–1228 in the Aup1 open reading frame, representing amino acids 213–409) was digested with BamHI and NdeI and ligated into a BamHI-NdeI-digested pET14b. To generate pDJB11, the IDP1 open reading frame was amplified from genomic DNA using the oligonucleotides 5′-ATACTAAAGCTTTACAACATGAGTATGTTATCTAGAAG (with an HindIII overhang) and 5′-ATACTACCCGGGGCCTCGATCGACTTGATTTC (with an SmaI overhang). The resulting PCR product was digested and cloned into an SmaI-HindII-digested pYES-mtGFP (32), generating an in-frame fusion between IDP1 and then an HindIII-XhoI fragment carrying IDP1-GFP was subcloned into pCu416 (33), generating a fusion protein under the control of the CUP1 promoter. To generate plasmid pYX-GFPATG8, the GFP-ATG8 reading frame was removed from plasmid pHAB142 (34) using EcoRI and SalI and cloned into an EcoRI-SalI-digested pYX142-mtGFP (32).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| HAY75 | MATα,leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Ref. 59 |
| TVY1 | MATα,leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pep4Δ::LEU2 | Ref. 60 |
| HAY809 | HAY75, aup1Δ::HIS5 S.p. | Ref. 22 |
| HAY977 | HAY75, RTG3-GFP::HIS5 S.p. | This study |
| HAY984 | HAY977,aup1Δ::KanR | This study |
| DJY1 | HAY75, aup1Δ::KanR | This study |
| DJY2 | HAY75, rtg2Δ::KanR | This study |
| DJY3 | HAY75, CIT2-HA::HIS5 S.p. | This study |
| DJY4 | DJY1, CIT2-HA::HIS5 S.p. | This study |
| DJY5 | HAY75,rtg3Δ::HIS5 S.p. | This study |
| HAY870 | HAY75, COX9-RFP::KanR | This study |
| HAY805 | HAY75,VPH1-RFP::KanR | Ref. 22 |
| DJY6 | HAY805, rtg3Δ::HIS5 | This study |
| DJY7 | HAY870, ura3-52::pRS306[GFP-ATG8] | This study |
| DJY8 | HAY75, RTG3-HA::HIS5 S.p. | This study |
| DJY9 | DJY1, RTG3-HA::HIS5 S.p. | This study |
| DJY10 | DJY1, DLD3-HA::HIS5 S.p. | This study |
| DJY11 | DJY1, DLD3-HA::HIS5 S.p. | This study |
Site-directed Mutagenesis
Site-directed mutagenesis was according to Kunkel (35) or using the Stratagene QuikChangeTM system (Stratagene, La Jolla, CA). To generate the R366A Aup1 mutant, plasmid pHAB162 was purified in single-stranded form from Escherichia coli strain CJ236 (Bio-Rad), and complementary strand synthesis was primed using the mutagenizing oligonucleotide TGCCGCGATCTGAAGCTTCGAAGCTGCCCTTC, which encodes the amino acid sequence PRSEASKLPF instead of PRSERSKLPF as well as a silent HindIII site. For making the D306A mutant, we employed the QuikChange reaction using oligonucleotide GATCGTTTGGTGCTGTCGCTGGGA and its reverse complement in the QuikChange protocol with pHAB162 as template.
Antibodies and Chemical Reagents
Chemicals were purchased from Sigma unless otherwise stated. Custom oligonucleotides were from Hy-Labs (Rehovot, Israel) or IDT (Bet Shemesh, Israel). Anti-HA and anti-aminopeptidase I (Ape1) antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-Tlg2 antibodies were as previously described (36). Anti-GFP antibody was from Invitrogen, and anti-protein A antibody was from Sigma.
Preparation of Whole Cell Extracts for Western Blot Analysis
Cells (10 A600 units) were treated with trichloroacetic acid at a final concentration of 10% and washed three times with acetone. The dry cell pellet was then resuspended in 100 μl of cracking buffer (50 mm Tris, pH 6.8, 3.6 m urea, 1 mm EDTA, 1% SDS) and vortexed in a Disruptor Genie (Scientific Instruments, Bohemia, NY) at maximum speed with an equal volume of acid-washed glass beads (425–600-μm diameter) for 30 min. Unlysed cells were removed by centrifugation at 13,000 × g for 5 min. Protein levels were determined using BCA (Pierce). Protein extracts (at the amounts (in mg) indicated in individual experiments) were mixed with 4× SDS loading buffer (200 mm Tris, pH 6.8, 40% glycerol, 4% SDS, 1 m β-mercaptoethanol). The samples (0.5 A600 unit/lane or as otherwise stated) were incubated at 70 °C for 5 min prior to loading on gels.
Fluorescence Microscopy
Typically, culture samples were placed on standard microscope slides (3 μl) and viewed using a Nikon E600 upright fluorescent microscope equipped with a 100× Plan Fluor objective, using a fluorescein isothiocyanate fluorescent filter (for viewing GFP fluorescence) or a UV filter (for viewing 4′,6-diamidino-2-phenylindole). To achieve statistically significant numbers of cells per viewing field in some pictures, high cell densities were achieved by sedimenting 1 ml of cells for 30 s at 500 × g and resuspending the pellet in 20 μl of medium.
Determination of Protein Carbonylation
To quantify protein carbonylation, 15 μg of protein extract was derivatized with dinitropenylhydrazine using the OxyblotTM kit (Thermo Scientific) and identified by Western blotting with the supplied anti-DNP antibody and electronic capture of the ECL signal. Quantification of the signal was carried out by subtracting the background of identical unconjugated samples followed by comparison with the supplied carbonylated standard.
Purification of Recombinant His6-tagged Aup1 Variants
E. coli strain BL21 harboring plasmids expressing Aup1 variants from the T7 promoter were induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. Cell pellets were frozen in 4 ml of Tris-NaCl (20 mm Tris, 0.5 m NaCl, pH 8.0) at −80 °C until use. Cells were disrupted by sonication on ice for 6 × 30 s with a Sonics Vibra-Cell 750-watt sonicator in 20 ml of binding buffer (20 mm Tris, pH 8.0, 0.5 m NaCl, 5 mm imidazole). All subsequent operations were carried out on ice or at 4 °C. Buffer volume was adjusted to 30 ml, and Triton X-100 was added to a final concentration of 0.3%. Extracts were cleared by centrifugation for 15 min at 14,000 × g. The supernatant was transferred to a 50-ml conical polypropylene tube, and 4 ml of pre-equilibrated 50% Ni2+-nitrilotriacetic acid-agarose (Qiagen) beads were added. Binding was carried out for 1 h under gentle agitation. The beads were washed once in 50 ml of binding buffer, resuspended in 6 ml of binding buffer, and transferred to a column. The column was washed with 3 volumes each of binding buffer followed by Tris-NaCl supplemented with 20 and 60 mm imidazole. His-tagged proteins were eluted with a step gradient of imidazole with the most significant fraction (later used in biochemical assays) being 60–80 mm imidazole. Protein content was determined by the BCA assay (Pierce).
