Abstract
Tumor necrosis factor (TNF) receptor-associated factor-2 (TRAF2) binds to cIAP1 and cIAP2 (cIAP1/2) and recruits them to the cytoplasmic domain of several members of the TNF receptor (TNFR) superfamily, including the TNF-TNFR1 ligand-receptor complex. Here, we define a cIAP1/2-interacting motif (CIM) within the TRAF-N domain of TRAF2, and we use TRAF2 CIM mutants to determine the role of TRAF2 and cIAP1/2 individually, and the TRAF2-cIAP1/2 interaction, in TNFR1-dependent signaling. We show that both the TRAF2 RING domain and the TRAF2 CIM are required to regulate NF-κB-inducing kinase stability and suppress constitutive noncanonical NF-κB activation. Conversely, following TNFR1 stimulation, cells bearing a CIM-mutated TRAF2 showed reduced canonical NF-κB activation and TNF-induced RIPK1 ubiquitylation. Remarkably, the RING domain of TRAF2 was dispensable for these functions. However, like the TRAF2 CIM, the RING domain of TRAF2 was required for protection against TNF-induced apoptosis. These results show that TRAF2 has anti-apoptotic signaling roles in addition to promoting NF-κB signaling and that efficient activation of NF-κB by TNFR1 requires the recruitment of cIAP1/2 by TRAF2.
Introduction
The inhibitor of apoptosis (IAP)7 family is composed of baculoviral IAP repeat-containing proteins, several of which also bear a RING domain that is capable of acting as a ubiquitin E3 ligase (1). cIAP1 and cIAP2 (cIAP1/2) are two RING-containing IAPs whose amplification or genetic mutation has been associated with cancers and may promote tumor cell survival (2–8). These highly conserved IAPs were initially identified as components of a TRAF2-containing complex bound to the cytoplasmic domain of TNFR2 (9), and they have subsequently been implicated in the regulation of signaling by several more receptors of the TNF superfamily (10–21). Although the BIR1 domain of cIAP1/2 mediates binding to TRAF2 (17, 22, 23), the complementary binding region of TRAF2 is unknown, and the relative roles of TRAF2 and cIAP1/2 E3 ligase activity in TNF superfamily signaling remain unclear.
Genetic deletion of TRAF2 in the mouse results in early postnatal lethality that is caused by increased NF-κB-mediated TNF production and increased cellular sensitivity to TNF killing (12, 24, 25). Most TRAF family members, including TRAF2, bear RING E3 ubiquitin ligase domains that are believed to catalyze Lys-63-linked ubiquitylation (26, 27). One of the proposed targets of TRAF2 RING E3 ligase activity is RIPK1 (28, 29), which is modified with Lys-63-linked polyubiquitin chains upon TNFR1 activation. This modified TNFR1-complexed RIPK1 has been proposed to serve as a platform for the recruitment and/or activation of TAB2-TAB3-TAK1 and NEMO-IKKα-IKKβ kinase complexes (30–32), which target the NF-κB inhibitor IκB for Lys-48-linked polyubiquitin-mediated proteasomal degradation, to initiate canonical p65-dependent NF-κB gene transcription.
As in the case of TRAF2, genetic deletion of cIAP1, or IAP antagonist-induced loss of cIAP1/2, also sensitizes cells to TNF killing (16, 19, 33–36). Also like TRAF2, both cIAP1 and cIAP2 have been reported to ubiquitylate RIPK1 (10, 15, 16). Consistent with the idea that cIAP1/2 play an important role in the activation of NF-κB by TNF, the removal of cIAP1/2 limits TNF from inducing NF-κB by the canonical pathway (10, 15, 16). Although it remains possible that both TRAF2 and cIAP1/2 play similar roles in modulating TNFR1 signaling through RIPK1 ubiquitylation, it is unknown whether cIAP1/2 act indirectly by activating the E3 ligase activity of TRAF2, if cIAP1/2 act in combination with TRAF2 to directly ubiquitylate RIPK1, or alternatively, whether cIAP1/2 are the major E3 ligases for RIPK1, and the role of TRAF2 is to recruit them to TNFR1.
Both TRAF2 or cIAP1/2 deletion cause constitutive noncanonical NF-κB activity, which occurs by stabilization of the NF-κB-inducing kinase NIK, IKKα activation, and processing of the noncanonical NF-κB subunit p100 to the active p52 form. Activation of noncanonical NF-κB can occur by proteasomal or lysosomal degradation of cIAP1, TRAF2, or TRAF3 and is mediated by ligand-induced activation of TNF superfamily receptors such as TNFR2, CD40, BAFFR, or FN14 (12, 13, 17, 20, 21, 37–44). On the basis of these recent studies (11, 12, 33), it appears that TRAF3 binding to NIK recruits the TRAF2-cIAP1/2 module and that cIAP1/2 mediate Lys-48-linked ubiquitylation of NIK, which reduces NIK to undetectable levels in resting cells and prevents spontaneous noncanonical NF-κB signals.
In this study we have identified a CIM in TRAF2, and we used TRAF2 CIM mutants defective in cIAP1/2 binding to determine how cIAP1/2 and TRAF2 coordinately suppress noncanonical NF-κB activation and regulate signaling from TNFR1.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, Constructs, and Lentiviral Infections
All cell lines were maintained at 37 °C, 10% CO2 in Dulbecco's modified Eagle's medium supplemented with 8% fetal bovine serum and were passaged twice weekly. Transient transfections using 1 μg of DNA per 10-cm tissue culture plate were performed with Effectene (Qiagen) using the manufacturer's protocols. The NF-κB lentiviral reporter vector pTRH1 mCMV NF-κB dscGFP was purchased from System Biosciences. Cre-recombinase and SV40 large T antigen were cloned into the lentiviral vector pFU. Mouse TRAF2 and the CIM and RING domain TRAF2 mutants were cloned into pEF-NFLAG and the 4-hydroxytamoxifen-inducible lentiviral vector pF 5× upstream activating sequence, which we have described in detail elsewhere (19, 45). Full-length cIAP1 and the BIR1 constructs were cloned into pEF-NHA. Complete sequence of all constructs can be obtained upon request. Lentiviral particles were generated as described previously (17, 19).
