Abstract
Members of the plexin family are unique transmembrane receptors in that they interact directly with Rho family small GTPases; moreover, they contain a GTPase-activating protein (GAP) domain for R-Ras, which is crucial for plexin-mediated regulation of cell motility. However, the functional role and structural basis of the interactions between the different intracellular domains of plexins remained unclear. Here we present the 2.4 Å crystal structure of the complete intracellular region of human plexin-B1. The structure is monomeric and reveals that the GAP domain is folded into one structure from two segments, separated by the Rho GTPase binding domain (RBD). The RBD is not dimerized, as observed previously. Instead, binding of a conserved loop region appears to compete with dimerization and anchors the RBD to the GAP domain. Cell-based assays on mutant proteins confirm the functional importance of this coupling loop. Molecular modeling based on structural homology to p120GAP·H-Ras suggests that Ras GTPases can bind to the plexin GAP region. Experimentally, we show that the monomeric intracellular plexin-B1 binds R-Ras but not H-Ras. These findings suggest that the monomeric form of the intracellular region is primed for GAP activity and extend a model for plexin activation.
Introduction
Plexins are single transmembrane receptors for guidance cues, called semaphorins, which regulate the motility and positional maintenance of certain cells. With this function, the receptors play critical roles in many developmental processes, including axon guidance, angiogenesis, and bone formation (1, 2). Moreover, plexins and their ligands are also involved in the regulation of the immune response, in cancer progression, and are thought to restrain tissue regeneration after injury (3, 4).
Plexins are unusual receptors in that they interact directly with Rho and Ras family small GTPases (5–7). An intracellular region that has high homology to Ras GTPase-activating proteins (GAPs)7 facilitates the hydrolysis of R-Ras-bound GTP. This deactivation of R-Ras leads to functional inhibition of integrins and to a loss of cell adhesion in response to semaphorins (5–8). Interestingly, no GAP activity of plexin-B1 was detected toward the R-Ras-homologous H-Ras (5), suggesting greater substrate specificity compared with the GAP protein p120GAP (9). How the plexin receptor is activated and specifically how the GAP function is regulated have been questions of considerable interest (10–12). A number of studies have pointed to a sequence segment that interrupts the GAP-homologous region and is capable of binding small Rho family GTPases. In the case of plexin-B1, this Rho GTPase binding domain (RBD) can associate with Rnd1, Rac1, and RhoD, which are thought to regulate plexin function. Specifically, in vitro studies in a number of laboratories have used the intracellular region of plexins expressed as two fragments, named C1 (containing the RBD and an N-terminal GAP-homologous segment) and C2 (C-terminal GAP segment). The studies suggest that such fragments are loosely associated. Moreover, the interaction between the RBD and Rnd1 or Rac1 appears to separate the two fragments (5–8, 13).
Structural biology has had a tremendous impact on our understanding of GTPase function and regulation (e.g. see Ref. 14). Representative structures for all of the major families of small GTPase-activating proteins are known, and also by using mutagenesis, the catalytic residues involved have been identified (15). However, the GAP domain is often surrounded by other protein segments that are known to participate in cell signaling events, such as an SH2 domain in chimerins (16), C2 in SynGAP (17), and a pleckstrin homology/lipid binding domain in p120GAP (18). Our understanding of how GAP activity is controlled is still limited, because not many structures that include regulatory domains have been determined to date.
Characterizing the structure of the intracellular region of human plexin-B1 promises to elucidate the mechanism by which the RBD can control receptor signaling and the function of the GAP domain. The NMR solution conformation (19, 20) and x-ray structure of the RBD of human plexin-B1 show that this domain forms a dimeric ubiquitin-like structure (21). GTPase association with the RBD domain occurs at a common interface that is adjacent to the dimerization region. These observations combined with biophysical studies suggest that Rho GTPase binding can destabilize a dimeric form of the intracellular region of plexins. On the extracellular side, it has been proposed from the dimeric crystal structure of semaphorin-3A that ligand binding to the semaphorin-homologous region of plexin would cause a conformational rearrangement in the dimeric form of the receptor (22). It is also known that Rac1 binding to the cytoplasmic plexin-B1 RBD increases ligand binding on the cell surface (23). Together, these studies led to the refinement of a model for plexin activation that involves the destabilization of a RBD-mediated intracellular region dimer and explained the observed synergy between ligand binding and GTPase-dependent regulation of these receptors (21).
Here, we present the 2.4 Å x-ray structure of the entire intracellular region of plexin-B1 (residues 1511–2135). The role of the different domains is investigated by a combination of biophysical, computational, and functional studies. The protein is monomeric and has a single GAP domain fold with the RBD placed on its side. A detailed comparison also with the structure of the isolated dimeric RBD region and with the RBD bound to Rnd1 suggests that intradomain conformational changes induced by Rho GTPase binding are small in this system. Furthermore, the structures of the RBD dimers provide a model for a dimeric intracellular structure. This model is incompatible with Rho GTPase binding, thus supporting the role of these interactions for interdomain changes. Functional studies in cells confirm the importance of a newly discovered protein segment (the “coupling loop”) that is designed to oppose dimerization of the RBD region as part of the receptor activation mechanism. The plexin GAP fold has high structural similarity to that of p120GAP. Our data suggest that the intracellular domain of plexin-B1, when in the monomeric state, is primed for GAP function even in the absence of Rho GTPases, leading to a critical extension of the current model for plexin activation.
EXPERIMENTAL PROCEDURES
Protein Constructs and Expression
The cDNA encoding the intracellular region of human plexin-B1 used for crystallization and in vitro studies (residues 1511–2135) was a gift from Dr. Manabu Negishi (Kyoto University). The DNA was PCR-amplified and inserted into pET19b vector (Invitrogen) using T4 ligase and into pFBOH-LIC vector (GenBankTM GI:134105591) using the In-Fusion kit (Clontech). His6-tagged recombinant plexin-B1 was expressed in Spodoptera frugiperda cells via baculovirus infection. Following cell lysis, proteins were purified using nickel-nitrilotriacetic acid resin in 50 mm Tris, pH 8.2, 500 mm NaCl, 5% glycerol, and 5 mm imidazole, and eluted with the same buffer containing 250 mm imidazole. Protein samples were further purified using a Superdex-200 column equilibrated with 50 mm Tris, pH 8.5, 500 mm NaCl, 4 mm MgCl2, 5% glycerol, and 1 mm TCEP. Further experimental details are available on the SGC web site. The plexin-B1 RBD, small Rho GTPases Rac1 (wild type), Rac1 Q61L, and Rnd1 were purified as described previously (20). H- and R-Ras were subcloned into pET19b and mutated at positions 12 (Gly → Val) and 87 (Gln → Leu), respectively, to generate constitutively active proteins. Rnd1, Rac1 Q61L, and R-Ras Q87L binding to the intracellular region of wild type plexin-B1 could be shown by gel filtration and by isothermal titration calorimetry. However, the doubly and triply mutated plexin-B1 proteins could not be obtained in sufficient amounts to carry out the same experiments.
