Abstract
The DNA polymerase processivity factor of the Epstein-Barr virus, BMRF1, associates with the polymerase catalytic subunit, BALF5, to enhance the polymerase processivity and exonuclease activities of the holoenzyme. In this study, the crystal structure of C-terminally truncated BMRF1 (BMRF1-ΔC) was solved in an oligomeric state. The molecular structure of BMRF1-ΔC shares structural similarity with other processivity factors, such as herpes simplex virus UL42, cytomegalovirus UL44, and human proliferating cell nuclear antigen. However, the oligomerization architectures of these proteins range from a monomer to a trimer. PAGE and mutational analyses indicated that BMRF1-ΔC, like UL44, forms a C-shaped head-to-head dimer. DNA binding assays suggested that basic amino acid residues on the concave surface of the C-shaped dimer play an important role in interactions with DNA. The C95E mutant, which disrupts dimer formation, lacked DNA binding activity, indicating that dimer formation is required for DNA binding. These characteristics are similar to those of another dimeric viral processivity factor, UL44. Although the R87E and H141F mutants of BMRF1-ΔC exhibited dramatically reduced polymerase processivity, they were still able to bind DNA and to dimerize. These amino acid residues are located near the dimer interface, suggesting that BMRF1-ΔC associates with the catalytic subunit BALF5 around the dimer interface. Consequently, the monomeric form of BMRF1-ΔC probably binds to BALF5, because the steric consequences would prevent the maintenance of the dimeric form. A distinctive feature of BMRF1-ΔC is that the dimeric and monomeric forms might be utilized for the DNA binding and replication processes, respectively.
Introduction
The Epstein-Barr virus (EBV),4 a human herpesvirus harboring a 172-kbp dsDNA genome, is associated with several B-cell and epithelial cell malignancies and can choose between two alternative life cycles, latent and lytic infection (1). The EBV genomes are replicated as circular plasmid molecules, using the cellular replication machinery of the host in the latent phase of the viral life cycle. On the other hand, after the induction of lytic viral replication, the EBV genome is amplified 100–1,000-fold by the viral replication machinery. The replication intermediates are large head-to-tail concatemers resulting from rolling-circle DNA replication initiated from oriLyt (2). The EBV replication machinery consists of seven viral gene products (3) as follows: the BZLF1 protein, an oriLyt-binding protein; the BALF5 protein, a DNA polymerase catalytic subunit; the BMRF1 protein, a polymerase processivity factor; the BALF2 protein, a single-stranded DNA-binding protein; and the BBLF4, BSLF1, and BBLF2/3 proteins, putative helicase, primase, and helicase-primase-associated proteins, respectively. It has been suggested that all of the proteins, except for the BZLF1 protein, work together at replication forks to synthesize the leading and lagging strands of the concatemeric EBV genome (2). The EBV DNA polymerase holoenzyme exhibits highly processive replication and possesses 3′-to-5′-exonuclease activity (4, 5). The BMRF1 protein interacts with the BALF5 polymerase catalytic subunit to form a holoenzyme, which enhances the BALF5 protein-associated polymerase processivity and exonuclease activities (6, 7), although the stoichiometry of this complex remains unknown. EBV DNA replication occurs at discrete sites in nuclei, called replication compartments, where all of the viral replication proteins are assembled (8). The BMRF1 protein is also referred to as EA-D (Early Antigen Diffused) and is used as a clinical marker for EBV infection. The BMRF1 protein is abundantly expressed in the cells, unlike the BALF5 polymerase catalytic subunit, and exhibits a homogeneous, as opposed to punctate, distribution throughout the replication compartments, completely coincident with the newly synthesized viral genome (8). Immunostaining data, together with the finding that almost all of the abundantly expressed BMRF1 molecules bind to DNA (8), indicated that the BMRF1 protein not only acts at viral replication forks as a polymerase processivity factor, but it also is widely distributed on the newly synthesized EBV genomic DNA.
In general, processivity factors are associated with their cognate DNA polymerases on the template during replication. These proteins, which are also known as “sliding clamps,” include proliferating cell nuclear antigen (PCNA) from eukaryotes (9, 10) and archaebacteria (11, 12), the β-subunit of Escherichia coli DNA polymerase III (13), and gp45 from the T4 (14) and RB69 bacteriophages (15). These proteins assemble as toroidal, ring-shaped structures, forming a central channel to accommodate the template DNA. In addition, they lack intrinsic DNA binding activity. However, the herpesvirus polymerase processivity factors display different molecular assemblies. The human cytomegalovirus (HCMV) processivity factor, UL44, forms a dimer in the crystal structure as well as in solution (16). The dimeric form possesses DNA binding activity, which is reduced by the introduction of a mutation preventing dimerization (16). In contrast, the herpes simplex virus type 1 (HSV-1) processivity factor, UL42, directly binds to DNA as a monomer (17). Although BMRF1 also possesses DNA binding activity (18), its stoichiometry for DNA binding remains unknown. Electron microscopy observations revealed that BMRF1 adopts a ring-shaped structure, which may contain six monomers (19). This is almost twice as large as the previously reported PCNA ring structure. Glutathione S-transferase pulldown assays also revealed that BMRF1 can form homo-oligomers (19).
The association between processivity factors and the herpes simplex virus or HCMV DNA polymerase is achieved through interactions with the C-terminal region of the DNA polymerase. For instance, the last 22 residues of the HCMV DNA polymerase catalytic subunit UL54 are necessary and sufficient for its interaction with the processivity factor UL44. A sequence alignment of the various DNA polymerase catalytic subunits revealed that EBV BALF5 lacks the corresponding C-terminal region, in contrast to HCMV UL54 or the HSV-1 UL30, implying that the BMRF1-BALF5 system adopts a different arrangement for the intermolecular interaction.