Phosphatase Assays
300 μl of reaction buffer (0.1% 2-mercaptoethanol, 1 mm EGTA, 50 mm imidazole (pH 7.2) 0.05 mg/ml bovine serum albumin, 0.2 mm 6,8-difluoro-4-methylumbelliferyl phosphate containing either 10 mm MnCl2 or 10 mm MgCl2) was added to 2 μg of purified recombinant His6-Aup1 variants or purified GFP control in 96-well microtiter plates. Reactions were incubated at 37 °C and either monitored at end point by stopping the reaction with 200 μl of 100 mm sodium borate, pH 10, after a 2-h incubation or as a continuous time series for kinetic analyses. Fluorescence was monitored with a Bio-Tek Synergy 4 plate reader with excitation at 358 nm and emission at 455 nm. Phosphatase activity was defined as arbitrary fluorescence units/μg of protein/min.
RESULTS
Caffeine Sensitivity of aup1Δ Cells Suggests RTG Involvement in the Regulation of Mitophagy
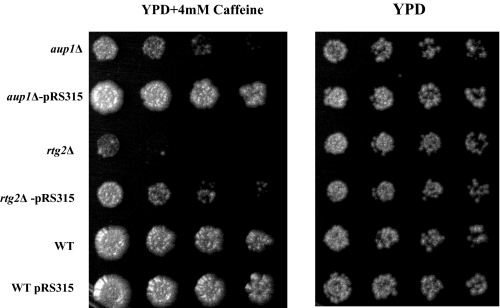
Mutants in AUP1 show increased sensitivity to rapamycin (23) and caffeine (24). Both of these compounds have been shown to cause inhibition of the Tor protein kinase (25, 26). We initially intended to screen for multicopy suppressors of these phenotypes. Surprisingly, however, we found that complementation of the leucine auxotrophy (see Table 1) but not nucleotide or other amino acid auxotrophies of these cells completely cured the caffeine sensitivity of aup1Δ cells without significantly affecting the isogenic wild type counterpart (Fig. 1): Transformation of the aup1Δ mutant with an empty vector carrying the LEU2 gene (pRS315) completely abolished caffeine sensitivity.
FIGURE 1.
The caffeine sensitivities of the aup1Δ and rtg2Δ are both suppressed by leucine prototrophy. Wild type (WT) (HAY75), aup1Δ (HAY809), and rtg2Δ (DJY2) were grown to stationary phase in SD medium, serially diluted (from an initial concentration of 6 × 106 cells/ml) in 1:10 steps, and spotted onto YPD + caffeine plates (4 mm). Plates were incubated at 26 °C and imaged. Bottom row of plates, replicate aliquots were serially diluted (from an initial concentration of 6 × 106 cells/ml) in 1:10 steps and spotted onto control YPD plates to demonstrate equivalent growth potential in the absence of caffeine.
During the course of this study, it was published that laboratory yeast strains show a defect in leucine uptake that is contingent on the metabolic status of the cells (37). This led us to hypothesize that caffeine may adversely affect leucine uptake. Since Tor regulates vacuolar degradation and transcription of amino acid transporters (38, 39), one possibility is that AUP1 deletion may affect leucine transport. The promoter region of at least one leucine transporter, Bap2, contains consensus sequences (GGTAC) for binding to RTG pathway transcription factors (see the YEASTRACT web site) (40). Because the RTG pathway carries signals from the mitochondria to the nucleus, this suggested a putative mode for AUP1-dependent effects on leucine uptake. This was reinforced by the fact that the caffeine sensitivity of an auxotrophic rtg2Δ mutant was also suppressed by the introduction of a wild type LEU2 gene (Fig. 1), consistent with the hypothesis that Aup1 and the RTG pathway may share common targets. Since the RTG pathway regulates nuclear gene expression in response to mitochondrial dysfunction (27–29), this result led us to try and determine whether any functional relationship exists between Aup1, the RTG pathway, and mitophagy.
Rtg3 Is Translocated to the Nucleus under Mitophagic Conditions in an Aup1-dependent Fashion
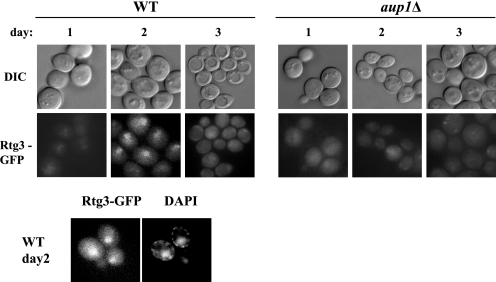
To better understand the nature of any possible interrelationship between mitophagy and the retrograde response, we followed the fluorescence of Rtg3-GFP during the course of mitophagy. Rtg3 is a transcription factor that responds to retrograde signaling by transport from the cytoplasm to the nucleus (28). As seen in Fig. 2, following a long term incubation in lactate medium (and in the presence of glutamate), conditions that induce massive mitophagy on day 3 of the incubation (22), we observe transport of Rtg3-GFP to the nucleus by the second day, as verified by 4′,6-diamidino-2-phenylindole staining (panels on the right). Interestingly, the nuclear localization of Rtg3-GFP was transient and reproducibly dissipated by the third day. These results suggest that the RTG pathway is active at around the same time that mitophagy is being induced. To test whether Aup1 plays a role in this translocation of Rtg3, we analyzed the behavior of Rtg3-GFP in an aup1Δ mutant under the same conditions. As shown in Fig. 2, the mutant is severely defective in nuclear mobilization of Rtg3-GFP in prolonged incubations in lactate medium. Thus, Rtg3 is transported to the nucleus under mitophagic conditions, in an Aup1-dependent fashion.
FIGURE 2.
Rtg3-GFP is translocated to the nucleus in an Aup1- dependent fashion, during growth on lactate medium. Isogenic wild type (WT) (HAY977) and aup1Δ mutants (HAY984) expressing Rtg3-GFP were grown in SL medium supplemented with 0.2% (w/v) glutamate for 3 days and imaged daily. 4′,6-Diamidino-2-phenylindole (DAPI) staining panels of day 2 wild type cells were added for verification of nuclear localization of the GFP signal. Scale bar, 5 μm. DIC, differential interference contrast.