Antibodies
The primary antibodies used are as follows: anti-FLAG (F-3165, Sigma); anti-β-actin (A-1978, Sigma); anti-RIPK1 (mouse, 610458, BD Transduction Laboratories; human, 551042, Pharmingen); anti-p65 (SC-372, Santa Cruz Biotechnology); anti-phospho-Ser-65 (3033, Cell Signaling); anti-phospho-Ser-32/36 IkBα (9246, Cell Signaling); anti-IκBα (9242, Cell Signaling); anti-TNFR1 (19139, Abcam); anti-TRAF2 (558890, Pharmingen; SC-876, Santa Cruz Biotechnology; AP1040, Calbiochem); anti-cIAP1 (ALX-803-335, Alexis Biochemicals); anti-cIAP pan (MAB3400, RnD); anti-cIAP2 (in-house, 16E6-3); anti-TRADD (SC-7868, Santa Cruz Biotechnology); anti-NIK (4994, Cell Signaling); and anti-p100/p52 (4882, Cell Signaling).
Generation of MEFs
Knock-out MEFs were generated from embryonic day 15 embryos using standard procedures and infected with SV40 large T antigen expressing lentivirus. TRAF2 conditional knock-out MEFs were similarly generated from TRAF2 LoxP/LoxP embryonic day 15 embryos. To delete TRAF2, the transformed MEFs were infected with a cre-expressing lentivirus (pFU Cre SV40 Hygro), and deletion was confirmed by PCR and Western blotting.
Death Assays
Cells were seeded on 12-well tissue culture plates at ∼70% confluency and allowed to adhere. Compound A (500 nm), described in Ref. 19, or human Fc-TNFα (60 ng/ml) was added to cells for 24 h, and cell death was measured by propidium iodide (PI) staining and flow cytometry. In each sample 10,000 events were measured, and the cell death (% PI-positive cells) was quantified.
Western Blotting and Immunoprecipitations
Immunoprecipitations were performed as described previously (17, 19, 46) with some minor alterations. Briefly, 4 × 107 MEFs were treated in the presence or absence of human FLAG-TNF at 1 μg/ml for 5 min. Cells were lysed in IP-lysis buffer (30 mm Tris-HCl, pH 7.4, 120 mm NaCl, 2 mm EDTA, 2 mm KCl, 1% Triton X-100, complete protease/inhibitor mixture; Roche Applied Science) at 4 °C for 30 min. The lysates were centrifuged at 15,000 × g for 30 min; 0.5 μg of FLAG-TNF was added to the nonstimulated (time = 0) control, and the TNFR1 complex was precipitated using M2 beads (Sigma) for 16 h. The beads were washed five times with IP-lysis buffer and eluted with 2× LDS buffer (NuPAGE, Invitrogen). Proteins were separated by SDS-PAGE (NuPAGE, Invitrogen) and analyzed by Western blotting. Membranes were stripped with 50 mm glycine, pH 2.3, and reprobed with other antibodies.
Immunofluorescence
MEFs grown overnight on glass coverslips were treated with Fc-TNF (60 ng/ml) for the indicated times, immediately fixed with 3.2% paraformaldehyde for 20 min, washed in phosphate-buffered saline, and permeabilized with 0.5% Triton X-100 for 5 min. Cells were blocked, incubated with p65 antibody, washed four times with phosphate-buffered saline, and then incubated with anti-rabbit AlexaFluor 488-conjugated secondary antibody (Invitrogen) and washed four times again. All blocking steps and antibody incubations were performed with phosphate-buffered saline containing 1% bovine serum albumin for 30 min. Cells were viewed on a Olympus BX50 fluorescence microscope and SPOT RT imaging software using a ×40 objective.
RESULTS
Identification of a CIM in TRAF2
The cIAP1 BIR1 domain and several key residues within this region have been shown to mediate cIAP1 binding to TRAF2 (17, 22, 23). However, the region of TRAF2 required for cIAP1 and cIAP2 binding has not been clearly defined because yeast two-hybrid experiments suggested the TRAF-N domain was important (9). Our own yeast two-hybrid analysis suggested that a TRAF2-N domain-containing fragment, residues 262–349, is sufficient for cIAP1/2 binding (Fig. 1, A and C, and data not shown). To verify the ability of this region to bind cIAP1 in vivo, we performed co-immunoprecipitation experiments in 293T cells. Immunoprecipitation of full-length FLAG-TRAF2 confirmed that it interacts with the cIAP1 BIR1 domain and can also bind two BIR1 domain truncation mutants lacking the first 22 and 39 residues (Fig. 1B) (17, 22, 23). Furthermore, a FLAG-TRAF2-N (amino acids 262–349) construct was sufficient by itself to immunoprecipitate hemagglutinin-tagged full-length cIAP1 or the cIAP1 BIR1 domain alone (Fig. 1C). Experiments using purified cIAP1 BIR1 and TRAF2-N domains demonstrated that the binding between these two regions was direct (Fig. 2D).
FIGURE 1.
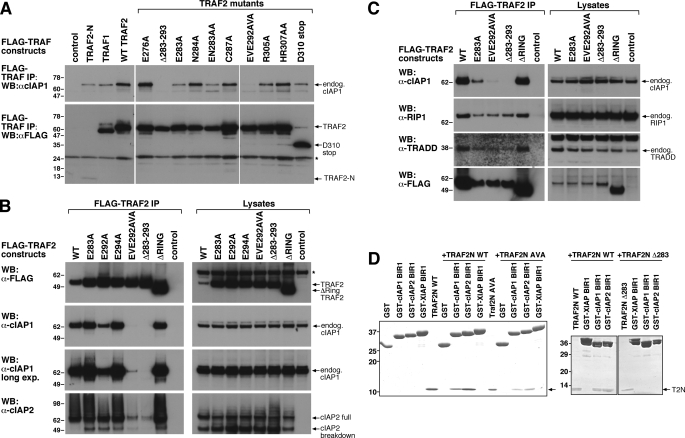
Identification of a TRAF2-CIM. A, schematic of TRAF2 domain structure and localization of the CIM. aa, amino acids. B and C, TRAF-N domain of TRAF2 interacts with the BIR1 domain of cIAP1. 293T cells were co-transfected with the indicated HA-cIAP1 constructs and FLAG-TRAF2 (B) or FLAG-TRAF2-N (C), and their interaction was examined by FLAG immunoprecipitation (IP) and Western blot (WB). m22 and m39 refer to BIR1 deletion constructs lacking the first 22 and 39 residues, respectively. D, conservation within the TRAF2-N domain. The TRAF-N domain of TRAF2 was aligned from the indicated organisms (accession numbers in parentheses) and compared with the same region of human TRAF1 (bottom line). Identical residues are shaded in black and conserved residues in gray. Residues mutated in TRAF2 to examine the effect on cIAP1/2 binding are highlighted with an asterisk, and the deletion (283–293) and truncation mutants (Asp-310-stop) are also indicated.