In this study, we determined the crystal structure of the C-terminal region-truncated construct (1–314 residues, WTΔC) of BMRF1. This construct exhibited the DNA binding and DNA polymerase processivity activities. The crystal structure and the functional assays demonstrated that the oligomerization state (monomer or dimer) is related to the DNA binding activity and processivity.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The EBV BMRF1 protein (GenBankTM accession number V01555) was prepared for crystallization as a truncated protein lacking the C-terminal 90 amino acids (1–314, BMRF1-WTΔC). A structure prediction analysis by the PSIPRED Protein Structure Prediction Server (20) indicated that no secondary structure elements were assigned to about 100 residues from the C terminus of the protein, and therefore, this region could negatively affect the crystallization. This truncation did not affect either the protein activities (DNA binding and processivity) (data not shown) or the physical interaction with BALF5 DNA polymerase (supplemental Fig. 1), as compared with the full-length protein. For structure determination, selenomethionine-labeled BMRF1-WTΔC, with an N-terminal histidine-tag, was expressed in the cell-free expression system (21). The protein was purified by chromatography on a HisTrap column (GE Healthcare) and was subjected to tobacco etch virus protease digestion. BMRF1 was subsequently purified through HiTrap Q and Superdex 75 gel filtration chromatography steps (GE Healthcare). The protein was concentrated in 20 mm Tris-HCl buffer (pH 8.0), containing 150 mm NaCl and 2 mm dithiothreitol, to a final concentration of 13.95 mg/ml. All mutants used for functional analyses (see below) were expressed with methionine instead of selenomethionine and were purified as described above. The samples for blue native PAGE were prepared without gel filtration; the samples were dialyzed against the final buffer (same as the gel filtration buffer) before electrophoresis.
Crystallization and Structure Determination
In the crystallization screening, small crystals appeared under several conditions. However, the crystals were not large enough to collect reflection data. The final crystallization conditions were found in the subsequent refinement process, including additive screening (Hampton Research). Crystals of EBV BMRF1 were grown in 1.25 m Li2SO4, 0.5 m (NH4)2SO4, 8% PEG400, 275 mm 2,6-dimethyl-4-heptyl-β-d-maltopyranoside, and 0.1 m HEPES buffer (pH 8.0) at 20 °C by the hanging drop vapor diffusion method. Three sets of x-ray diffraction data at different wavelengths (peak, 0.97907 Å; edge, 0.97947 Å; and high remote, 0.96400 Å) were collected at beamline BL44B2 of SPring-8 (Harima, Japan) with an ADSC Quantum-315 CCD detector under cryogenic conditions with Paratone-N. The diffraction data were processed and scaled with the HKL2000 program package (22). The positions of the selenomethionine atoms were analyzed by SHELXC/D (23). The three-dimensional structure of BMRF1 was determined by the multiple anomalous dispersion method at 2.9 Å resolution using SOLVE (24) with 46 selenomethionine positions, which were determined by SHELXD. Subsequently, density modification was conducted with RESOLVE (25). The model building was performed with O (26). A total of eight protein molecules was located in the asymmetric unit. The structures were refined with CNS (27) without restraints by noncrystallographic symmetry during the refinement process. The calculated electron density for the C terminus, around residues 300–314, was not clear, and thus this region was omitted from the following refinement processes. The final model was assessed by PROCHECK, in the CCP4 suite (28). The Ramachandran plot revealed that 88.8% of the residues are in the most favored regions, with 11.2% in the additionally allowed regions in molecule A. The data collection and refinement statistics are summarized in Table 1. The ribbon and molecular surface models in the figures were depicted by PyMol (29).
TABLE 1.
Crystal parameters, data collection, and refinement statistics
| Crystal characteristics | |||
| Space group | C2221 | ||
| Unit cell parameters | a = 125.7 Å, b = 191.6 Å, c = 371.7 Å | ||
| Molecules in asymmetric unit | 8 | ||
| Peak | Edge | Remote | |
| MADa | |||
| Wavelength | 0.97907 Å | 0.97947 Å | 0.96400 Å |
| Resolution range | 48.4 to 2.90 Å | 48.4 to 2.90 Å | 47.9 to 2.90 Å |
| Redundancy | 5.0 | 5.0 | 5.0 |
| Unique reflections | 98,550 | 98,545 | 98,552 |
| Completenessb | 99.6% (100.0%) | 99.6% (100.0%) | 99.6% (100.0%) |
| I/σ(I) | 15.4 (4.4) | 19.2 (4.3) | 17.2 (3.8) |
| Rsymc | 0.092 (0.365) | 0.082 (0.371) | 0.088 (0.411) |
| Figure of merit (FOM) | |||
| Before/after solvent modification | 0.37/0.69 | ||
| Refinement statistics | |||
| Resolution range | 48.4–2.90 Å | ||
| Unique reflections | 98,516 | ||
| R-factor/free R-factord | 0.206 (0.288)/0.251 (0.328) | ||
| No. of protein atoms | 17,968 | ||
| No. of ion atomse | 9 | ||
| No. of water molecules | 121 | ||
| r.m.s.d. from ideal geometry | |||
| Bond lengths | 0.008 Å | ||
| Bond angles | 1.50° | ||
| Average isotropic B-value | 47.90 Å2 | ||
a MAD means multiple anomalous dispersion method.
b Numbers in parentheses refer to the highest resolution shell 3.00 to 2.90.
c Rsym = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h).
d R-factor = Σh‖Fobs| − |Fcalc‖/Σh|Fobs|. Free R-factor was calculated using 10% of reflections omitted from refinement. Numbers in parentheses refer to the highest resolution shell 3.08 to 2.90.
e Chloride ions.
Ultracentrifugation
The protein samples were purified as above, and the final buffer, including 5 mm 2-mercaptoethanol instead of dithiothreitol, was used for gel filtration chromatography to avoid UV absorbance by dithiothreitol. Sedimentation equilibrium experiments were performed in a Beckman Optima XL-I instrument with six-channel centerpieces, using a Beckman An-50Ti rotor. The mutants (C95E and C95E/H141F) appeared to be unstable with the high centrifugal force and started to precipitate during the ultracentrifugation. The protein concentrations loaded into the cells were 0.87, 0.44, and 0.22 mg/ml for the wild type, 0.82, 0.41, and 0.20 mg/ml for the C206E mutant, and 0.77, 0.38, and 0.19 mg/ml for the C95E/H141F mutant. Equilibrium distributions were analyzed at 9,000, 10,000, and 12,000 rpm after 16 h at each speed; the measurement temperature was 4 °C, and the wavelength for measurements was 280 nm. For the molecular weight analysis, a partial specific volume of 0.73 cm3/g and a solution density of 1.0067 g/cm3 were used.