Rtg3 Phosphorylation Is Altered in aup1Δ Cells
Rtg3 has been shown to be a phosphoprotein that undergoes changes in phosphorylation concomitant with nuclear translocation (41–43). This can be observed as a shift in the mobility of the protein in SDS-PAGE (42). We verified that in our hands, Rtg3 is a phosphoprotein in cells growing in lactate medium and that changes in Rtg3 mobility are due to protein phosphorylation, as shown by phosphatase treatment of tagged Rtg3 purified from lactate-grown cells (supplemental Fig. 1) (42).
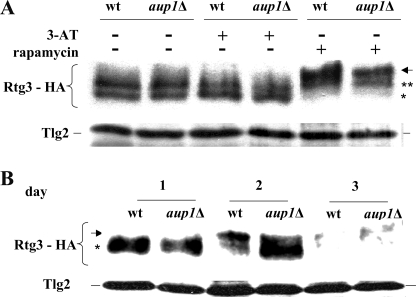
We proceeded to analyze the effect of deleting AUP1 under several physiological states. Rtg3 is known to be hyperphosphorylated in response to rapamycin and dephosphorylated in response to 3-aminotriazole (3-AT) (41, 42). However, despite their opposite effects on Rtg3 phosphorylation, both 3-AT and rapamycin treatments correlate with nuclear translocation of the protein (41–43), implying that the phosphorylation state of Rtg3 can be uncoupled from nuclear transport, as a function of the physiological status of the cell. As shown in Fig. 3A, administration of rapamycin to cells expressing HA-tagged Rtg3 leads to an upshift of the protein band, as previously reported, whereas 3-AT causes an increase in mobility. These responses appear to occur normally in aup1Δ mutants. To understand whether this change in mobility is correlated with the effects of AUP1 deletion on Rtg3 nuclear translocation in lactate, we followed Rtg3-HA during prolonged incubation in lactate medium. As seen in Fig. 3B, the Rtg3 band shifts to lower mobility on the second day of the incubation in minimal lactate medium in wild type cells, mirroring nuclear transport of the protein as well as the time course of mitophagy. This shift is clearly defective in aup1Δ mutants, in which Rtg3-HA remains mostly in the lower band through the experiment.
FIGURE 3.
Aup1 affects Rtg3 phosphorylation during growth on lactate. A, wild type (wt) (DJY8) and aup1Δ (DJY9) cells expressing Rtg3-HA were grown in SD medium supplemented with 0.2% glutamate at 26 °C to an A600 of 0.5 and challenged with 50 mm 3-AT or 0.2 μg/ml rapamycin as indicated. Cells were incubated for 30 min, and 20 μg of protein extracted from each sample were processed for Western blotting. B, cells expressing Rtg3-HA (DJY8) were grown in synthetic lactate medium supplemented with 0.2% glutamate. At each time point, 10 A600 units of cells were sampled, and 20 μg of protein from each sample were analyzed by immunoblotting. The arrow denotes the migration distance of the hyperphosphorylated form in each experiment, whereas asterisks denote the faster migrating (hypophosphorylated) species of Rtg3-HA.
Aup1 Is Required for Effective Up-regulation of Known RTG Pathway Target Proteins in Response to a Known Retrograde Signaling Stimulus
If Aup1 functions upstream of the RTG signaling cascade, we expect that regulation of known RTG responses will be affected in aup1Δ mutants. Inhibition of histidine biosynthesis by the imidazoleglycerol-phosphate dehydratase (product of the HIS3 gene) inhibitor 3-AT is known to cause translocation of Rtg3 into the nucleus and transcription of RTG pathway target genes by activating the RTG signal cascade. Importantly, this signaling pathway is distinct from the GAAC and Tor signaling pathways (44), although inhibition of Tor can mimic some aspects of retrograde signaling due to mechanistic overlaps.
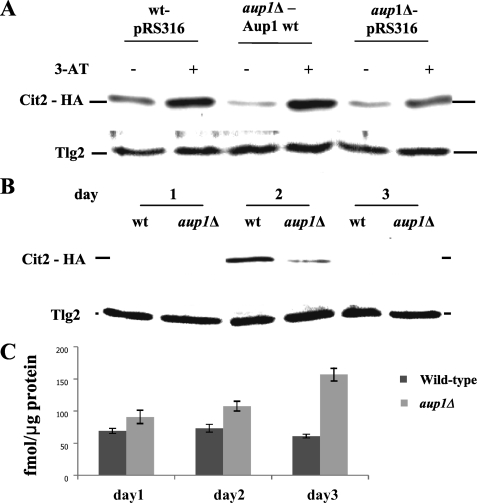
To test whether the RTG pathway is defective in aup1Δ cells, we C-terminally tagged a known RTG target gene, CIT2 (27, 28), with the influenza virus HA epitope and followed the levels of the protein in response to 3-AT. As seen in Fig. 4A, aup1Δ cells showed a defect in the up-regulation of Cit2-HA in response to 3-AT relative to isogenic wild type cells. The same result was also observed with a second RTG target, Dld3-HA (27, 28) (see supplemental Fig. 2). In line with previous observations that mechanistically distinguished between the RTG pathway and Tor-regulated RTG gene expression (44), we find that aup1Δ cells have no defects in up-regulation of Dld3-HA or Cit2-HA in response to rapamycin and that these cells also show normal nuclear translocation of Rtg3-GFP in response to rapamycin. Furthermore, we did not observe any effects on Aup1-HA levels in cells lacking RTG3, both in glucose and in lactate as carbon source (data not shown). Thus, our data support a specific role for Aup1 in the regulation of Rtg3 and Rtg3-dependent transcription in response to the characterized stimuli that define the retrograde signaling pathway.
FIGURE 4.