FIGURE 2.
TRAF2 residues 292EVE294 and 283–293 are important for cIAP1 and cIAP2 binding to TRAF2. A, determination of TRAF2 residues required for cIAP1 binding. 293T cells were transfected with the indicated FLAG-TRAF2 constructs for 48 h and then immunoprecipitated (IP) from cell lysates. Binding of endogenous (endog.) cIAP1 to FLAG-TRAF2 was determined by Western blot (WB). The asterisk indicates IgG light chain cross-reactivity. B, mutation of both TRAF2 Glu-292 and Glu-294 is required to reduce cIAP1 and cIAP2 binding. 293T cells were transfected with the indicated FLAG-TRAF2 constructs in addition to cIAP2. FLAG-TRAF2 complexes were immunoprecipitated, and TRAF2 binding to endogenous cIAP1 and ectopically expressed cIAP2 was determined by Western blot. Asterisk indicates a nonspecific band. C, FLAG-TRAF2 CIM mutants showing reduced cIAP1/2 binding interact with RIPK1 and TRADD. 293T cells were transfected with FLAG-TRAF2 constructs and immunoprecipitated as in A, and binding to RIPK1 and TRADD was determined by Western blot. D, TRAF2-N domain binds directly to the cIAP1 BIR1 domain. The indicated glutathione S-transferase proteins were incubated with purified WT or mutant TRAF2-N domain protein as indicated, and following glutathione S-transferase (GST) immunoprecipitation, binding of the TRAF2-N domain (T2N) was detected by Coomassie staining.
Although the crystal structure of truncated TRAF2 is available, only the C-terminal part of the TRAF-N domain is present (47, 48) making it difficult to identify the potential interaction interface between TRAF2-N and cIAP1 BIR1. We therefore examined the conservation of the TRAF2-N domain from a number of organisms to predict which residues might be important for the TRAF2-cIAP1 interaction (Fig. 1D). TRAF1 has also been shown to interact with cIAP1/2 (9, 49) and therefore was also included in the alignment. This analysis revealed a cluster of residues spanning amino acids 283–294 that were highly conserved across all TRAF2 species examined and also TRAF1 (Fig. 1D). Importantly, this region showed little conservation with TRAF3 or Drosophila TRAF2 (supplemental Fig. S1A and S1B), none of which bind cIAP proteins. We therefore created several alanine point substitutions between or near mouse TRAF2 residues 283–294, a TRAF2 deletion spanning residues 283–293, and a TRAF2 truncation by introducing a stop codon at residue Asp-310 (Fig. 1D).
TRAF2 Residues 283–293 Are Required for Binding to cIAP1 and cIAP2
To determine which residues of TRAF2 are important for binding cIAP1 and cIAP2, we transiently transfected the indicated FLAG-TRAF2 constructs (Figs. 1D and 2A) into 293T cells and examined their binding to endogenous cIAP1 and overexpressed cIAP2 by FLAG immunoprecipitation and Western blot. As before, TRAF2-N (amino acids 262–349) and wild type (WT) TRAF2 bound cIAP1, and as expected, TRAF1 also immunoprecipitated cIAP1. The TRAF2 deletion Δ283–293 did not bind detectable amounts of cIAP1 or cIAP2, and the double alanine point substitution EVE292AVA mutant showed drastically reduced binding to both cIAP1 and cIAP2 (Fig. 2, A–D). Single alanine point substitution of either Glu-292 or Glu-294 was less disruptive of cIAP1 and cIAP2 binding than mutation of both residues in the EVE292AVA mutant (Fig. 2B). TRAF2 E283A (and E283A/N283A) showed slightly reduced cIAP1 binding, but the other TRAF2 mutants, including the TRAF2 Asp-310 truncation mutant and a ΔRING mutant lacking the first 87 amino acids, all interacted with cIAP1 like wild type TRAF2 (Fig. 2, A and B). Similar decreases in TRAF2 mutant binding to cIAP1 were also observed in other cell types, such as D645 glioma cells (supplemental Fig. S1C). These mutations are unlikely to affect TRAF2 trimerization because the cIAP1/2-interacting motif was not present in the trimeric TRAF2 crystal structure (47, 48). Consistent with the idea that the TRAF2 mutations do not affect TRAF2 structure dramatically, the TRAF2 mutants with reduced binding to cIAP1/2 interacted with endogenous TRADD and RIPK1 to the same extent as the wild type protein (Fig. 2C) (50). Experiments using purified cIAP1 or cIAP2 BIR1 domain proteins with purified wild type, EVE292AVA, or Δ283–293 TRAF2-N domain proteins confirmed that BIR1 binding to TRAF2-N was direct and that mutant TRAF2-N binding was reduced (EVE292AVA) or abolished (Δ283–293) (Fig. 2D). The CIM of TRAF2 therefore spans residues 283–294 and mediates the direct interaction between the TRAF-N domain of TRAF2 and the BIR1 domain of cIAP1.
Functional TRAF2 CIM and RING Domains Are Required to Inhibit Noncanonical NF-κB
In unstimulated WT cells, TRAF3 binding to NIK recruits a TRAF2-cIAP1/2 complex to allow cIAP1/2-mediated NIK degradation and inhibition of spontaneous noncanonical NF-κB (11, 12, 33, 51). Deletion of TRAF2 or cIAP1 therefore results in constitutive noncanonical NF-κB signaling (Fig. 3, A and C) (17, 19, 52). Complementation of TRAF2 knock-out MEFs with the EVE292AVA TRAF2 mutant, which is still able to bind low amounts of cIAP1/2 (Fig. 2, B and D), was able to partially restore NF-κB activity in TRAF2 knock-out cells to the basal levels observed in WT cells (Fig. 3, B, and C), although the TRAF2 Δ283–293 CIM mutant completely failed to do so, as measured by increased NIK levels, p100 processing to p52, and NF-κB GFP reporter activity (Fig. 3, B and C). The TRAF2 RING domain mutant also failed to reduce TRAF2 knock-out NIK levels and p100 processing (Fig. 3C) despite the fact that it binds cIAP1 and cIAP2 (Fig. 2B). This shows that cIAP1/2-mediated Lys-48-linked ubiquitylation and degradation of NIK rely on TRAF2 as an adaptor protein and that the TRAF2 RING domain is also essential for the ability of cIAP1/2 to degrade NIK.