Blue Native PAGE
Blue native PAGE was performed by employing a discontinuous gel system. The separating gel (12% (w/v) polyacrylamide) was composed of 50 mm imidazole buffer (pH 7.0), containing 67 mm 6-aminocaproic acid and 16% glycerol, and the stacking gel was prepared with 6% (w/v) polyacrylamide. The cathode buffer was prepared at pH 7.0 with imidazole and 50 mm Tricine, supplemented with 0.02% Coomassie Brilliant Blue G-250. The anode buffer was 50 mm imidazole, pH 7.0. The sample buffer was composed of 20 mm Tris-HCl buffer (pH 7.4), containing 0.1 m EDTA, 50 mm NaCl, 10% glycerol, and 10% CBG buffer stock (5% CBG, 500 mm 6-aminocaproic acid, and 100 mm imidazole buffer (pH 7.0)). Protein samples were mixed with an equal volume of the sample buffer and incubated for 30 min before loading. Electrophoresis was performed at 100 V for the stacking gel and 250 V for the separating gel at 20 °C. NativeMarkTM standard (Invitrogen) was used as a molecular weight standard.
dsDNA Probe for DNA Binding Assay
The radiolabeled plasmid DNA, pCR2.1 (3.9 kbp) (Invitrogen), used for filter-binding experiments was prepared by digesting the plasmid DNA with the XbaI restriction enzyme and then filling the overhanging ends with the Klenow fragment of E. coli DNA polymerase I (New England Biolabs, Inc.) in the presence of 20 μm each of dGTP, dCTP, and dTTP, 1 μm dATP, and 1.67 μm [α-32P]dATP (6,000 Ci/mmol). The reaction mixture was incubated at 25 °C for 20 min and then was heated to 75 °C for 20 min to inactivate the enzyme. The unreacted nucleotides were removed by centrifugation through a Chroma Spin-100 spin column (Clontech).
Nitrocellulose Filter Binding Assay
Filter binding assays were performed by a modification of the double-filter method described previously (18, 30, 31). The DNA-protein complexes were trapped on the alkali-treated nitrocellulose filters (18), and the remaining unbound DNA was trapped on the DE81 filter (Whatman) placed under the nitrocellulose filter. The assay mixture (40 μl) contained 40 ng of the 3′-end-labeled linear dsDNA and the indicated amounts of each EBV BMRF1 mutant protein in the DNA binding buffer, consisting of 20 mm Tris-HCl buffer (pH 8.0), 5 mm MgCl2, 20 mm NaCl, and 1 mm dithiothreitol. The mixture was incubated at 30 °C for 5 min. The reactions were then diluted 10-fold in the DNA binding buffer and applied to the alkali-treated nitrocellulose/DE81 filter stack soaked in binding buffer, using a multichannel filtration manifold (Millipore). The filters were immediately washed once with DNA binding buffer and twice with washing buffer, consisting of 20 mm Tris-HCl buffer (pH 7.6) containing 5 mm MgCl2, and then were dried at 80 °C. The radioactivity was measured with a liquid scintillation counter (Aloka). Apparent Kd values were calculated by a saturation isotherm analysis, using the concentrations of BMRF1 protein that resulted in half-maximal binding, as described previously (16, 32).
Preparation of the BALF5 Polymerase Catalytic Subunit
The EBV DNA polymerase catalytic subunit (BALF5) was purified from total extracts of recombinant baculovirus AcBALF5-infected Sf21 cells, essentially as described previously with some modifications (6). The cells infected with AcBALF5 (14.2 g wet weight) were subjected to Dounce homogenization. BALF5 was purified by chromatography on columns of heparin-agarose, HiTrap heparin HP, and FPLC Mono S HR5/5. The fractions containing BALF5 were pooled as fraction IV. Fraction IV (200 ng/μl) had substoichiometric levels of the EBV polymerase catalytic subunit and was used as the purified BALF5 fraction.
Polymerase Processivity Assay
An oligodeoxynucleotide with the sequence CACAATTCCACACAAC, complementary to nucleotides 6170–6185 of M13mp18 single-stranded DNA (33), was purchased from New England Biolabs, Inc. To form singly primed M13 single-stranded DNA, the synthetic 16-mer DNA was annealed at a molar ratio of 20:1 to M13mp18 single-stranded DNA in 20 mm Tris-HCl buffer (pH 8.0), containing 5 mm MgCl2 and 0.3 m NaCl. The hybridization mixture was incubated at 90 °C for 5 min, allowed to cool to room temperature for 1 h, and then incubated for 1 h at 30 °C. The primed M13 single-stranded DNA was separated from the excess primer by centrifugation through a Chroma Spin-100 spin column (Clontech).
The reaction mixture (15 μl) contained each of the purified EBV BMRF1 mutants (100 ng), 200 ng of the purified BALF5 protein, 20 μg of singly primed M13 single-stranded DNA (0.5 μg as a circle), 0.5 mm ATP, 10 μm dATP, and 50 μm each of dGTP and dTTP in the reaction buffer (50 mm Tris-HCl buffer (pH 8.0), 3 mm MgCl2, 100 mm NaCl, 1 mm dithiothreitol, 100 μg of bovine serum albumin per ml, 10% glycerol). The mixtures were preincubated at room temperature for 5 min. To start the reaction, a 5-μl aliquot of the reaction buffer containing 50 μm dCTP and 0.33 μm [α-32P]dATP (6,000 Ci/mmol) was added, and the replication reaction was incubated at 35 °C for 10 min. The reactions were stopped by placing the mixtures on ice and adding 5 μl of alkaline loading buffer, containing 2 mm EDTA, 50 mm NaOH, 2.5% glycerol, and 0.025% bromocresol green. The samples were heated to 95 °C for 2 min and then rapidly chilled to 0 °C. The samples were then loaded onto a 1.0% (w/v) alkaline-agarose gel containing 50 mm NaOH and 1 mm EDTA (30). After electrophoresis at 50 V for 6.5 h, the gel was washed in 7% (v/v) trichloroacetic acid and then dried on DE81 paper under vacuum. The dried gel was exposed to a Fuji imaging plate and was analyzed by the Fuji Image Analyzer BAS 2500.