A, Aup1 is required for efficient up-regulation of Cit2 in response to 3-AT and during mitophagy. Wild type (wt) and aup1Δ cells expressing Cit2-HA (DJY3 and DJY4, top) and harboring either empty vector or a vector expressing wild type Aup1 were grown overnight in SD medium lacking histidine and supplemented with 0.2% glutamate to an A600 of 0.5. The cells were then challenged with 50 mm 3-AT for 30 min, and 10 A600 units of cells were sampled at each time point and processed for immunoblotting. 20 μg of protein were loaded per lane, and the filters were probed with anti-HA antibodies to detect Cit2 and with anti-Tlg2 antibodies as load control. B, the RTG pathway was induced during stationary phase mitophagy in an Aup1-dependent fashion. Wild type (DJY3) and aup1Δ (DJY4) cells expressing Cit2-HA were grown in lactate as described in the legend to Fig. 3B, and samples (20 μg of protein) were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody at each time point. Anti-Tlg2 immunoblotting was used as load control. C, oxidative damage accumulates in aup1Δ cells between the second and third days of the incubation, relative to wild type cells. Wild type (HAY75) and aup1Δ (HAY809) cells were grown on lactate medium and sampled as in B. Protein extracts were prepared, and 15 μg of each sample were conjugated to DNPH as described under “Experimental Procedures.” Anti-DNP antibody signals (fmol of DNP incorporated/μg of total protein) were quantified by SDS-PAGE followed by Western blotting. Bars, S.E. (n = 3).
One question raised by these results is whether the induction of stationary phase mitophagy in lactate-grown cells correlates with Aup1-dependent induction of Cit2-HA. We therefore followed the levels of Cit2-HA in lactate-grown cells over the course of mitophagy induction through a 3-day incubation. As seen in Fig. 4B, Cit2-HA is induced during the second day of the incubation, and this induction is strongly repressed in aup1Δ mutants. This result implies that Aup1-dependent regulation of Cit2-HA expression occurs just before our physical detection of mitophagy, between the second and third days of the incubation (see Ref. 22; also see Fig. 5). Recently, it was shown that activation of mitophagy is induced by cellular redox stress (45). We therefore analyzed the accumulation of oxidized proteins in wild type and aup1 mutants under these conditions. For this, we used a protein carbonylation assay that is an accepted standard for protein oxidation damage (see “Experimental Procedures”). As shown in Fig. 4C, wild type cells maintain a constant level of carbonylated proteins when grown on lactate as carbon source (60–70 fmol of carbonylated residues/μg of protein), whereas aup1Δ cells show a clear, 2.5-fold absolute increase in the accumulation of oxidized proteins that spikes on the third day (91 fmol/μg on day 1, 107 fmol/μg on day 2, and 157 fmol/μg on day 3) relative to the wild type cells. Thus, inactivation of the Aup1-dependent RTG pathway correlates with the accumulation of oxidized proteins.
FIGURE 5.
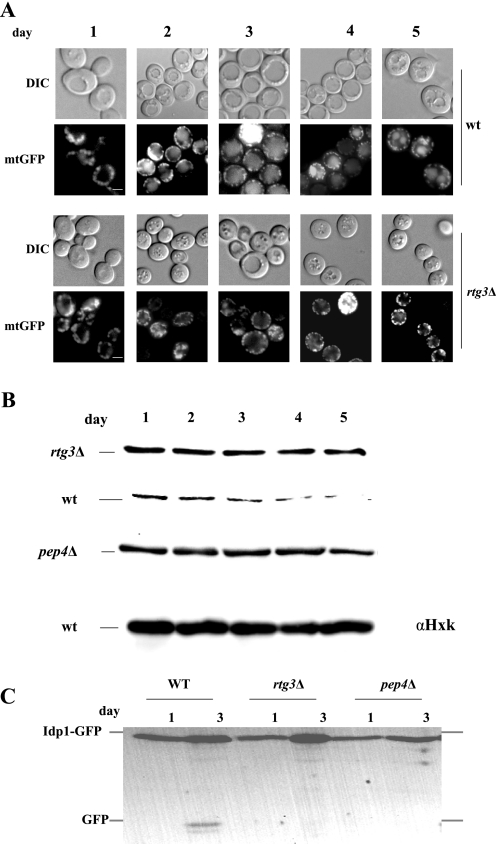
Rtg3 is essential for stationary phase mitophagy. A, AUP1 (HAY805) and aup1Δ (DJY6) cells expressing mitochondrial matrix-targeted GFP (mtGFP) were incubated in synthetic lactate medium for 5 days. Cells were analyzed by fluorescence microscopy daily, and mitophagy was scored as appearance of the GFP signal in the vacuolar lumen. Scale bar, 5 μm. B, wild type (wt) (HAY75), aup1Δ (DJY10), and pep4Δ (TVY1) cells were incubated as in A, and 10 A600 units were sampled daily for anti-aconitase immunoblotting. 20 μg of protein were analyzed per lane. Anti-hexokinase immunoblotting is shown as load control. C, wild type (HAY75), aup1Δ (DJY1), and pep4Δ (TVY1) cells expressing Idp1-GFP were grown as above, and 20 μg of protein were analyzed by immunoblotting with anti-GFP antibody at each time point. Scale bar, 5 μm. DIC, differential interference contrast.
Rtg3 Is Essential for Mitophagy but Not for Starvation-induced Macroautophagy and the Cvt Pathway
The effects of AUP1 deletion on the induction of RTG pathway targets could be interpreted in at least two ways. First, it is possible that Aup1 independently regulates multiple processes, including mitophagy and the RTG response. On the other hand, a more parsimonious explanation would be that the RTG pathway itself plays a role in stationary phase mitophagy. In this case, Aup1 may be an upstream regulator of the Rtg pathway in a linear chain that controls mitophagy. To further understand the relationship between mitophagy and the RTG pathway, we tested the effect of deleting RTG3 on stationary phase mitophagy, using three independent assays. In cells expressing a mitochondrial matrix-targeted GFP, we previously reported that GFP fluorescence was transported to the vacuolar lumen in long term stationary phase cultures grown on lactate as carbon source (22). This effect began on the third day of the incubation and paralleled the Pep4-dependent degradation of aconitase, a mitochondrial matrix protein, allowing us to suggest that a mitophagic process is taking place en masse under these conditions. As seen in Fig. 5A, rtg3Δ cells show a defect in the transport of mitochondrially targeted GFP to the vacuolar lumen, even after 4 and 5 days in lactate medium. In contrast, wild type cells show vacuolar fluorescence by the third day, as we previously reported. This is paralleled by a defect in aconitase degradation under the same conditions (Fig. 5B). A third assay for mitophagy relies on Western blotting using anti-GFP antibodies for release of free GFP from chimeras of mitochondrial protein tagged with GFP in a vacuole-dependent fashion (46). To this end, we tagged the mitochondrial NADP-specific isocitrate dehydrogenase (Idp1), a protein not essential for respiratory growth, with a C-terminal GFP tag (see “Experimental Procedures”). As seen in Fig. 5C, Idp1-GFP undergoes clipping by the third day of the incubation, in a Pep4-dependent fashion, confirming the autophagic nature of the clipping event. Importantly, mutants lacking Rtg3 show a defect in the release of free GFP, consistent with our other assays.