Crystallization and Crystal Structure Determination
Crystals of the plexin-B1 intracellular region were grown in 16% polyethylene glycol 2000 MME, 0.1 m Tris, pH 8.0, 0.2 m trimethylamine N-oxide in a hanging drop setup at a 1 μl of protein (0.1 mm)/1-μl well solution ratio. To provide phase information, a mercury derivative was made by soaking the plexin-B1 crystal in the well solution supplemented with 1 mm thiomersal for 6 h before flash freezing in liquid nitrogen. For crystallization of the RBD·Rnd1 complex, His tag-removed plexin-B1 RBD and Rnd1 GTPase were first mixed in a 1:1 molarity ratio in 50 mm Tris buffer (pH 7.5) containing 150 mm NaCl, 5 mm DTT, and 5 mm MgCl2. 0.5 μl of protein complex (0.4 mm) was then mixed with 0.5 μl of well solution containing 20% polyethylene glycol 3350, 0.2 m CaCl2. Crystallization was set up using sitting drop vaporization against the well solution. All crystals grew to a mountable size in 2–3 days. Further experimental details are available on the SGC web site. Diffraction data were collected at the synchrotron and are detailed in Table 1. Data were processed with the HKL2000 suite (24). The structure of the intracellular region of plexin-B1 was solved by single wavelength anomalous diffraction, as implemented in SOLVE. Initial phases were improved by density modification with the program RESOLVE (25), and an initial model was built in O (26). Using MOLREP (27), the model was replaced into the refinement data set. Further improvement was performed by iteration of coordinate and temperature factor refinement, geometry validation, and manual rebuilding with REFMAC (28), MOLPROBITY (29), and COOT (30), respectively. The structure of the complex was solved by the molecular replacement method using structures of Rnd1 (Protein Data Bank code 2CLS) and plexin-B1 RBD (Protein Data Bank code 2R2O) as starting models. Both refined models exhibit reasonable stereochemistry (Table 1).
TABLE 1.
Data collection and refinement statistics of the intracellular plexin structure (3HM6) and of the RBD·RND1 complex (2REX)
| Data collection | Intracell. PLXNB1 mercury derivative | Intracell. PLXNB1 for refinement | Plexin-B1 RBD·Rnd1 complex |
|---|---|---|---|
| Beamline | NSLS X25 | NSLS X25 | CHESS F1 |
| Wave length | 0.9999 Å | 0.9999 Å | 0.9179 Å |
| No. of images | 850 × 0.3° | 600 × 0.3° | 360 × 0.5° |
| Space group | P3121 | P3121 | C2 |
| Unit cell | a = 75.34 Å, c = 210.8 Å | a = 74.4 Å, c = 214.4 Å | a = 150.1 Å, b = 71.7 Å, c = 101.9 Å, β = 128.4° |
| Resolution range (highest shell) (Å) | 30.00–2.50 (2.59–2.50) | 30.00–2.40 (2.49–2.40) | 30.00–2.30 (2.38–2.30) |
| Rsym | 7.8% (>100%) | 4.5% (57.7%) | 6.0% (23.1%) |
| I/σ | 34.0 (1.3) | 48.8 (3.3) | 22.2 (3.5) |
| Redundancy | 7.5 (4.4) (Bijvoet mates counted separately) | 9.8 (7.6) | 3.7 (3.2) |
| Completeness | 99.8% (99.0%) | 99.9% (99.9%) | 97.3% (84.3%) |
| Refinement | 3HM6 | 2REX |
|---|---|---|
| Resolution range (Å) | 28.08–2.40 | 20.00–2.30 |
| No. of atoms refined (protein/others) | 4115 (4078/37) | 4434 (4291/143) |
| No. of unique HKLs | 26,208 | 36,863 |
| B-factors: protein/others/Wilson (Å2) | 63.7/48.8/58.9 | 40.3/26.4/40.4 |
| Rwork/Rfree | 0.227/0.262 | 0.215/0.248 |
| Root mean square deviation of bond lengths/angles | 0.013 Å/1.2° | 0.017 Å/1.4° |
| Ramachandran plot favored/outliers | 96.5%/0.2% (single, preproline outlier: Leu-1981) | 96.8%/0.4% (outliers: Pro-97 in chains B and D) |
| Resolution range (Å) | 28.08–2.40 | 20.00–2.30 |
| No. of atoms refined (protein/others) | 4115 (4078/37) | 4434 (4291/143) |
| No. of unique HKLs | 26,208 | 36,863 |
| B-factors: protein/others/Wilson (Å2) | 63.7/48.8/58.9 | 40.3/26.4/40.4 |
| Rwork/Rfree | 0.227/0.262 | 0.215/0.248 |
| Root mean square deviation bond lengths/angles | 0.013 Å/1.2° | 0.017 Å/1.4° |
| Ramachandran plot favored/outliers | 96.5%/0.2% (single, preproline outlier: Leu-1981) | 96.8%/0.4% (outliers: Pro-97 in chains B and D) |
Molecular Modeling
The plexin·Ras, and p120GAP·Ras complexes were all built based on the p120GAP·H-Ras structure (Protein Data Bank code 1WQ1). For this, the crystal structure of plexin was superposed on that of p120GAP, using the homologous regions of the two proteins. Because no GTP-bound structure is available, R-Ras was homology-modeled using MODELLER (31), based on the structure of H-Ras in complex with p120GAP. The equilibration of the structures using molecular dynamics (NAMD with the CHARMM27 potential function) is described in the supplemental material.