RESULTS
Structure of the BMRF1 Protein
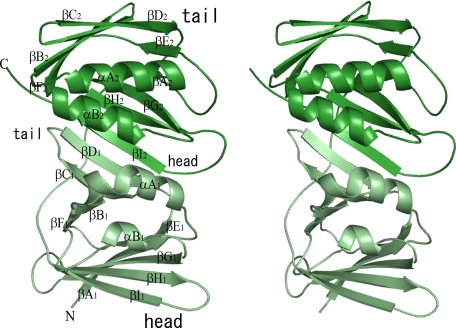
Each protein molecule contained a total of 18 β-strands and 4 α-helices. These secondary structural elements are designated as reported previously (16). The crystal structure of EBV BMRF1 consists of two subdomains, which possess similar topology. Self-structural alignment by MATRAS (34) suggested that these subdomains are residues 1–148 and 159–299. The subdomains superimpose with an r.m.s.d. of 3.5 Å among 121 common C-α carbons and are related by a pseudo 2-fold axis (Fig. 1). The structural similarity between the two domains covers almost the entire length of the protein, but one distinguishable difference is a 16-residue insertion (residues 213–228) between βD2 and βE2 in the C-terminal subdomain. This region forms a loop structure, which is stabilized by a hydrophobic interaction between Phe222 and the protein core. Each subdomain contains two anti-parallel β-sheets. The contact surface of the subdomains forms interstrand hydrogen bonds between βD1 and βI2, which means that the subdomains are arranged in a “head-to-tail” manner (Fig. 1). As a member of the polymerase processivity factors, BMRF1 exhibits a similar fold to those of the other factors. A structural homology analysis revealed that the r.m.s.d. values upon superimposition with human PCNA, the herpes simplex virus UL42, and HCMV UL44 are 4.5 Å (for 243 common Cα carbons), 3.9 Å (251 Cα carbons), and 3.2 Å (245 Cα carbons), respectively, although the sequence identities are as low as 8–9%.
FIGURE 1.
Molecular structure of BMRF1. The crystal structure of a monomer of EBV BMRF1 is drawn as a ribbon model. The subdomains (N- and C-terminal) are colored pale green and deep green, respectively. The designations of the secondary elements are composed of three parts as follows: the first, the type of secondary structure α or β; the second, the order of elements A–I; and the third, the first or second subdomain, subscript 1 or 2.
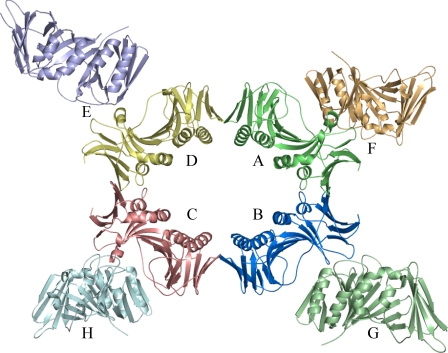
The eight molecules of BMRF1 in the asymmetric unit are labeled from A to H. Ring formation is observed with molecules A–D (Fig. 2). The side of this molecular ring is flanked by the other four molecules. Although the crystal packing interactions differ between the ring-forming and -flanking molecules, their structures lack remarkable structural differences. The r.m.s.d. values for each pair of molecules are in the range of 0.388–0.899 Å. Ring formation has also been observed in the structure of human PCNA (10) as a trimer and E. coli/Streptococcus pyogenes β-subunit as a dimer (35). The β-subunits of E. coli and S. pyogenes consist of three subdomains, and therefore, the ring includes a total of six subdomains. In contrast, BMRF1 forms an eight-subdomain ring. The molecular contact surfaces within this ring form continuous β-sheets in “head-to-head” (βI1-βI1′) and “tail-to-tail” manner (βD2–βD2′) (“′” means the neighboring molecule) (supplemental Fig. 2). The head-to-head contact surface area was larger (∼930 Å2) than the tail-to-tail contact area (340 Å2). In addition to these main chain-main chain interactions, disulfide bonds (Cys95–Cys95′ and Cys206–Cys206′) were observed on both contact surfaces. The head-to-head contact is similar to that in the dimeric form of UL44. However, human PCNA adopted the head-to-tail contact for the ring formation, unlike BMRF1.
FIGURE 2.
Molecular packing of BMRF1 in the crystal asymmetric unit. The EBV BMRF1 molecules in the asymmetric unit are depicted as ribbon models. Each monomer is differently colored.
Mutant Preparation
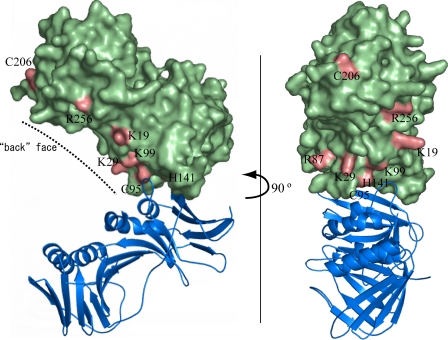
In the structures of polymerase processivity factors, various oligomerization states, including the monomeric state, have been reported so far. The structure of EBV BMRF1 revealed a unique oligomerization manner in the crystal packing, consisting of the tetrameric ring flanked by four molecules. On the other hand, a sedimentation equilibrium analysis after purification of the WTΔC protein indicated that it forms a dimer (see below). The charge distribution on the surface of the BMRF1 structure revealed that many positively charged amino acid residues are located inside the tetrameric ring. This positively charged region may interact with DNA, as supported by the recent report for the structural homolog of BMRF1, HSV-1 UL42 (36), although the functional unit of UL42 for DNA binding is the monomer. To investigate the oligomerization manner and the DNA-binding site, several mutants were prepared as follows: C95E, H141F, C95E/H141F, and C206E for the dimer interface, and K19E, K29E, R87E, K99E, and R256E for the putative DNA binding region. These mutants are mapped on the molecular surface, as shown in Fig. 3.
FIGURE 3.
Mutated amino acid residues of EBV BMRF1. The mutated amino acid residues are displayed in pink on the green surface model. The partner molecule forming a homodimer is drawn as a blue ribbon model. The dashed line indicates the back face of the molecule.
Analysis of the Oligomerization State
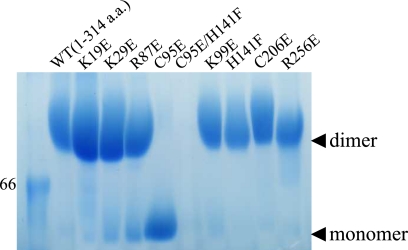
To analyze the oligomerization state of EBV BMRF1 in solution, analytical ultracentrifugation experiments were performed for the WTΔC protein and the C95E, C95E/H141F, and C206E mutants, which involve the intermolecular contacts. Sedimentation equilibrium analysis revealed that the measured molecular mass of BMRF1-WTΔC was 63.2 kDa, in good agreement with the calculated value for the dimer (67.5 Da). The mutant C206E was also observed as a dimer (64.3 kDa, measured; 67.6 Da, calculated). The other mutants seemed to be unstable in the strong centrifugal force. Sedimentation velocity experiments suggested that the proteins started to aggregate during ultracentrifugation, and thus their molecular masses could not be correctly estimated. When the protein samples were concentrated, we used Amicon centrifugal filters. To avoid aggregation, the protein samples were kept at a relatively low concentration (∼2 mg/ml), with short centrifugation durations or lower acceleration, and then were subjected to blue native PAGE. As shown in Fig. 4, the protein band of the C95E mutant corresponded to the monomer, and the other mutants and the wild type migrated as dimers, except for the C95E/H141F double mutant, which did not migrate.