FIGURE 3.
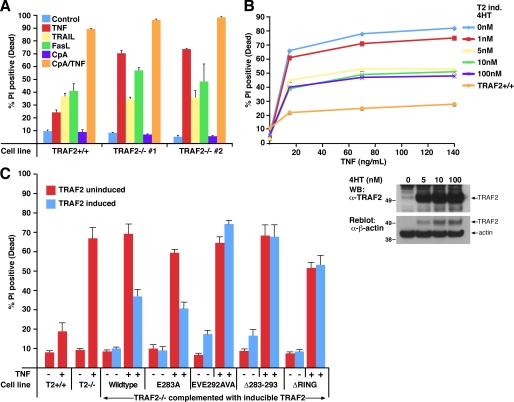
The TRAF2 CIM region 283–293 and RING domain are important for repression of constitutive noncanonical NF-κB activity caused by TRAF2 deletion. A, TRAF2 deletion results in constitutive NF-κB activity that is further enhanced by TNF stimulation. TRAF2 was deleted from parental TRAF2LoxP/LoxP conditional knock-out MEFs containing a lentiviral NF-κB GFP reporter by infection with a lentivirus harboring a Cre recombinase-expressing plasmid. TRAF2 parental WT and knock-out NF-κB GFP reporter cells were stimulated with TNF for 24 h, and reporter activity was measured by flow cytometry. Blue coloring, WT unstimulated cells; red coloring, TRAF2−/− unstimulated cells; green coloring, TNF-stimulated cells. B, WT but not CIM-mutated TRAF2 restores basal NF-κB GFP reporter activity in TRAF2−/− cells. TRAF2 expression in the TRAF2 conditional knock-out MEFs described in A was restored with the indicated WT and mutant constructs. Basal NF-κB reporter activity was measured by flow cytometry 72 h post-restoration of TRAF2 expression. Yellow coloring, parental WT MEFs; light blue coloring, TRAF2−/− MEFs; red coloring, uninduced TRAF2−/− cells reconstituted with the indicated inducible constructs; blue coloring, induced TRAF2−/− cells reconstituted with the indicated inducible constructs. The experiments shown are representative of results obtained on three independent occasions. C, TRAF2 deletion results in constitutive noncanonical NF-κB activity that is not reduced to normal levels by expression of TRAF2 CIM Δ283–293 or RING domain mutants. TRAF2−/− cells described in A were infected with the indicated inducible lentiviral TRAF2 constructs, and TRAF2 expression was induced in independent clones. 72 h post-induction, cells were harvested and analyzed by Western blot (WB) for the indicated proteins.
The TRAF2 CIM Is Required for Resistance to TNF-induced Cell Death
TRAF2 knock-out MEFs immortalized with the SV40 large T antigen are highly susceptible to cell death induced by TNF treatment alone (Fig. 4A) (17). These TRAF2 knock-out cells showed no significant difference in their ability to be killed by TNF-related apoptosis-inducing ligand and only a slight, potentially insignificant, difference in their ability to be killed by FasL (Fig. 4A), suggesting TRAF2 primarily provides protection against TNF-induced apoptosis. Similarly, SV40 large T immortalized WT MEFs that have been treated with the IAP antagonist compound A to remove cIAP1 and cIAP2 are killed by TNF treatment within 24 h (Fig. 4A) (19). Because cIAP1/2 are recruited by TRAF2, these data indicate that either cIAP1/2 or TRAF2, or both, confers protection against TNF killing but do not distinguish between these possibilities.
FIGURE 4.
TRAF2 CIM residues 292EVE294 and 283–293 are required for TRAF2-mediated protection against TNF-induced death. A, immortalized TRAF2−/− cells are sensitized to TNF, but not FasL or TNF-related apoptosis-inducing ligand, -induced apoptosis. SV40 large T immortalized TRAF2 conditional knock-out MEFs were infected with Cre recombinase to delete TRAF2. Parental TRAF2LoxP/LoxP and TRAF2−/− MEFs were treated with TNF (60 ng/ml), FasL (10 ng/ml), TNF-related apoptosis-inducing ligand (TRAIL) (1 μg/ml), or compound A (500 nm) as indicated for 24 h, and cells death was quantified by PI staining and flow cytometry. Error bars are S.E. of at least three independent experiments. B, restoration of TRAF2 expression in TRAF2−/− cells restores TNF resistance. TRAF2−/− MEFs were infected with a lentiviral construct containing 4-hydroxytamoxifen-inducible WT murine TRAF2. TRAF2 expression was induced by the indicated doses of 4-hydroxytamoxifen (4HT) (Western blot (WB), bottom), and cell resistance to TNF (60 ng/ml) killing after 24 h was assessed by PI staining and flow cytometry. C, TRAF2 292EVE294 and Δ283–293 mutants do not protect against TNF killing in TRAF2−/− MEFs. TRAF2−/− cells were infected with the indicated 4-hydroxytamoxifen-inducible lentiviral TRAF2 constructs, and TRAF2 expression was induced for 48 h (see Fig. 3C for TRAF2 expression levels). Cells were then treated with TNF (60 ng/ml) for a further 24 h, and cell death was compared with their uninduced counterparts by PI staining and flow cytometry. Error bars are S.E. of 3–5 independent experiments for several different clones.
Reintroduction of inducible WT TRAF2, or the E283A TRAF2 mutant that is able to bind cIAP1/2, into TRAF2 knock-out MEFs almost fully restored protection against TNF killing (Fig. 4, B and C). However, induction of the TRAF2 CIM Δ283–293, EVE292AVA, or RING domain mutants in TRAF2 knock-out MEFs failed to prevent TNF-induced cell death (Fig. 4C). These data demonstrate that cIAP1/2 binding to TRAF2 is crucial to prevent apoptosis induced by TNF and suggest that any interaction cIAP1/2 may have with RIPK1 (14) is not sufficient for TNF resistance in the absence of their binding TRAF2. Significantly, these results indicate that the RING domain of TRAF2 is also required for TNF resistance (Fig. 4C), even though this construct retains the ability to bind both cIAP1 and cIAP2 (Fig. 1B). Therefore, both TRAF2 and cIAP1/2 play important roles in protecting cells from TNF-induced death, and they must do so bound to each other.