Biophysical Experiments
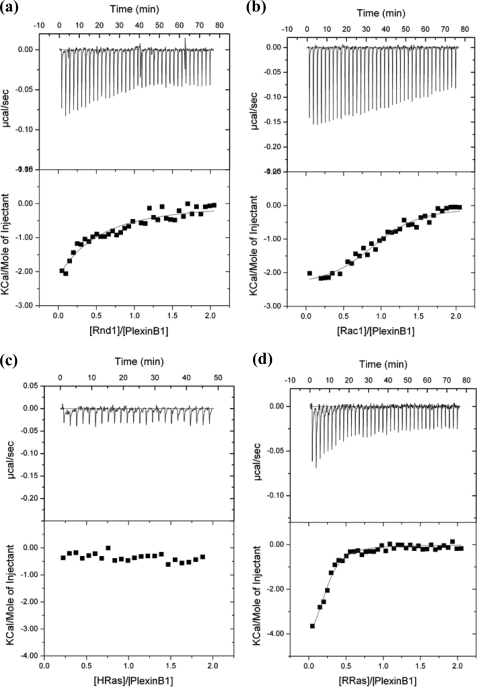
See supplemental Fig. S4 for confirmation that the intracellular protein is a monomer by gel filtration and binds Rnd1, Rac1, and R-Ras GTPases. Interaction studies of full-length intracellular plexin-B1 with GTPases (Rac1/Rnd1/R-Ras/H-Ras) were, furthermore, carried out using isothermal titration calorimetry (Microcal iTC200, GE Healthcare). All of the proteins were extensively exchanged into Tris buffer (20 mm Tris, pH 7.5, 1 mm Tris(2-carboxyethyl)phosphine, 50 mm sodium chloride, 4 mm magnesium chloride) prior to experiments. The primary ligand, plexin-B1, was placed under temperature control into the heat-sensitive cell (volume 200 μl), and the secondary ligand (Rac1 Q61L, Rnd1, R-Ras or H-Ras) was placed into a syringe immersed into the cell (capacity 40 μl). The concentration of the GTPase was 500 μm, and the concentration of plexin-B1 was kept at 40 μm. The presence of ∼1 mm AlF4− (mixing 2 mm AlCl3 and 20 mm NaF) did not substantially affect the binding affinity, which was also similar to that obtained with constitutively active R- and H-Ras mutants. The data were corrected by subtracting a titration of GTPase with buffer solution and were fitted to a model that gives the binary equilibrium association constant (Ka = 1/Kd, where Kd is equal to [plexin][GTPase]/[plexin·GTPase] (Table 2). All experiments were carried out at 10 °C two or three times. The error given in parentheses is estimated at 10–20% for Kd and ∼10% for ΔH, due to protein concentration and modest binding affinity.
TABLE 2.
GTPase Binding data of the intracellular plexin-B1 region derived from ITC measurements (Fig. 4)
Uncertainty is given in parentheses.