FIGURE 4.
Blue native PAGE. Ten samples were applied on the gel. The numbers and WT at the top of the gel indicate the mutated position and the wild type (WTΔC), respectively. The marker band corresponds to 66 kDa. Monomer and dimer positions are indicated by black arrowheads.
DNA Binding Activity of the BMRF1 Mutant Proteins
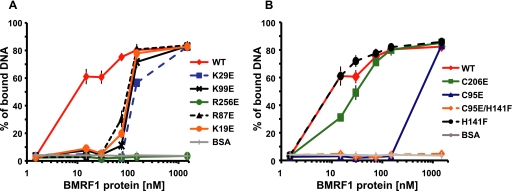
To investigate the effects of glutamic acid substitutions of the EBV BMRF1 mutants on the DNA binding activity, we measured the relative affinities of the WTΔC and mutant proteins for a 3.9-kbp dsDNA, using a filter binding assay (Fig. 5). An apparent Kd of ∼8 nm was calculated for WTΔC. The mutations that substituted glutamic acid for the positively charged amino acid residues located inside the ring reduced the DNA binding activity (Fig. 5A); the K29E, K99E, R87E, and K19E mutants displayed apparent Kd values in the range of 80–200 nm. The DNA binding activity of the mutant R256E was the same as the negative control, bovine serum albumin. Thus, each glutamic acid substitution for a conserved, positively charged residue inside the ring affected the affinity for DNA. The C95E and C95E/H141F mutants failed to form dimers (Fig. 4) and exhibited drastically decreased DNA binding activities (apparent Kd values of 500–600 nm and higher for C95E and C95E/H141F, respectively) (Fig. 5B). The C206E and H141F mutants could form dimers as efficiently as the WTΔC, as revealed by the blue native PAGE analysis (Fig. 4), and retained the full ability to bind to DNA, with apparent Kd values of 10–20 and 8–9 nm, respectively (Fig. 5B). These results suggest that dimer formation is necessary for BMRF1 to bind to DNA by itself.
FIGURE 5.
DNA binding assays for the BMRF1 wild type (WT) and mutants. Each of the purified EBV BMRF1 wild type (WTΔC) and mutant proteins, in which mutations were located in the putative DNA binding region (A) and in the dimer interface (B), was incubated with 32P-labeled dsDNA (3.9 kbp). Bovine serum albumin (BSA) was used as a negative control. Protein-bound and free DNAs were quantified by a filter binding assay (details under “Experimental Procedures”), and the ratios of the protein-bound DNA to total DNA were plotted with the indicated protein concentrations. Each plot shows the average of data from three independent experiments, together with the standard deviations.
Mutations of the BMRF1 Protein Affect Its Ability to Increase the Polymerase Processivity of the EBV Polymerase Catalytic Subunit
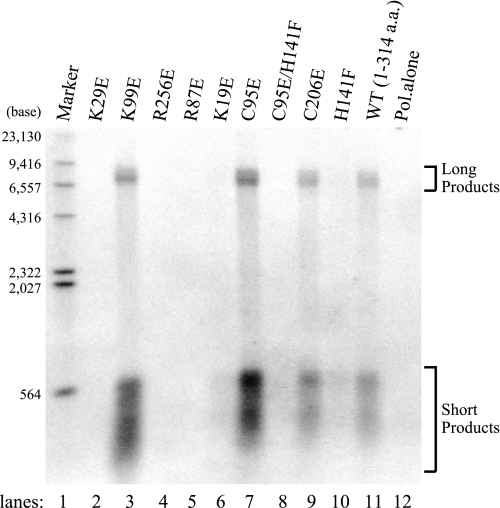
EBV BMRF1 is a polymerase processivity factor that increases the polymerization rate and the processivity of the EBV polymerase catalytic subunit, BALF5 (6). To determine the effect of the BMRF1 mutants on the polymerase processivity of the EBV DNA polymerase holoenzyme on a long single-stranded DNA template, the holoenzyme was reconstituted onto singly primed M13mp18 single-stranded DNA circles by using stoichiometric levels of BALF5 along with the BMRF1 mutants. The BMRF1 proteins lack intrinsic polymerization capability (6). DNA synthetic processivity was measured by using the singly primed M13 template with an excess molar ratio of primer-template to polymerase, so that each polymerase molecule was bound to a primer terminus. dGTP and dTTP are needed to prevent the removal of the DNA primer by the 3′-to-5′-exonuclease activity of BALF5. Synchronous DNA synthesis by the reconstituted forms of the holoenzyme was initiated upon the addition of the remaining deoxynucleoside triphosphates, and DNA products were analyzed by electrophoresis on an alkaline-agarose gel (Fig. 6). Under our reaction conditions, short products appeared in addition to the full-length products, indicating pausing sites on the circular M13 single-stranded DNA (Fig. 6). We presume that these pausing sites represented sites with substantial secondary structures on the single-stranded DNA template under the reaction conditions (37, 38). With substoichiometric levels of the EBV polymerase catalytic subunit alone, very little DNA synthesis, even of the short products, was detected under the reaction conditions (Fig. 6, lane 12). The addition of the BMRF1-WTΔC, however, resulted in the accumulation of short products as well as completed products from the 7.2-kb M13mp18 template. The K29E, R256E, and R87E mutants and the C95E/H141F double mutant abolished the ability to increase the BALF5 polymerase processivity (Fig. 6, lanes 2, 4, 5, and 8, respectively). The K19E and H141F mutants generated a slight accumulation of medium and short products, although they formed few completed products (Fig. 6, lanes 6 and 10, respectively). The K99E, C95E, and C206E mutants acted as polymerase processivity factors that were as good as the WTΔC (Fig. 6, lanes 3, 7, 9, and 11, respectively). It should be noted that all of the mutants with decreased binding affinity for DNA are not defective in the ability to increase the polymerase processivity, in contrast to the observations for UL42 (39, 40) and UL44 (41). In particular, the C95E mutant, which is defective in dimer formation as well as in DNA binding activity, efficiently increased the polymerase processivity, strongly suggesting that the monomeric form of the BMRF1 protein interacts with the BALF5 polymerase catalytic subunit to function as a polymerase processivity factor.