TNF-induced RIPK1 Modification Is Dependent on a Functional TRAF2-cIAP1/2 Interaction, but the TRAF2 RING Domain Is Dispensable
Recent work has suggested that cIAP1/2 act as the E3 ubiquitin ligases to ubiquitylate RIPK1 following TNFR1 activation (14–16). However, it has also been proposed that the E3 ubiquitin ligase activity of TRAF2 mediates ubiquitylation of RIPK1 following TNFR1 signaling (28, 29). Combined deletion of TRAF2 and TRAF5 (TRAF2/TRAF5 DKO) significantly prevented RIPK1 ubiquitylation in response to TNF stimulation and dramatically reduced cIAP1/2 binding to TNFR1 (Fig. 5). Thus, the phenotype of the TRAF2/TRAF5 knock-out cells is consistent with both models and cannot distinguish between either. Restoration of WT TRAF2 expression in TRAF2/TRAF5 DKO MEFs restored RIPK1 ubiquitylation and cIAP1/2 recruitment to TNFR1, as expected (Fig. 5). However, TRAF2/5 DKO cells reconstituted with the TRAF2 CIM Δ283–293 mutant did not recruit cIAP1 and did not restore RIPK1 ubiquitylation in response to TNF (Fig. 5). Reintroduction of TRAF2 ΔRING, which is still able to bind cIAPs (Fig. 2) but lacks E3 ligase activity, also restored RIPK1 ubiquitylation and cIAP1/2 recruitment (Fig. 5). These data are consistent with the idea that cIAP1/2 are the most important E3 ligases for RIPK1, and the data fit the model whereby if TRAF2 does not recruit cIAP1/2 then RIPK1 fails to undergo TNF-induced ubiquitylation and associates with FADD and caspase-8 to induce cell death. Intriguingly, although TNF-induced RIPK1 ubiquitylation (Fig. 5) and NF-κB function (see below) are retained upon loss of the TRAF2 RING domain, these cells are still susceptible to TNF killing (Fig. 4C).
FIGURE 5.
TRAF2 CIM is important for TNF-induced RIPK1 ubiquitylation, but the TRAF2 RING domain is dispensable. The indicated MEFs were stimulated with FLAG-TNF (1 μg/ml) for 5 min, and the TNFR1 signaling complex precipitated with anti-FLAG antibody was described under “Experimental Procedures.” The eluted TNFR1 protein complex was examined by Western blot (WB) for the indicated proteins. To precipitate unstimulated TNFR1 (time = 0), FLAG-TNF was added to cell lysates. IP, immunoprecipitation.
The TRAF2 CIM, but Not the TRAF2 RING Domain, Is Required for Efficient TNF-induced NF-κB Activation
As described previously (24, 53), TNF-induced NF-κB activation in TRAF2 knock-out cells was similar to wild type cells, as determined by NF-κB reporter assays and Western blotting for p65 phosphorylation and IκBα (Figs. 3A and 6A). Interestingly, we consistently observed that cIAP1−/−, TRAF2−/−, and TRAF2/TRAF5 DKO MEFs that have been immortalized with the SV40 large T antigen have increased levels of basal phosphorylated p65 when compared with WT cells (Fig. 6 A and B) (17, 19). However, this does not correlate with increased basal nuclear p65 levels in any of these cell lines (Figs. 6, C and D) (17).
FIGURE 6.
TRAF2 CIM is important for efficient TNF-induced NF-κB activation, but the TRAF2 RING domain is dispensable. A, TNF induction of NF-κB in TRAF2 knock-out MEFs. WT and TRAF2−/− cells were treated with TNF (60 ng/ml) for the indicated times, and cell lysates were analyzed by Western blot (WB). B, restoration of the normal NF-κB response to TNF in TRAF2/TRAF5 double knock-out MEFs requires the TRAF2 CIM but not the TRAF2 RING domain. The indicated cell types were treated with TNF (60 ng/ml) for the time periods shown, and cell lysates were analyzed by Western blot. C, TNF-induced translocation of p65 into the nucleus requires the TRAF2 CIM but not the TRAF2 RING domain. The indicated MEF cell lines were stimulated with TNF (60 ng/ml) for 20 min, and p65 localization was analyzed by immunostaining and fluorescence microscopy. DKO, TRAF2/TRAF5 double knock-out. D, delayed p65 nuclear translocation in TRAF2/TRAF5 double knock-out MEFs is prevented by IAP antagonist depletion of cIAP1/2. WT and TRAF2/TRAF5 double knock-out MEFs were treated with 60 ng/ml TNF for the indicated times with or without the IAP antagonist, compound A, and p65 localization was examined by immunofluorescence microscopy.
TRAF2 and TRAF5 are redundant with respect to TNF-induced activation of NF-κB, because deletion of both genes is required to prevent it (54). To examine the role of the TRAF2-cIAP1/2 interaction in TNF induction of NF-κB, we therefore complemented TRAF2/TRAF5 DKO MEFs with TRAF2 WT, CIM mutant, and RING domain deletion constructs. TRAF2/TRAF5 DKO cells were significantly impaired in TNF-induced p65 phosphorylation and IκBα degradation and translocation of p65 to the nucleus (Fig. 6, B and C), and complementation with WT TRAF2 restored all these NF-κB-associated functions (Fig. 6, B and C). Although the TRAF2 CIM mutant EVE292AVA, which retains weak cIAP1/2 binding (Fig. 2, B and D), partially restored TNF-induced NF-κB function (Fig. 6. B and C), the TRAF2 Δ283–293 CIM mutant was unable to restore either IκBα degradation or nuclear translocation of p65 in response to TNF (Fig. 6, B and C).
It has recently been suggested that TNF-induced Lys-63-linked ubiquitylation of TRAF2 on lysine 31 regulates binding of TAB2/TAB3 to TRAF2 and is required for proper TNF-induced IKK and NF-κB activity (55). If this model is correct, then complementation of TRAF2/TRAF5 DKO cells with a ΔRING TRAF2 mutant, which lacks residues 1–87, should fail to restore TNF-induced NF-κB function. However, we observed that TRAF2/TRAF5 DKO MEFs complemented with ΔRING TRAF2 restored TNF-induced p65 phosphorylation, IκB degradation, and translocation of p65 to the nucleus to a similar level as observed when TRAF2/TRAF5 DKO MEFs were complemented with WT TRAF2 (Fig. 6, B and C).