| GTPase | Plexin-B1 |
|
|---|---|---|
| Kd | ΔH | |
| μm | kcal/mol | |
| H-Ras + AlF4 | No interaction | |
| R-Ras | 26 (2.7) | −3.8 (0.4) |
| R-Ras + AlF4 | 22 (2.9) | −1.9 (0.2) |
| Rac1 Q61L | 43 (4.5) | −2.2 (0.3) |
| Rnd1 | 35 (7) | −1.7 (0.2) |
Functional Cell-based Collapse Assay
Cellular collapse assays were performed as previously described (32). Briefly, COS cells and NIH-3T3 fibroblasts were transfected to express plexin-B1 wild type or mutant receptors. Plexin expression in cell protein lysates was analyzed by Western blotting, according to standard protocols, using anti-plexin-B1 antibody IC-2. In parallel, the same COS-transfected cells were seeded on glass coverslips for the functional assay. After a 1-h incubation with 5 nm Sema4D, the cells were subjected to immunofluorescence analysis with anti-plexin-B1 antibody EC-6.9. F-actin was stained with fluorescein-labeled phalloidin (Sigma) to provide counterstaining. Digital images were acquired, using a Zeiss Axioskop microscope equipped with a Bio-Rad confocal imaging system. Collapsed cells were identified as having a diameter equal to or less than 30 μm.
RESULTS
Characteristics of the Structure of the Intracellular Region of Plexin-B1
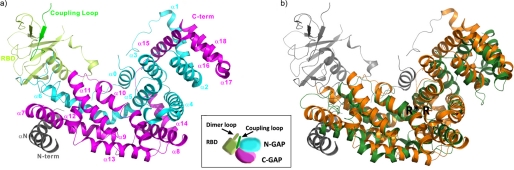
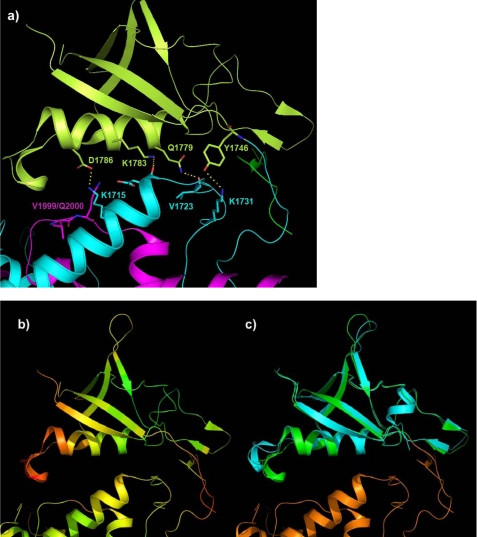
We crystallized and solved the structure of the intracellular plexin-B1 region (see Table 1 and “Experimental Procedures”). Fig. 1a shows a global view of the structure. The core GAP structure is composed of one segment that precedes and the other that succeeds the RBD, forming an intertwined structure (as viewed, α2-α5 is on top of the C-terminal segment; other parts of the N-terminal segment are placed behind it). The sequence and structural similarity between plexin and Ras GAPs is shown in supplemental Fig. S1 and Fig. 1b, respectively. Especially regions of the plexin-B1 GAP structure that are at or near the interface with the GTPase substrate in the p120GAP·H-Ras complex (Protein Data Bank code 1WQ1) are highly similar.
FIGURE 1.
Structure of the intracellular region of plexin-B1 and its comparison with p120GAP. a, the GAP domain has been split into an N- and C-terminal portion (cyan and magenta, respectively) by the RBD (yellow). The RBD dimerization loop forms a β-sheet with a coupling loop (green), residues 1879–1890. The latter sequence is located between the RBD and C-terminal segment of the GAP domain. A portion of the N-terminal chain, residues 1511–1562, forms a helical structure (gray). The inset shows the organization of the intracellular region in schematic diagram form. b, superposition of the plexin GAP domain (orange) with p120GAP (dark green). The structures are closely superimposable (Cα root mean square deviation of 3.3 Å for 280 matching residues). Plexin structural elements that are additional to the p120GAP domain are shown in gray. The location of the catalytic Arg-1677 and supporting Arg-1984 is indicated by R and R*, respectively.
Density also delineates a portion of the N-terminal segment as a long helical backbone, comprising 26 of the 52 residues prior to Ile1563, the first residue for which side chain density is discernable. The RBD is placed to the side of the structure, making contacts with the N-terminal, and more weakly, with the C-terminal GAP segment, as detailed below. Density (see supplemental Fig. S2) also defines a loop region just C-terminal to the RBD that folds back to make contact with the RBD as well as with the N-terminal GAP segment. We refer to this region as the coupling loop.