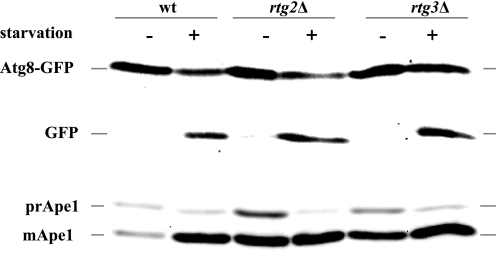
To determine whether these results reflect a general role of the retrograde signaling pathway in autophagy, we assayed starvation-induced macroautophagy in mutants and wild type cells by following the ability of cells to deliver GFP-Atg8 into the vacuolar lumen in response to nitrogen starvation.
Atg8 is a ubiquitin-like protein that undergoes conjugation to phosphatidylethanolamine and is essential for autophagosome expansion (47–50). During macroautophagy, Atg8 is trapped inside autophagosomes and delivered into the lumen of the vacuole. Upon arrival in the vacuole, the Atg8 moiety of GFP-Atg8 chimeras is degraded, releasing free GFP, which can be detected either by fluorescence or by immunoblotting (51–53). The data, as presented in Fig. 6 (top), show that Rtg3 is not required for starvation-induced macroautophagy. We also assayed the Cvt pathway by testing the same extracts for Ape1 maturation. The Cvt pathway is a related, selective autophagic process that specifically delivers prApe1 from the cytosol to the vacuole lumen (54). Ape1 is synthesized as a 62-kDa precursor, and vacuolar delivery results in its clipping to a 50-kDa mature form. As seen in Fig. 6 (bottom), rtg3Δ and rtg2Δ cells are not defective in Ape1 maturation. It is important to note, however, that extracts from these cells reproducibly contained a much higher amount of Ape1 (more than 10-fold) relative to isogenic wild type cells under nutrient-rich conditions, and this may reflect a role for Rtg3 in regulating APE1 expression.
FIGURE 6.
Rtg2 and Rtg3 are not required for starvation-induced autophagy or for the Cvt pathway. Wild type (wt) (HAY75), rtg2Δ (DJY2), and rtg3Δ (DJY5) cells expressing GFP-Atg8 from the TPI promoter (plasmid pYX-GFPATG8) were grown to midlog phase in SD medium, washed, and resuspended in SD-N or in SD for 2 h. Samples (10 A600 units) were then taken and analyzed by immunoblotting with anti-GFP and anti-Ape1 antibodies.
Mitophagic Regulation Is Kinetically Distinct from Starvation-induced Macroautophagy
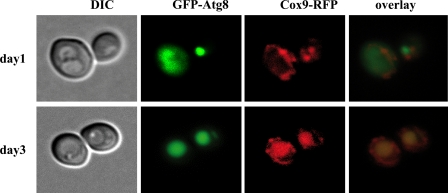
Macroautophagy, the well studied autophagic response to starvation, is thought to be a non-selective catabolic pathway that is also induced through Tor-related signaling mechanisms. One possible interpretation of our data is that at long incubation time points, the cells are starving, and the mitophagic phenomenon merely reflects this general free-for-all degradation of cytosolic components. To test this hypothesis, we closely compared the time course for macroautophagy with the onset of mitophagy, in cells expressing GFP-Atg8 as well as a mitochondrial RFP tag (Cox9-RFP). This fact has been used as an assay for macroautophagy because GFP-Atg8 fluorescence can be shown to shift from a cytosolic localization in cells growing in rich medium to the vacuolar lumen in starved cells (55). As seen in Fig. 7, lactate-grown cells exhibit vacuolar GFP fluorescence following a 24-h incubation, about 2 days before significant amounts of RFP fluorescence are accumulated in the vacuole. Since autophagic trafficking of GFP-Atg8 is temporally separate from mitophagy, this result distinguishes between the kinetics of general starvation-induced macroautophagy and the kinetics of mitophagy as regulated through the Aup1-RTG axis and is consistent with the existence of a specific regulatory mechanism distinct from general macroautophagy.
FIGURE 7.
The regulation of mitophagy and starvation-induced macroautophagy can be uncoupled. Wild type cells co-expressing GFP-Atg8 and Cox9-RFP (strain DJY7) were grown in synthetic lactate medium for 3 days. Cells were imaged daily by fluorescence microscopy. DIC, differential interference contrast.
A Novel RXKL Motif, Conserved between Aup1 and Mammalian PP1K Family Proteins, Is Important for Aup1 Function
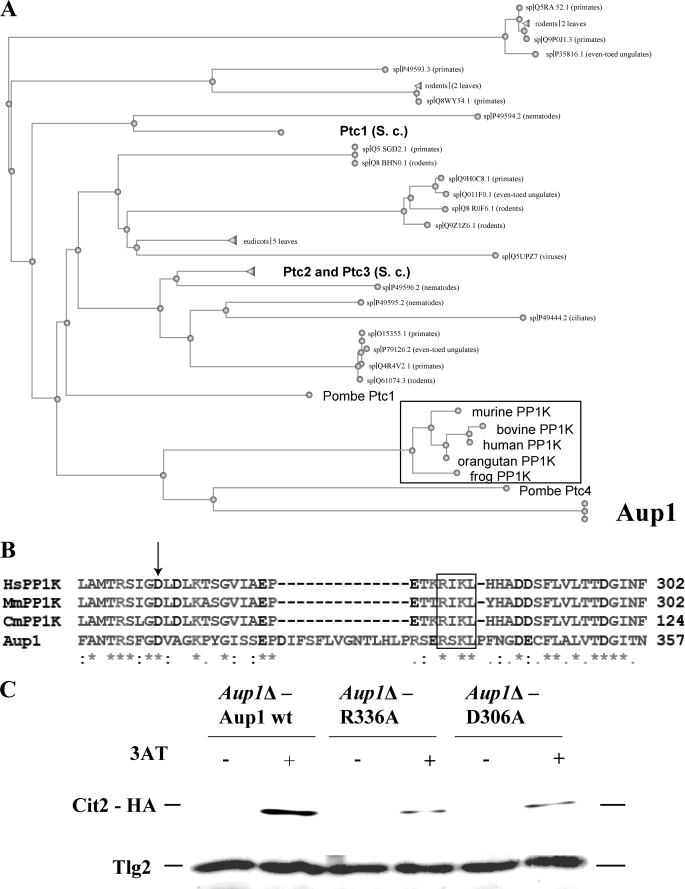
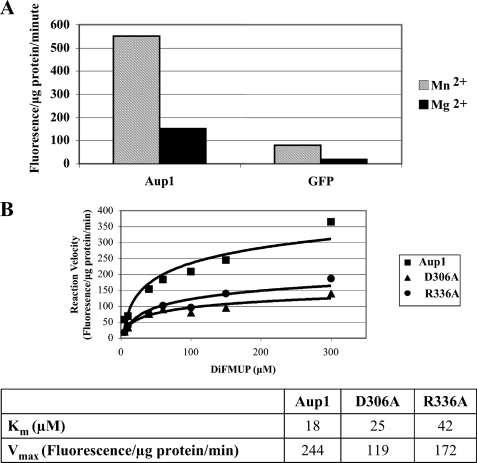
Aup1 is part of a subfamily of eukaryotic PP2C homologues that are predicted to have a mitochondrial localization. As shown in the dendrogram of Fig. 8A, Aup1 has a higher similarity to the mammalian PP1K family of phosphatase homologs than to other yeast PP2C homologues. We identified several conserved motifs that fall into two categories. In the first category are residues conserved across all PP2C homologs, including Aup1, whereas the second category contains residues that are specific to Aup1 and PP1K subfamily members, such as MmPP1K, HsPP1K, and CmPP1K, and do not appear in the other PP2C homologs. An example of residues that fall in the first category are aspartate residues that are predicted by threading analysis to correspond to catalytically important Mn2+ binding sites (Fig. 8B, arrow). As an example of the second, we chose the RXKL box motif that we identified as specific to the PP1K/Aup1 family (Fig. 8B, box). As shown in Fig. 8C, both the PP1K-specific Arg336 and the universally conserved Asp306 are required for efficient 3-AT induction of Cit2-HA. In addition, the R336A mutant had an effect on the Rtg3-HA phosphorylation pattern identical to the effect observed for aup1Δ (supplemental Fig. 3). This loss of activity could be due to a loss of phosphatase activity but may also be due to other issues. To determine whether the loss of biological activity in the D306A and R336A correlates with loss of phosphatase activity, we tried to purify recombinant His6-tagged Aup1 from bacterial inclusion bodies. This approach was previously reported to yield active phosphatase upon renaturation (56). In our hands, however, the same construct yielded no phosphatase activity. However, Aup1 is unusual in that it contains two iterations of the PP2C motif. To obtain active phosphatase, we found it necessary to express the C-terminal Aup1 PP2C iteration (amino acids 213–409) alone. Within this context, the N-terminally truncated recombinant product had clear Mn2+- dependent phosphatase activity (Fig. 9A). We then compared the activities of the wild type protein with purified D306A and R336A versions (Fig. 9B). Both mutants showed significantly reduced levels of phosphatase activity, consistent with the biological effect on Cit2 induction. This result is consistent with the RXKL box being a functionally important motif in Aup1 and, by extension, in its apparent mammalian orthologs. However, the effects of the R336A mutant on phosphatase activity is due to a significant increase in Km (18–42 mm), whereas Vmax is more weakly affected (1.4-fold). In contrast, the D306A mutation has a smaller effect on Km (18–25 μm) and a stronger effect on Vmax (2-fold).
FIGURE 8.
An RXKL motif, conserved between Aup1 and mammalian PP1K proteins, is essential for Aup1 function. A, comparative evolutionary tree for BLAST comparison of Aup1 (in bold, at the bottom; Swissprot designation PP2C-6) against the Swissprot data base. Framed in black are the vertebrate PP1K homologs. Three S. cerevisiae proteins that appear in the comparison are also in bold: Ptc1, Ptc2, and Ptc3. The single non-vertebrate PP2C homolog that shows a higher similarity to Aup1 is Schizosaccharomyces pombe Ptc4, which is an ortholog of the PP1K/Aup1 family. Expansion of the search to the non-redundant data base (which includes unannotated sequences) results in a major expansion of the fungal ortholog clade represented by the single S. pombe Ptc4 node in this picture and a complete loss of S. cerevisiae representatives, suggesting that there are no paralogs of Aup1 within the S. cerevisiae genome and that the PP1K/Aup1 family is a distinct branch within the PP2C protein superfamily. B, identification of a conserved RXKL motif that defines the PP1K family of phosphatase homologs. Human (Hs), mouse (Mm), and dog (Cm) PP1K virtual translation products were aligned with the Aup1 protein sequence using ClustalW. The arrow denotes aspartate 306 in Aup1, which is predicted by threading analysis to participate in the coordination of a catalytically important manganese ion. The RXKL motif is denoted by the black frame. C, aspartate 306 and arginine 336 are essential for Aup1 function. Mutant aup1Δ (HAY 809) cells expressing Cit2-HA were transformed with plasmids encoding wild type, R336A, and D306A variants of AUP1 (see “Experimental Procedures”) expressed under control of the endogenous promoter and tested for their ability to induce the Rtg pathway response gene CIT2 in response to 3-AT. Cells were grown overnight in SD medium lacking histidine and supplemented with 0.2% glutamate to an A600 of 0.5. The cells were then challenged with 50 mm 3-AT for 30 min, and 10 A600 units of cells were sampled at each time point and processed for immunoblotting. 20 μg of protein were loaded per lane, and the filters were probed with anti-HA antibodies and with anti-Tlg2 antibodies as load control.
FIGURE 9.
Arginine 336 of Aup1, within the RXKL motif, is required for phosphatase activity. A, the purified C-terminal PP2C motif iteration (amino acids 213–419) shows PP2C-type phosphatase activity. 2 μg of purified Aup1 (amino acids 213–419) or purified GFP control were assayed for phosphatase activity in the presence of Mg2+ or Mn2+, as detailed under “Experimental Procedures.” B, steady state kinetic analysis of purified Aup1 (amino acids 213–419) versus the D306A and R336A variants. Results are representative of three independent determinations.
DISCUSSION
The RTG pathway was originally identified as a cellular signaling pathway that is induced in cells with a defective electron transport chain (27, 29). It is also induced in response to inhibition of amino acid biosynthetic enzymes, such as His3 and Gln1, apparently due to the central role of the tricarboxylic acid cycle in generating the carbon backbone in the biosynthesis of amino acids (44). In the present study, we have linked the RTG pathway with stationary phase mitophagy, a process that occurs under physiological conditions in an otherwise unperturbed yeast culture. We suggest that Aup1, a mitochondrial intermembrane space protein, functions to regulate the Rtg3 transcription factor. This functional relationship implies that the mitophagy defect of aup1Δ cells can itself be explained through the fact that Rtg3 itself is required for stationary phase mitophagy.