Although rapid TNF-induced nuclear translocation of p65 is mostly lost in TRAF2/TRAF5 DKO MEFs when compared with WT cells, some p65 nuclear localization could be detected after 60 min of TNF stimulation (Fig. 6D). Because immunoprecipitated TNFR1 from TRAF2/TRAF5 DKO cells still bound low levels of cIAP1/2 (Fig. 5), we speculated that the late nuclear translocation of p65 in TRAF2/TRAF5 DKO cells observed by immunofluorescence could be caused cIAP1/2 function. Consistent with this idea, when we depleted cIAP1/2 from TRAF2/TRAF5 DKO cells using the IAP antagonist compound A (19), p65 translocation to the nucleus failed to occur, even 60 min after TNF stimulation (Fig. 6D).
Deletion of TRAF2 Results in Enhanced TNF-induced c-FLIP Degradation
Previous work has demonstrated that in the absence of NF-κB signaling in TRAF2/TRAF5 DKO or p65 knock-out cells, the lack of TNF-induced c-FLIPL expression sensitizes to TNFR1 killing (56). Although it has been proposed that TRAF2 knock-out cells are also sensitive to TNF due to reduced basal levels of c-FLIPL (57), our Western blots suggested that both unstimulated TRAF2−/− and TRAF2/TRAF5 DKO MEFs contain almost equivalent levels of c-FLIPL when compared with WT cells (Fig. 7). However, when stimulated with TNF, both TRAF2−/− and TRAF2/TRAF5 DKO cells displayed a dramatic loss of c-FLIPL when compared with WT cells (Fig. 7). Hence, despite the normal NF-κB activation in TRAF2−/− MEFs, c-FLIPL is still efficiently depleted upon TNF stimulation, and this probably accounts, at least partially, for their increased sensitivity to TNFR1-induced death.
FIGURE 7.
Deletion of TRAF2 increases c-FLIPL loss upon TNF stimulation. The indicated MEF cell lines were stimulated with TNF for 0, 1, or 2 h, and cell lysates were examined by Western blot (WB) for the proteins shown.
DISCUSSION
Our study identifying and characterizing the role of the cIAP1/2-interacting motif of TRAF2 reveals that a key role of TRAF2 is in the recruitment of cIAP1/2 into signaling platforms. One of these is the NIK complex that is essential for noncanonical NF-κB activation. TRAF2 is a key molecule in the regulation of NIK stability as loss of TRAF2 results in NIK stabilization, activation, and constitutive noncanonical NF-κB signaling (17, 52, 58). Using TRAF2 CIM and RING mutants, we complemented TRAF2 knock-out cells and found that restoration of the normal low level of NIK and reduction of spontaneous NF-κB activity required both the TRAF2 CIM and RING domains. These findings are consistent with recent reports showing that repression of noncanonical NF-κB activity requires a complex of TRAF3, TRAF2, and cIAP1/2, with each component performing a distinct function (11, 12, 19, 33). TRAF3 interacts with NIK and also binds the TRAF2-cIAP1/2 module, thereby bringing cIAP1/2 into the proximity of NIK, resulting in cIAP1/2-mediated Lys-48-linked ubiquitylation of NIK and its proteasomal degradation (11, 12, 33). These studies and our work here demonstrate that in addition to cIAP1/2, both the TRAF2 and TRAF3 RING domains are required for NIK degradation.
Our data also show that TNF-induced nuclear translocation of the canonical NF-κB subunit p65 is significantly delayed in TRAF2/TRAF5 DKO cells, which agrees with previously reported NF-κB electrophoretic mobility gel shift assays and in vitro IKK activity assays performed on TRAF2/TRAF5 DKO cells (54, 55). cIAP1/2 have also been implicated in canonical NF-κB signaling following TNFR1 activation, and both cIAP1/2 and TRAF2 have been suggested to be the E3 ligases required for ubiquitylation of RIPK1 (10, 14–16, 28, 29). Ubiquitylated RIPK1 is believed to be the key signaling platform required for correct TNFR1-induced NF-κB, and a TRAF2 CIM mutant was unable to restore either RIPK1 ubiquitylation or normal NF-κB signaling in response to TNF. Recently, however, we have shown that RIPK1 knock-out MEFs are able to induce NF-κB in response to TNF.8 We therefore believe that RIPK1 ubiquitylation serves as a marker for TNF-induced activation of NF-κB but is not a requirement. Although our work here and recent studies therefore suggest a role for cIAP1/2 in normal canonical NF-κB responses initiated by TNF stimulation (10, 15, 16), it remains unclear what their key targets might be given that RIPK1 itself is dispensable for TNF-induced NF-κB.
The ubiquitylation of RIPK1 by cIAP1/2 has been shown to prevent it associating with FADD and caspase-8, thereby limiting TNFR1-induced cell death (10, 15, 16). Consistent with this model, cIAP1 knock-out MEFs have increased RIPK1 recruitment to TNFR1 and increased sensitivity to TNF killing compared with wild type MEFs (10, 19). Similarly, IAP antagonist treatment that depletes cIAP1/2 causes a loss of RIPK1 modification following TNF stimulation (10, 16). Following TNFR1 activation, we observed increased amounts of RIPK1 bound to TNFR1 and decreased RIPK1 ubiquitylation in TRAF2/TRAF5 DKO cells. TRAF2/TRAF5 DKO cells reconstituted with either WT TRAF2 or a TRAF2 ΔRING mutant, but not a TRAF2 CIM mutant, regained RIPK1 ubiquitylation. These results show that TRAF2-dependent TNF-induced RIPK1 ubiquitylation requires an intact TRAF2-cIAP1/2 interaction. Combined with the fact that the TRAF2 RING domain was dispensable for RIPK1 ubiquitylation, our data support the model that cIAP1/2 are the critical E3 ligases required for RIPK1 ubiquitylation.