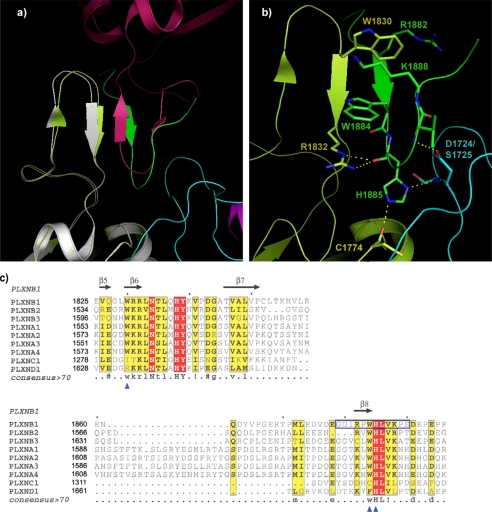
A Coupling Loop Replaces RBD Dimerization Contacts
Previously, we presented the structure of the RBD dimer, associated through a dimerization loop, residues 1818–1833 (21), shown partly in Fig. 2a. Superimposed is the full-length intracellular structure, which appears as a monomer (no structural oligomers are indicated in this crystal, as shown in supplemental Fig. S3). Importantly, in this structure, the place of the symmetrical dimerization loop is taken by the coupling loop (residues 1879–1890), which is just C-terminal of the RBD. Key interactions of the dimerization loops in the structure of the RBD dimer, such as a π-cation interaction between Trp-1830 and Arg-1832′ (21), are replaced in the intracellular protein with a similar interaction between the dimerization loop Arg-1832 and the coupling loop Trp-1884, as shown in Fig. 2b. The side chain of Arg-1832 is also localized by a hydrogen bond with the backbone carbonyl group of His-1885. This residue is located in a sharp turn of the coupling loop, making contact with both the RBD and with the RBD N-terminal GAP domain. Key residues of the coupling loop (WHLV(K/R)) are absolutely conserved between different plexin-A and -B family members (Fig. 2c), suggesting that this region has an important structural, if not functional, role.
FIGURE 2.
The coupling loop displaces the RBD dimerization loop. a, region showing the coupling loop (green) and the dimerization loop (gray) in a comparison of the structure of the intracellular protein RBD (pale green) with that of the dimeric RBD (dark red). Antiparallel main chain hydrogen bonding between dimerization loop strands in the RBD dimer (strands in gray and dark red) have been replaced by a parallel arrangement between the dimerization and coupling loop of the same molecule (strands in pale green and green). b, details of interactions between the coupling loop, RBD, and N-terminal GAP segment. Hydrogen bonds involving side chains are shown. c, the coupling loop (boxed) is highly conserved in plexin-A and -B family members. Alignment of part of the RBD region from the nine human plexins is shown. Sites used for the mutagenesis study of the dimerization loop (β6) and coupling loop (β8) are indicated (blue triangles).
Functional Studies of Plexin-B1 Mutants Suggest a Role for the Coupling Loop
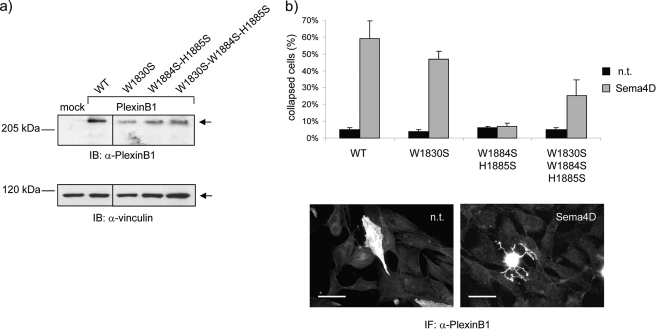
To assess the functional importance of the coupling loop and its interaction with the RBD dimerization loop, we designed several mutations in the full-length receptor mammalian expression vector. W1830S is known to disrupt the dimerization of free RBD domains in vitro (19), whereas W1884S/H1885S is expected to destabilize RBD-coupling loop and RBD-GAP interactions, increasing RBD dimerization. The wild type and mutant receptors were expressed at comparable levels in transfected COS cells (Fig. 3a).