Our data demonstrate that Aup1 is required for Rtg3/Rtg1 nuclear translocation during growth on lactate as a carbon source (Fig. 2), and this is sufficient to explain the involvement of Aup1 in mitophagy (22), since we find that the RTG response plays a role in the regulation of mitophagy (Fig. 5). In addition, we find that efficient up-regulation of Cit2 and Dld3 in response to 3-AT also requires Aup1 (Fig. 4), suggesting that Aup1 plays some role in the regulation of the RTG pathway during growth on glucose as well.
Stationary Phase Mitophagy Is Distinctly Regulated
Although the existence of mitochondrial quality control mechanisms by autophagic pathways has received significant attention recently, it has not been clear to what extent it is linked to starvation responses because these two effects are linked by a reciprocal cause and effect relationship; the mitochondrial citric acid cycle takes part in nitrogen assimilation through the amination of oxaloacetate and α-ketoglutarate, and therefore mitochondrial malfunction may lead to effects on nitrogen balance. On the other hand, starvation may affect mitochondrial metabolism by altering the function of the tricarboxylic acid cycle. One can monitor the starvation state of the cells through the induction of standard autophagic responses. Cells undergoing starvation-induced autophagy can be distinguished by the transport of GFP-Atg8 to the vacuolar lumen (48, 51, 52). It is worthwhile to note that Atg8 is not a passive cargo molecule but rather a factor required for autophagosome expansion (47) and that transport of GFP-Atg8 to the vacuolar lumen is indicative of general macroautophagy (52). The fact that GFP-Atg8 is transported to the vacuole lumen in very early stages of our experiment (day 1) at a time point that is distinctly separate from the transport of mitochondrial RFP to the vacuolar lumen (days 2–3) argues that there are distinct mitophagy-specific regulatory elements. Such elements may represent either a kinetic ordering or a different input mechanism altogether. We suggest that Aup1 and the RTG pathway elements represent such a mechanism because they are not required for general autophagy (Fig. 6). In agreement with our results, Deffieu et al. (45) showed that oxidative stress induces mitophagy but does not induce general macroautophagy. The fact that aup1Δ cells hyperaccumulate oxidized proteins between the second and third days of incubation on lactate (Fig. 4C) may suggest that Aup1 functions to transduce a signal from specific hyperoxidized protein(s) to activate the RTG response.
Aup1 Is the S. cerevisiae Representative of the Mammalian PP1K Phosphatase Family
A comprehensive bioinformatic analysis of Aup1 reveals that there are no paralogs of Aup1 in S. cerevisiae, and the closest relatives are apparent orthologs in other yeast species and fungi. As seen in Fig. 8A, apparent mammalian orthologs also exist. We have identified a novel motif, the RXKL motif, which is a hallmark of the Aup1/PP1K subfamily (Fig. 8B). Threading analysis using the Phyre server (57) (available on the World Wide Web) revealed that this motif is predicted to be at the surface of the protein, not far from one of the two active site Mn2+ ions (58). The RIKL motif of the human PP1K homologue (Protein Data Bank code 2iq1) shows the motif to be located at the surface of the protein but juxtaposed at the other side of the β-sandwich cleft that also contains the binucleate metal coordination site. Such a location can either directly affect substrate accessibility to the active site or have an allosteric effect on the active site. We have experimentally demonstrated that the RXKL motif in Aup1 is functionally significant because mutation of arginine 336 to a leucine results in loss of biological function (Fig. 8C) as well as a clear reduction of in vitro phosphatase activity in recombinant versions of Aup1 (Fig. 9B).
The decrease in in vitro phosphatase activity of Aup1 mutants as measured in our assays is likely to be a drastic underestimation. Since our “wild type” construct is a truncated, single iteration version of native Aup1 that is assayed using an artificial substrate, we can assume that our base-line activity is much lower than the actual native activity. Nonetheless, mutations at Arg336 and Asp306 yield clear and distinct effects on phosphatase activity that are consistent with the observed biological effects.
We suggest that Aup1 functions to induce a developmental transition, mediated through the RTG signaling pathway, that allows or facilitates mitophagy. Clearly, additional components will need to be identified in order to understand the specific mechanism by which defective mitochondria are targeted for degradation. In addition, functional and biochemical studies of the mammalian members of the PP1K protein family will be required to reveal whether the physiological role of these proteins is also conserved between yeast and higher eukaryotes.
Acknowledgments
We thank Dr. Roger Tsien for the RFP clone used in this study, Dr. Ophry Pines for anti-Aco1 antibody, Dr. Orit Levius for technical assistance, and Galia Lerech for help with graphics.
This work was supported by Israel Science Foundation Grant 1016/07 (to H. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- GFP
- green fluorescent protein
- Cvt
- cytoplasm to vacuole targeting
- HA
- Hemophilus influenza hemagglutinin
- RFP
- red fluorescent protein
- PP2C
- protein phosphatase 2 C
- PP1K
- protein phosphatase 1K
- RTG
- retrograde signaling pathway
- 3-AT
- 3-aminotriazole.