Although we show that TRAF2 must recruit cIAP1/2 to properly activate NF-κB, TRAF2 is not just a passive adaptor, because the RING domain of TRAF2 is required for imparting resistance to induction of apoptosis by TNF. Our results with TRAF2−/− cells and those reconstituted with a TRAF2 ΔRING mutant demonstrate an apparently normal NF-κB induction combined with sensitivity to TNF killing. Although decreased cIAP1/2 recruitment into the TNFR1 signaling complex in TRAF2−/− MEFs may lead to less RIPK1 ubiquitylation and theoretically promote TNF-induced cell death, our data show that TRAF2 ΔRING restores cIAP1/2 binding to TNFR1 and RIPK1 ubiquitylation and yet fails to restore TNF resistance. The almost normal canonical NF-κB response on the background of increased noncanonical NF-κB signaling in TRAF2−/− MEFs (see Ref. 17 and this work) may render them sensitive to TNF, but this remains to be tested. Alternatively, an enhancement in c-FLIPL degradation may render TRAF2−/− cells more susceptible to TNFR1-induced death. To this end, we observed a dramatic decrease in c-FLIPL levels in TRAF2−/− cells upon TNF stimulation, which is likely to contribute to their enhanced TNF sensitivity. Although the data fits current models where active caspase-8 has been shown efficiently cleave c-FLIPL (59), it remains to be determined how c-FLIPL levels are regulated by the presence of TRAF2.
Our results define a TRAF2-c1AP1/2-interacting motif and provide independent support for recent studies with IAP antagonist compounds that demonstrate a fundamental role for cIAP1/2 in regulating TNF signaling. Reconstitution of TRAF2 and TRAF2/TRAF5 DKO cells with a TRAF2 ΔRING mutant has, however, raised new questions. This mutant was able to recapitulate the ability of wild type TRAF2 to activate canonical NF-κB in response to TNF in TRAF2/TRAF5 DKO cells, but it was unable to protect TRAF2 knock-out cells from the cytotoxic activity of TNF. It will therefore be interesting to determine what is the additional cytoprotective function of TRAF2.
Acknowledgments
We thank Hiroyasu Nakano for TRAF2/TRAF5 double knock-out MEFs; David Baltimore for pFU and lentiviral packaging constructs; Jurg Tschopp for the generous gifts of materials; and Robert Brink (Garvan Institute) for critically reading the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
L. Wong and J. Silke, submitted for publication.
- IAP
- inhibitor of apoptosis
- NF-κB
- nuclear factor-κ-light-chain-enhancer of activated B cells
- TNF
- tumor necrosis factor
- TNFR1
- tumor necrosis factor receptor-1
- TRAF
- TNF receptor-associated factor
- RIPK1
- receptor-interacting protein-1
- RING
- really interesting new gene
- TAK1
- TGF-β-activated kinase-1
- WT
- wild type
- GFP
- green fluorescent protein
- MEF
- mouse embryo fibroblast
- PI
- propidium iodide
- DKO
- double knock-out
- CIM
- cIAP1/2-interacting motif
- NIK
- NF-κB-inducing kinase.
REFERENCES
- 1.Vaux D. L., Silke J. (2005) Nat. Rev. Mol. Cell Biol. 6, 287–297 [DOI] [PubMed] [Google Scholar]
- 2.Imoto I., Yang Z. Q., Pimkhaokham A., Tsuda H., Shimada Y., Imamura M., Ohki M., Inazawa J. (2001) Cancer Res. 61, 6629–6634 [PubMed] [Google Scholar]
- 3.Tanimoto T., Tsuda H., Imazeki N., Ohno Y., Imoto I., Inazawa J., Matsubara O. (2005) Cancer Lett. 224, 141–151 [DOI] [PubMed] [Google Scholar]
- 4.Dai Z., Zhu W. G., Morrison C. D., Brena R. M., Smiraglia D. J., Raval A., Wu Y. Z., Rush L. J., Ross P., Molina J. R., Otterson G. A., Plass C. (2003) Hum. Mol. Genet. 12, 791–801 [DOI] [PubMed] [Google Scholar]
- 5.Imoto I., Tsuda H., Hirasawa A., Miura M., Sakamoto M., Hirohashi S., Inazawa J. (2002) Cancer Res. 62, 4860–4866 [PubMed] [Google Scholar]
- 6.Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S. T., Luk J. M., Wigler M., Hannon G. J., Mu D., Lucito R., Powers S., Lowe S. W. (2006) Cell 125, 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keats J. J., Fonseca R., Chesi M., Schop R., Baker A., Chng W. J., Van Wier S., Tiedemann R., Shi C. X., Sebag M., Braggio E., Henry T., Zhu Y. X., Fogle H., Price-Troska T., Ahmann G., Mancini C., Brents L. A., Kumar S., Greipp P., Dispenzieri A., Bryant B., Mulligan G., Bruhn L., Barrett M., Valdez R., Trent J., Stewart A. K., Carpten J., Bergsagel P. L. (2007) Cancer Cell 12, 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Annunziata C. M., Davis R. E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., Dave S., Hurt E. M., Tan B., Zhao H., Stephens O., Santra M., Williams D. R., Dang L., Barlogie B., Shaughnessy J. D., Jr., Kuehl W. M., Staudt L. M. (2007) Cancer Cell 12, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothe M., Pan M. G., Henzel W. J., Ayres T. M., Goeddel D. V. (1995) Cell 83, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 10.Varfolomeev E., Goncharov T., Fedorova A. V., Dynek J. N., Zobel K., Deshayes K., Fairbrother W. J., Vucic D. (2008) J. Biol. Chem. 283, 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W., Korneluk R. G., Cheng G. (2008) Nat. Immunol. 9, 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P. H., Keats J. J., Wang H., Vignali D. A., Bergsagel P. L., Karin M. (2008) Nat. Immunol. 9, 1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotin-Mleczek M., Henkler F., Samel D., Reichwein M., Hausser A., Parmryd I., Scheurich P., Schmid J. A., Wajant H. (2002) J. Cell Sci. 115, 2757–2770 [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Du F., Wang X. (2008) Cell 133, 693–703 [DOI] [PubMed] [Google Scholar]