FIGURE 3.
Expression and functional analysis of plexin-B1 mutants in mammalian cells confirms the importance of the coupling loop and its relationship to dimerization. a, COS cells were transfected to express either wild type (WT) or mutated Plexin-B1 receptors. Total lysates of cellular proteins were analyzed by immunoblotting (IB) using an antibody against the intracellular domain of the plexin. b, the same cells as described above were seeded on glass coverslips and treated with Sema4D to trigger the typical collapse response (32). By immunofluorescence analysis, the plexin-B1-expressing cells were subsequently identified, and the fraction of collapsed cells was scored based on the typical morphological aspects (see “Experimental Procedures” for details; representative images at the bottom). Scale bar, 40 μm.
When functionally tested in response to plexin-B1 ligand Sema4D, the dimerization mutant W1830S was highly competent to elicit cytoskeletal remodeling and morphological changes, leading to the typical collapse phenotype (Fig. 3b) (32). Notably, cells expressing this mutant did not undergo collapse in the absence of the ligand, indicating that RBD monomerization is not sufficient per se to elicit the functional response. Importantly, in the case of the coupling loop mutant W1884S/H1885S, neither COS (Fig. 3b) nor NIH-3T3 fibroblast cells (not shown) expressing this variant plexin-B1 receptor underwent cell collapse in response to Sema4D, indicating that the loop is required for receptor function. Intriguingly, when the W1884S/H1885S mutations in the coupling loop were added to the W1830S mutation, the functional competence of the receptor was significantly restored (Fig. 3b). This indicates that, although the coupling loop may normally be required to disrupt RBD dimerization in response to ligand binding, receptor activation may be achieved independently from coupling loop function if the intracellular protein region is already in a monomeric state. Supplemental Fig. S4 shows that the plexin intracellular protein is a stable monomeric species by gel filtration and is able to bind Rnd1 as well as Rac1. Isothermal titration calorimetry confirms these interactions and also demonstrates binding of R-Ras (Fig. 4).
FIGURE 4.
Isothermal titration calorimetry measurements of the binding affinity between the intracellular plexin-B1 region and various GTPases. Shown are representative raw data (top) as well as the peak integrated and concentration-normalized enthalpy change (bottom) for Rac1 Q61L (a), Rnd1 (b), H-Ras (c), and R-Ras (d). Table 2 shows fitted data.
Contacts between the RBD and the GAP Domain
Hydrogen bonding and salt bridges exist between the RBD and the N-terminal GAP segment (Fig. 2, a and b), but several, mostly non-polar, interactions are also made with the C-terminal GAP segment (Fig. 5a). The side chains involved in hydrogen bond-salt bridge interactions are absolutely conserved in the plexin-A and -B family (supplemental Fig. S6). Although the electron density for most side chains is clear in this region of the structure, the B-factors are noticeably high, especially toward the C terminus of helix αR in the RBD structure and at the RBD N terminus as well as at the termini of the coupling loop and for part of the dimerization loop (Fig. 5b). The same regions of the RBD structure show conformational differences when they are part of the dimeric form (as shown in Fig. 5c). They are also known to be flexible in solution (33). Other regions of the RBD structure show remarkably little structural variation, suggesting that the contacts with the GAP domain can be accommodated without significant conformational changes in the RBD structure.
FIGURE 5.
RBD-GAP interactions are supplemented by interactions with the coupling loop. a, details of a second region of RBD-GAP interactions (see also Fig. 2b). The color scheme is that of Fig. 1a. Hydrogen bonds involving side chains are shown. b, crystallographic B-factors of the RBD and surrounding region (increasing B-factor colored green to red). c, superposition of the RBD region of the intracellular plexin-B1 structure (green) with the x-ray structure of the isolated domain dimer lobes (blue).