FIGURE 6.
Polymerase processivity by the BMRF1 wild type (WT) and mutants. Long chain DNA synthesis was measured and visualized by alkaline-agarose gel electrophoresis of the products that incorporated radiolabeled dATP with a singly primed M13 single-stranded DNA (7.2 kb) as a primer-template by the EBV BALF5 polymerase (Pol) catalytic subunit. Reactions were conducted in the presence of the EBV BMRF1 mutants (lanes 2–10) or the wild type (WTΔC) (lane 11), or in the absence of the BMRF1 protein (lane 12). Replication assays were performed as described under “Experimental Procedures.” The replication products were visualized by autoradiography after electrophoresis on a 1.0% alkaline-agarose gel. Molecular size markers are heat-denatured 5′-terminally labeled HindIII fragments of λ DNA (lane 1). a.a., amino acids.
DISCUSSION
Head-to-Head Dimer Formation of BMRF1
Although the crystal structure indicated the formation of an interesting tetrameric ring, the electrophoresis and sedimentation assays suggested that the main component of EBV BMRF1 in solution is dimeric. In the ring structure, two contact surfaces may be involved in dimerization. On both surfaces, the β-strands form a continuous sheet structure, βI1–βI1′ and βD2–βD2′, respectively. The blue native PAGE analysis revealed that the C95E mutant migrated at the monomer position, whereas the C206E mutant moved at the dimer position, implying that the monomer-monomer contact surface includes the region around Cys95. The buried area calculations reasonably supported this result (see “Results”). Overall, we concluded that BMRF1 dimerizes in a head-to-head manner, connecting the βI1 strands to each other.
A topological comparison among the DNA polymerase processivity factors shows that these proteins share a common overall structure. However, each protein appears to form various multimers. The PCNA proteins form ring-shaped trimers with head-to-tail contacts (42). HSV-1 UL42 stably exists as a monomer (17), whereas HCMV UL44 forms a head-to-head contacting C-shaped dimer in the crystal structure (16). The BMRF1 dimer can be superimposed on the UL44 dimer with an r.m.s.d. value of 3.5 Å.
At the dimer interface of UL44, the hydrophobic interactions significantly contribute to dimer formation. The mutations substituting Ala for Phe121 and Leu86/Leu87 increased the Kd value by 10–100-fold (16). In addition to these residues, Leu93 and Met123 are also involved in hydrophobic interactions. The hydrophobic contacts in UL44 appear to be stable in solution, and therefore, its existence as a monomer may be energetically unfavorable. The residues forming the hydrophobic contacts are conserved in the β-herpesvirus family (16). By contrast, in BMRF1, hydrophilic residues (Arg93, Glu100, Tyr132, and Ser139) are located at the dimer interface, forming a hydrogen-bonding network. The hydrophilic environment around βI1 is rather similar to that of monomeric UL42, rather than dimeric UL44. It is likely that BMRF1 can transform to the monomeric form in solution without serious energetic destabilization. Actually, the monomeric forms were found in the same crystal lattice (molecules E–H in Fig. 2). Kaposi sarcoma herpesvirus processivity factor-8 also functions as a dimer (43) and shares sequence similarity with BMRF1 (27% with aligned residues). The hydrophilic residues at the dimer interface in BMRF1 (Arg93, Glu100, Tyr132, and Ser139) are also well conserved in processivity factor-8 (Ser90, Glu97, Tyr132, and Thr139). One of the characteristics of the polymerase processivity factors in γ-herpesvirus may be that they dimerize through hydrophilic interactions.
According to an electron microscopy study (19), a ring structure with a 55-Å diameter was observed for intact BMRF1, which includes six molecules within the ring. The crystal structure of BMRF1-WTΔC showed an elliptic ring (long axis ∼50 Å, short axis ∼40 Å). Assuming the head-to-head dimer structure, these rings may have the same interface arrangement between dimers. It is interesting that BMRF1 is able to adopt different oligomeric states based on the dimer as the functional unit, although the functional relevances of the rings are still unknown for both cases.
BMRF1 Binds dsDNA on Its Concave Surface as a Dimer
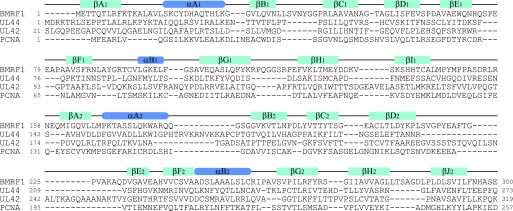
The crystal structure of the E. coli polymerase β-subunit revealed that dsDNA binds to the inside of the ring structure, through basic amino acid residues (44). Although there is no complex structure between DNA and a virus polymerase processivity factor available thus far, site-directed mutation analyses of HSV-1 UL42 supported the proposal that protein-DNA interactions occur with the positively charged amino acid residues on the “back” face (40). The mutational analyses in EBV BMRF1 also demonstrated that substitutions of positively charged residues reduce the DNA-binding affinity (see under “Results”). Furthermore, the mutation disrupting dimer formation, C95E, significantly reduced the DNA binding activity (Fig. 5B). This means that dimer formation, as well as the positively charged amino acid residues on the back face of BMRF1, is required for DNA binding. An analysis of the dimeric processivity factor HCMV UL44 also indicated that its DNA-binding affinity is related to dimer formation (16). BMRF1 and UL44 form a concave surface, directing the back face inside the C-shape. The basic concave surface is very important for DNA binding by the dimerized proteins, BMRF1 and UL44. In addition, UL44 possesses the basic loop (163–174 residues), although its electron density was not identified in the crystal structure (16). A modeling calculation suggested that this loop can interact with DNA and wrap around the open side of the C-shaped clamp. UL44 consequently might bind to DNA in a similar manner as PCNA, which surrounds DNA by ring formation (32). However, because this loop is an insertion specific for UL44 (Fig. 7), the combination of the C-shaped clamp and the wrapping loops is uniquely adapted for DNA binding by UL44.
FIGURE 7.
Structure-based sequence alignment of polymerase processivity factors. The sequences of three viral processivity factors (EBV BMRF1, HSMV UL44, and HSV-1 UL42) and human PCNA are aligned. The secondary structure elements are depicted by green bars (β-strands) and blue bars (α-helices).