- 15.Mahoney D. J., Cheung H. H., Mrad R. L., Plenchette S., Simard C., Enwere E., Arora V., Mak T. W., Lacasse E. C., Waring J., Korneluk R. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 17.Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., Yeoh G., Brink R., Vaux D. L., Silke J. (2008) J. Cell Biol. 182, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y., Conze D. B., Hanover J. A., Ashwell J. D. (2007) J. Biol. Chem. 282, 7777–7782 [DOI] [PubMed] [Google Scholar]
- 19.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 20.Wu C. J., Conze D. B., Li X., Ying S. X., Hanover J. A., Ashwell J. D. (2005) EMBO J. 24, 1886–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X., Yang Y., Ashwell J. D. (2002) Nature 416, 345–347 [DOI] [PubMed] [Google Scholar]
- 22.Varfolomeev E., Wayson S. M., Dixit V. M., Fairbrother W. J., Vucic D. (2006) J. Biol. Chem. 281, 29022–29029 [DOI] [PubMed] [Google Scholar]
- 23.Samuel T., Welsh K., Lober T., Togo S. H., Zapata J. M., Reed J. C. (2006) J. Biol. Chem. 281, 1080–1090 [DOI] [PubMed] [Google Scholar]
- 24.Yeh W. C., Shahinian A., Speiser D., Kraunus J., Billia F., Wakeham A., de la Pompa J. L., Ferrick D., Hum B., Iscove N., Ohashi P., Rothe M., Goeddel D. V., Mak T. W. (1997) Immunity 7, 715–725 [DOI] [PubMed] [Google Scholar]
- 25.Nguyen L. T., Duncan G. S., Mirtsos C., Ng M., Speiser D. E., Shahinian A., Marino M. W., Mak T. W., Ohashi P. S., Yeh W. C. (1999) Immunity 11, 379–389 [DOI] [PubMed] [Google Scholar]
- 26.Pineda G., Ea C. K., Chen Z. J. (2007) Adv. Exp. Med. Biol. 597, 80–92 [DOI] [PubMed] [Google Scholar]
- 27.Shi C. S., Kehrl J. H. (2003) J. Biol. Chem. 278, 15429–15434 [DOI] [PubMed] [Google Scholar]
- 28.Lee T. H., Shank J., Cusson N., Kelliher M. A. (2004) J. Biol. Chem. 279, 33185–33191 [DOI] [PubMed] [Google Scholar]
- 29.Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 30.Wu C. J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. (2006) Nat. Cell Biol. 8, 398–406 [DOI] [PubMed] [Google Scholar]
- 31.Ea C. K., Deng L., Xia Z. P., Pineda G., Chen Z. J. (2006) Mol. Cell 22, 245–257 [DOI] [PubMed] [Google Scholar]
- 32.Li H., Kobayashi M., Blonska M., You Y., Lin X. (2006) J. Biol. Chem. 281, 13636–13643 [DOI] [PubMed] [Google Scholar]
- 33.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 34.Petersen S. L., Wang L., Yalcin-Chin A., Li L., Peyton M., Minna J., Harran P., Wang X. (2007) Cancer Cell 12, 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaither A., Porter D., Yao Y., Borawski J., Yang G., Donovan J., Sage D., Slisz J., Tran M., Straub C., Ramsey T., Iourgenko V., Huang A., Chen Y., Schlegel R., Labow M., Fawell S., Sellers W. R., Zawel L. (2007) Cancer Res. 67, 11493–11498 [DOI] [PubMed] [Google Scholar]
- 36.Li L., Thomas R. M., Suzuki H., De Brabander J. K., Wang X., Harran P. G. (2004) Science 305, 1471–1474 [DOI] [PubMed] [Google Scholar]
- 37.Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004) J. Biol. Chem. 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- 38.Hostager B. S., Catlett I. M., Bishop G. A. (2000) J. Biol. Chem. 275, 15392–15398 [DOI] [PubMed] [Google Scholar]
- 39.Chan F. K., Lenardo M. J. (2000) Eur. J. Immunol. 30, 652–660 [DOI] [PubMed] [Google Scholar]
- 40.Arch R. H., Gedrich R. W., Thompson C. B. (2000) Biochem. Biophys. Res. Commun. 272, 936–945 [DOI] [PubMed] [Google Scholar]
- 41.Duckett C. S., Thompson C. B. (1997) Genes Dev. 11, 2810–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown K. D., Hostager B. S., Bishop G. A. (2001) J. Exp. Med. 193, 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupoux A., Cartier J., Cathelin S., Filomenko R., Solary E., Dubrez-Daloz L. (2009) Blood 113, 175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L., Soetandyo N., Wang Q., Ye Y. (2008) Biochim. Biophys. Acta 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callus B. A., Ekert P. G., Heraud J. E., Jabbour A. M., Kotevski A., Vince J. E., Silke J., Vaux D. L. (2008) Cell Death Differ 15, 213–215 [DOI] [PubMed] [Google Scholar]
- 46.Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park Y. C., Burkitt V., Villa A. R., Tong L., Wu H. (1999) Nature 398, 533–538 [DOI] [PubMed] [Google Scholar]
- 48.Ye H., Park Y. C., Kreishman M., Kieff E., Wu H. (1999) Mol. Cell 4, 321–330 [DOI] [PubMed] [Google Scholar]
- 49.Uren A. G., Pakusch M., Hawkins C. J., Puls K. L., Vaux D. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4974–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu H., Huang J., Shu H. B., Baichwal V., Goeddel D. V. (1996) Immunity 4, 387–396 [DOI] [PubMed] [Google Scholar]
- 51.Silke J., Brink R. (2009) Cell Death Differ., in press [DOI] [PubMed] [Google Scholar]
- 52.Grech A. P., Amesbury M., Chan T., Gardam S., Basten A., Brink R. (2004) Immunity 21, 629–642 [DOI] [PubMed] [Google Scholar]
- 53.Lee S. Y., Reichlin A., Santana A., Sokol K. A., Nussenzweig M. C., Choi Y. (1997) Immunity 7, 703–713 [DOI] [PubMed] [Google Scholar]
- 54.Tada K., Okazaki T., Sakon S., Kobarai T., Kurosawa K., Yamaoka S., Hashimoto H., Mak T. W., Yagita H., Okumura K., Yeh W. C., Nakano H. (2001) J. Biol. Chem. 276, 36530–36534 [DOI] [PubMed] [Google Scholar]
- 55.Li S., Wang L., Dorf M. E. (2009) Mol. Cell 33, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakajima A., Komazawa-Sakon S., Takekawa M., Sasazuki T., Yeh W. C., Yagita H., Okumura K., Nakano H. (2006) EMBO J. 25, 5549–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guiet C., Silvestri E., De Smaele E., Franzoso G., Vito P. (2002) Cell Death Differ. 9, 138–144 [DOI] [PubMed] [Google Scholar]
- 58.Gardam S., Sierro F., Basten A., Mackay F., Brink R. (2008) Immunity 28, 391–401 [DOI] [PubMed] [Google Scholar]
- 59.Hughes M. A., Harper N., Butterworth M., Cain K., Cohen G. M., MacFarlane M. (2009) Mol. Cell 35, 265–279 [DOI] [PubMed] [Google Scholar]