REFERENCES
- 1.Abeliovich H. (2007) Autophagy 3, 275–277 [DOI] [PubMed] [Google Scholar]
- 2.Wallace D. C. (2005) Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abeliovich H., Klionsky D. J. (2001) Microbiol. Mol. Biol. Rev. 65, 463–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky D. J. (2005) Curr. Biol. 15, R282–R283 [DOI] [PubMed] [Google Scholar]
- 5.Klionsky D. J., Emr S. D. (2000) Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky D. J. (2005) J. Cell Sci. 118, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onodera J., Ohsumi Y. (2004) J. Biol. Chem. 279, 16071–16076 [DOI] [PubMed] [Google Scholar]
- 8.Wang C. W., Klionsky D. J. (2003) Mol. Med. 9, 65–76 [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn W. A., Jr., Cregg J. M., Kiel J. A., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. (2005) Autophagy 1, 75–83 [DOI] [PubMed] [Google Scholar]
- 10.Farré J. C., Subramani S. (2004) Trends Cell Biol. 14, 515–523 [DOI] [PubMed] [Google Scholar]
- 11.Sakai Y., Oku M., van der Klei I. J., Kiel J. A. (2006) Biochim. Biophys. Acta 1763, 1767–1775 [DOI] [PubMed] [Google Scholar]
- 12.Kvam E., Goldfarb D. S. (2007) Autophagy 3, 85–92 [DOI] [PubMed] [Google Scholar]
- 13.Kraft C., Deplazes A., Sohrmann M., Peter M. (2008) Nat. Cell Biol. 10, 602–610 [DOI] [PubMed] [Google Scholar]
- 14.Bernales S., McDonald K. L., Walter P. (2006) PLoS Biol. 4, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernales S., Schuck S., Walter P. (2007) Autophagy 3, 285–287 [DOI] [PubMed] [Google Scholar]
- 16.Elmore S. P., Qian T., Grissom S. F., Lemasters J. J. (2001) FASEB J. 15, 2286–2287 [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Enriquez S., He L., Lemasters J. J. (2004) Int. J. Biochem. Cell Biol. 36, 2463–2472 [DOI] [PubMed] [Google Scholar]
- 18.Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kissová I., Deffieu M., Manon S., Camougrand N. (2004) J. Biol. Chem. 279, 39068–39074 [DOI] [PubMed] [Google Scholar]
- 20.Nowikovsky K., Reipert S., Devenish R. J., Schweyen R. J. (2007) Cell Death Differ. 14, 1647–1656 [DOI] [PubMed] [Google Scholar]
- 21.Priault M., Salin B., Schaeffer J., Vallette F. M., di Rago J. P., Martinou J. C. (2005) Cell Death Differ. 12, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 22.Tal R., Winter G., Ecker N., Klionsky D. J., Abeliovich H. (2007) J. Biol. Chem. 282, 5617–5624 [DOI] [PubMed] [Google Scholar]
- 23.Xie M. W., Jin F., Hwang H., Hwang S., Anand V., Duncan M. C., Huang J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7215–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakumoto N., Mukai Y., Uchida K., Kouchi T., Kuwajima J., Nakagawa Y., Sugioka S., Yamamoto E., Furuyama T., Mizubuchi H., Ohsugi N., Sakuno T., Kikuchi K., Matsuoka I., Ogawa N., Kaneko Y., Harashima S. (1999) Yeast 15, 1669–1679 [DOI] [PubMed] [Google Scholar]
- 25.Reinke A., Chen J. C., Aronova S., Powers T. (2006) J. Biol. Chem. 281, 31616–31626 [DOI] [PubMed] [Google Scholar]
- 26.Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N. R., Hall M. N. (1993) Cell 73, 585–596 [DOI] [PubMed] [Google Scholar]
- 27.Liao X., Butow R. A. (1993) Cell 72, 61–71 [DOI] [PubMed] [Google Scholar]
- 28.Liu Z., Butow R. A. (2006) Annu. Rev. Genet. 40, 159–185 [DOI] [PubMed] [Google Scholar]
- 29.Parikh V. S., Morgan M. M., Scott R., Clements L. S., Butow R. A. (1987) Science 235, 576–580 [DOI] [PubMed] [Google Scholar]
- 30.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 31.Abeliovich H., Zhang C., Dunn W. A., Jr., Shokat K. M., Klionsky D. J. (2003) Mol. Biol. Cell 14, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westermann B., Neupert W. (2000) Yeast 16, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 33.Labbé S., Thiele D. J. (1999) Methods Enzymol. 306, 145–153 [DOI] [PubMed] [Google Scholar]
- 34.Winter G., Hazan R., Bakalinsky A. T., Abeliovich H. (2008) Autophagy 4, 28–36 [DOI] [PubMed] [Google Scholar]
- 35.Kunkel T. A. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abeliovich H., Grote E., Novick P., Ferro-Novick S. (1998) J. Biol. Chem. 273, 11719–11727 [DOI] [PubMed] [Google Scholar]
- 37.Cohen R., Engelberg D. (2007) FEMS Microbiol. Lett. 273, 239–243 [DOI] [PubMed] [Google Scholar]
- 38.Beck T., Schmidt A., Hall M. N. (1999) J. Cell Biol. 146, 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt A., Beck T., Koller A., Kunz J., Hall M. N. (1998) EMBO J. 17, 6924–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Helden J., André B., Collado-Vides J. (2000) Yeast 16, 177–187 [DOI] [PubMed] [Google Scholar]
- 41.Dilova I., Powers T. (2006) FEMS Yeast Res. 6, 112–119 [DOI] [PubMed] [Google Scholar]
- 42.Komeili A., Wedaman K. P., O'Shea E. K., Powers T. (2000) J. Cell Biol. 151, 863–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekito T., Thornton J., Butow R. A. (2000) Mol. Biol. Cell 11, 2103–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giannattasio S., Liu Z., Thornton J., Butow R. A. (2005) J. Biol. Chem. 280, 42528–42535 [DOI] [PubMed] [Google Scholar]
- 45.Deffieu M., Bhatia-Kissová I., Salin B., Galinier A., Manon S., Camougrand N. (2009) J. Biol. Chem. 284, 14828–14837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanki T., Klionsky D. J. (2008) J. Biol. Chem. 283, 32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abeliovich H., Dunn W. A., Jr., Kim J., Klionsky D. J. (2000) J. Cell Biol. 151, 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 49.Lang T., Schaeffeler E., Bernreuther D., Bredschneider M., Wolf D. H., Thumm M. (1998) EMBO J. 17, 3597–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 51.Huang W. P., Scott S. V., Kim J., Klionsky D. J. (2000) J. Biol. Chem. 275, 5845–5851 [DOI] [PubMed] [Google Scholar]
- 52.Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheong H., Klionsky D. J. (2008) Methods Enzymol. 451, 1–26 [DOI] [PubMed] [Google Scholar]
- 54.Klionsky D. J., Cueva R., Yaver D. S. (1992) J. Cell Biol. 119, 287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klionsky D. J., Cuervo A. M., Seglen P. O. (2007) Autophagy 3, 181–206 [DOI] [PubMed] [Google Scholar]
- 56.Ruan H., Yan Z., Sun H., Jiang L. (2006) FEMS Yeast Res. 7, 209–215 [DOI] [PubMed] [Google Scholar]
- 57.Kelley L. A., MacCallum R. M., Sternberg M. J. (2000) J. Mol. Biol. 299, 499–520 [DOI] [PubMed] [Google Scholar]
- 58.Das A. K., Helps N. R., Cohen P. T., Barford D. (1996) EMBO J. 15, 6798–6809 [PMC free article] [PubMed] [Google Scholar]
- 59.Abeliovich H., Darsow T., Emr S. D. (1999) EMBO J. 18, 6005–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerhardt B., Kordas T. J., Thompson C. M., Patel P., Vida T. (1998) J. Biol. Chem. 18, 15818–15829 [DOI] [PubMed] [Google Scholar]