It has been proposed that DNA binding by either a monomer or dimer depends on the basic amino acid residues on the back face. UL42 (monomeric processivity factor) contains many arginine residues on the back face, whereas UL44 (dimeric) contains lysine residues (16). This difference could be interpreted in terms of the difference in the DNA-binding affinity between arginine and lysine residues (16). The guanidinium group of the arginine residue allows it to form a stronger interaction with the negatively charged phosphate groups of DNA than the lysine residue. Thus, Arg-rich UL42 is able to maintain sufficient affinity for DNA as a monomer, and Lys-rich UL44 dimerizes to bind DNA to compensate for its weaker interaction than arginine. The back face of BMRF1 contains about 10 basic amino acid residues: Lys12, Lys19, Lys29, Arg87, Lys99, Lys170, Lys177, Arg180, Lys228, and Arg256. BMRF1 possesses a Lys-rich back face, and therefore, it may bind to DNA as a dimer.
The structural analyses of UL44 and the UL44-UL54 C-terminal fragment complex revealed that the complex structure exhibits a wider C-shaped opening, as a result of the peptide fragment binding (45). In comparison, as a dimer, the present BMRF1 structure is more similar to unliganded UL44 than the UL44-peptide complex. The distances between the corresponding Cα positions on αB2 (253–253′ for BMRF1 and 231–231′ for the UL44-peptide complex; where ′ indicates neighboring molecule in the dimer) are 38 Å (BMRF1) and 44 Å (UL44), respectively. The location of αA2 is also different from that in the UL44-peptide complex. αA2 of the UL44-peptide complex is inclined against the center axis of the C-shape, which allows the inserted basic loop to interact with the DNA. On the other hand, αA2 of BMRF1 is relatively horizontal to the center axis. It was reported that BMRF1 is abundantly expressed in lytic infected cells and shows a homogeneous, rather than punctate, distribution in the replication compartments when viral DNA is newly synthesized (8). From these observations, we speculate that BMRF1 protects the synthesized viral DNA from nuclease attack or histone assembly by occupying the surface of the DNA molecules. The size difference and the helical rearrangements in the C-shaped open part, between the unliganded form of BMRF1/UL44 and the liganded UL44, may reflect two different states. In the unliganded form, these proteins may remain bound to protect the viral DNA. The liganded form may correspond to the replication state in the HCMV replication system, although the EBV replication system adopts a different subunit architecture during the replication process (see below).
BMRF1 Interacts with the Polymerase Catalytic Subunit BALF5 as a Monomer
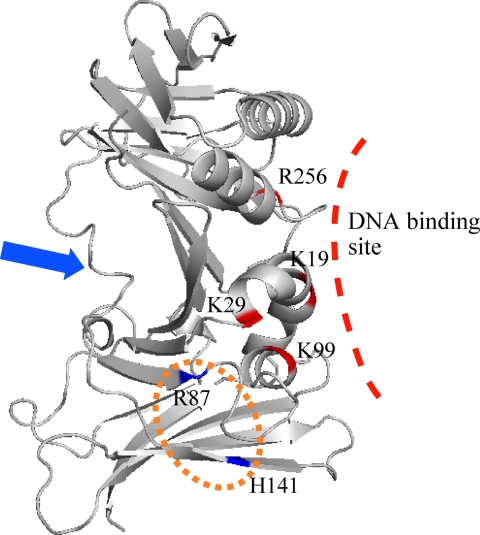
Mutational analyses revealed that polymerase processivity was not affected negatively by the C95E mutation, which disrupts dimer formation. Therefore, these results indicated that the monomeric form of EBV BMRF1 functions in the DNA replication process as a polymerase processivity factor. Although the H141F mutation disrupts neither dimer formation nor DNA binding (Fig. 5B), it abolished the polymerase processivity activity (Fig. 6, lane 10). This histidine residue is located at the dimer interface and is buried between the two molecules, when BMRF1 forms a dimer. The water-accessible surface of the His141 residue is less than 10 Å2. The R87E mutant also retains the DNA binding ability to some extent (apparent Kd values of 80 nm) (Fig. 5A) and the dimerization ability (Fig. 4), but it lacks the polymerase processivity activity (Fig. 6, lane 5). This mutation is also located near the dimer interface. These mutations might inhibit the association with the polymerase catalytic subunit, BALF5, and/or reduce the stability of BALF5-bound DNA. Other mutants, which did not stimulate polymerase activity (K19E, K29E, and R256E), could hardly bind to DNA. The reduced polymerase activity might be caused by low affinity for DNA. These possibilities cannot be distinguished from our results. The putative regions in BMRF1 involved in DNA and BALF5 binding are summarized in Fig. 8.
FIGURE 8.
Putative binding site for BALF5 on BMRF1. The mutated amino acid residues are displayed in blue or red on the ribbon model (EBV BMRF1 monomer). The binding site for HSV-1 UL42 and HCMV UL44 in the extreme C-terminal region of the polymerase catalytic subunit is indicated by the blue arrow. The putative EBV BALF5-binding site on BMRF1 is shown in an orange circle.
The C-terminal region of the polymerase catalytic subunit is critically important for interactions between the polymerase catalytic subunit and the processivity factor in the complexes of HSV-1 UL42-UL30 (46) and HCMV UL44-UL54 (47). Although the structure of the whole complex between the polymerase catalytic subunit and the processivity factor is still unavailable, the complex structures of processivity factors with the extreme C-terminal fragments of the polymerase catalytic subunit have been solved (45, 48). These structures revealed the key interactions underlying polymerase complex formation. In both cases, the C-terminal flexible region binds to the opposite side of the back face. These interactions have also been detected in the bacteriophage RB69 sliding clamp and polymerase system (15). On the other hand, the secondary structure prediction suggested that the extreme C-terminal region of BALF5 is almost fully structured. Furthermore, the putative BMRF1·BALF5 contact region includes at least the BMRF1 dimer interface. However, in the case of UL44-UL54, the C-terminal fragment is sufficient to form a complex (47), and its interaction region does not involve the dimer interface. This fact suggests that BMRF1 and UL44 utilize different contact surfaces with the polymerase catalytic subunit, although both can form dimers in solution. The DNA-binding affinity is related to the binding of the C-terminal fragment of UL54, implying that UL44 retains its dimeric form during DNA replication (45). In contrast, BMRF1 could be transformed from the dimer to the monomer during the replication process. Ultimately, the EBV BMRF1-BALF5 system might utilize a different binding manner from other systems, such as HSV-1 UL42-UL30 and HCMV UL44-UL54, with respect to the binding part of the polymerase and the interacting region of the processivity factor.
Acknowledgments
We thank Mio Inoue, Hiroaki Hamana, Yoshiko Ishizuka-Katsura, Nobuko Maoka, Satoshi Morita, and Yuri Tomabechi for technical assistance with vector and sample preparations. We also thank the beamline scientists Taiji Matsu, Hiroaki Nakajima, Yoshiaki Kawano, and Takaaki Hikima for help in data collection at RIKEN beamline BL44B2 of SPring-8.
This work was supported by Special Coordination Funds for Promoting Science and Technology and the RIKEN Structure Genomics/Proteomics Initiative in the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science, and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. 1 and 2, and an additional reference.
The atomic coordinates and structure factors (code 2Z0L) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- EBV
- Epstein-Barr virus
- PCNA
- proliferating cell nuclear antigen
- dsDNA
- double-stranded DNA
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- r.m.s.d.
- root mean square deviation
- HCMV
- human cytomegalovirus
- HSV-1
- herpes simplex virus, type 1.
REFERENCES
- 1.Tsurumi T., Fujita M., Kudoh A. (2005) Rev. Med. Virol. 15, 3–15 [DOI] [PubMed] [Google Scholar]
- 2.Tsurumi T. (2001) Curr. Top. Microbiol. Immunol. 258, 65–87 [DOI] [PubMed] [Google Scholar]
- 3.Fixman E. D., Hayward G. S., Hayward S. D. (1995) J. Virol. 69, 2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsurumi T. (1991) Virology 182, 376–381 [DOI] [PubMed] [Google Scholar]
- 5.Tsurumi T. (1991) Biochem. J. 280, 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsurumi T., Daikoku T., Kurachi R., Nishiyama Y. (1993) J. Virol. 67, 7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsurumi T., Daikoku T., Nishiyama Y. (1994) J. Virol. 68, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daikoku T., Kudoh A., Fujita M., Sugaya Y., Isomura H., Shirata N., Tsurumi T. (2005) J. Virol. 79, 3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. (1994) Cell 79, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 10.Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. (1996) Cell 87, 297–306 [DOI] [PubMed] [Google Scholar]
- 11.Chapados B. R., Hosfield D. J., Han S., Qiu J., Yelent B., Shen B., Tainer J. A. (2004) Cell 116, 39–50 [DOI] [PubMed] [Google Scholar]
- 12.Matsumiya S., Ishino S., Ishino Y., Morikawa K. (2003) Protein Sci. 12, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnouf D. Y., Olieric V., Wagner J., Fujii S., Reinbolt J., Fuchs R. P., Dumas P. (2004) J. Mol. Biol. 335, 1187–1197 [DOI] [PubMed] [Google Scholar]
- 14.Moarefi I., Jeruzalmi D., Turner J., O'Donnell M., Kuriyan J. (2000) J. Mol. Biol. 296, 1215–1223 [DOI] [PubMed] [Google Scholar]
- 15.Shamoo Y., Steitz T. A. (1999) Cell 99, 155–166 [DOI] [PubMed] [Google Scholar]
- 16.Appleton B. A., Loregian A., Filman D. J., Coen D. M., Hogle J. M. (2004) Mol. Cell 15, 233–244 [DOI] [PubMed] [Google Scholar]
- 17.Randell J. C., Coen D. M. (2004) J. Mol. Biol. 335, 409–413 [DOI] [PubMed] [Google Scholar]
- 18.Tsurumi T. (1993) J. Virol. 67, 1681–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makhov A. M., Subramanian D., Holley-Guthrie E., Kenney S. C., Griffith J. D. (2004) J. Biol. Chem. 279, 40358–40361 [DOI] [PubMed] [Google Scholar]
- 20.McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 21.Kigawa T., Yamaguchi-Nunokawa E., Kodama K., Matsuda T., Yabuki T., Matsuda N., Ishitani R., Nureki O., Yokoyama S. (2002) J. Struct. Funct. Genomics 2, 29–35 [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23.Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 27.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 29.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 30.McEntee K., Weinstock G. M., Lehman I. R. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weisshart K., Chow C. S., Coen D. M. (1999) J. Virol. 73, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanisch-Perron C., Vieira J., Messing J. (1985) Gene 33, 103–119 [DOI] [PubMed] [Google Scholar]
- 33.Kawabata T. (2003) Nucleic Acids Res. 31, 3367–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argiriadi M. A., Goedken E. R., Bruck I., O'Donnell M., Kuriyan J. (2006) BMC Struct. Biol. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komazin-Meredith G., Santos W. L., Filman D. J., Hogle J. M., Verdine G. L., Coen D. M. (2008) J. Biol. Chem. 283, 6154–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villani G., Fay P. J., Bambara R. A., Lehman I. R. (1981) J. Biol. Chem. 256, 8202–8207 [PubMed] [Google Scholar]
- 37.Tsurumi T., Kobayashi A., Tamai K., Yamada H., Daikoku T., Yamashita Y., Nishiyama Y. (1996) Virology 222, 352–364 [DOI] [PubMed] [Google Scholar]
- 38.Chow C. S., Coen D. M. (1995) J. Virol. 69, 6965–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Randell J. C., Komazin G., Jiang C., Hwang C. B., Coen D. M. (2005) J. Virol. 79, 12025–12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loregian A., Sinigalia E., Mercorelli B., Palù G., Coen D. M. (2007) Nucleic Acids Res. 35, 4779–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruck I., O'Donnell M. (2001) Genome Biol. 2, REVIEWS3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X., Lin K., Ricciardi R. P. (2004) J. Biol. Chem. 279, 28375–28386 [DOI] [PubMed] [Google Scholar]
- 43.Georgescu R. E., Kim S. S., Yurieva O., Kuriyan J., Kong X. P., O'Donnell M. (2008) Cell 132, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komazin-Meredith G., Petrella R. J., Santos W. L., Filman D. J., Hogle J. M., Verdine G. L., Karplus M., Coen D. M. (2008) Structure 16, 1214–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Appleton B. A., Brooks J., Loregian A., Filman D. J., Coen D. M., Hogle J. M. (2006) J. Biol. Chem. 281, 5224–5232 [DOI] [PubMed] [Google Scholar]
- 46.Digard P., Bebrin W. R., Weisshart K., Coen D. M. (1993) J. Virol. 67, 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loregian A., Appleton B. A., Hogle J. M., Coen D. M. (2004) J. Virol. 78, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuccola H. J., Filman D. J., Coen D. M., Hogle J. M. (2000) Mol. Cell 5, 267–278 [DOI] [PubMed] [Google Scholar]