RBD·Rnd1 Complex Structure and Incompatibility of the Dimeric Intracellular Structure with Rho GTPase Binding
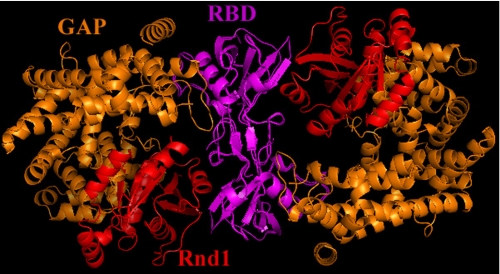
In this report, we also present a structure of the plexin-B1 RBD complexed with the small GTPase Rnd1 at 2.3 Å resolution (Table 1). The details of the RBD·Rnd1 interface will be discussed elsewhere, but of particular interest here is the effect of GTPase binding on the RBD structure. This binding takes place on the side almost opposite to the RBD-GAP interface discussed so far, and only small changes are observed within the RBD conformation upon association with GTPase (supplemental Fig. S5). Since the region that makes contact with the GAP domain is not significantly affected by Rnd1 GTPase binding, the RBD dimer structure was used to model a dimeric form of the whole intracellular protein by superposition (Fig. 6). In the case of Rnd1 (and other Rho GTPases) binding to such a putative dimer of the whole intracellular region, it is clear that there are considerable steric clashes between the GTPase bound to one plexin molecule and the GAP domain of the other, second plexin. Thus, the dimeric intracellular protein cannot bind Rho GTPases. However, the equilibrium will shift to the Rho- GTPase bound monomeric forms if that structure is more stable than the unbound dimeric form.
FIGURE 6.
A model for the dimeric form of the intracellular protein does not allow Rho GTPase binding. Shown is the superposition of the monomeric intracellular structure with the model of an intracellular dimer based on the RBD dimer bound to Rnd1 (Protein Data Bank code 2REX). The GTPase binds to the RBD region previously described by solution NMR studies (21), which is located roughly opposite to its interface with the GAP domain. The model shows that in a dimer, the Rnd1/Rac1 protein would clash with the second GAP domain.
Characterization of Models for Plexin-B1·Ras GTPase Complexes
Since the region of the plexin-B1 GAP structure that would bind Ras GTPases is highly similar to that in p120GAP, we have used the p120GAP·H-Ras bound structure to model the complex between plexin-B1 and R-Ras as well as H-Ras. We also modeled p120GAP·R-Ras. All four protein complexes were then relaxed over a 5-ns molecular dynamics simulation (see supplemental material). Experimentally, it was shown in cell-based assays that R-Ras binds to plexin-B1 and is a substrate for its GAP function, whereas H-Ras is not (5). By contrast p120GAP works on both R- and H-Ras (9). All of the structures equilibrate to within 2–3 Å main chain root mean square deviation from their starting conformations. Overall, the contacts made by R-Ras and H-Ras with plexin-B1 are in the same regions as those seen in the other Ras GAP·GTPase complexes (34). However, some are predicted to differ in detail (supplemental Fig. S6), and the residues involved are not conserved between plexin and the other GAP proteins (p120GAP, SynGAP) (supplemental Fig. S1). They are for the most part highly conserved between the nine members of the human plexin family (supplemental Fig. S6). Although electron density is seen for only part of the critical side chains, such as Arg-1677 in plexin-B1 MODELLER is able to place them correctly in the starting structure. However, differences between the structures become apparent during the 5-ns equilibration simulation. Supplemental Fig. S7 shows critical residues surrounding the Ras-bound GTP nucleotide in the four simulations, and supplemental Fig. S8 shows distances of interest that evolve differently in the simulations. More significant for the H-Ras simulation is that this Ras protein appears to be leaving the binding pocket, increasing the distances between the Arg finger and GTP.
Binding of GTPases
Experimentally, it can be shown using isothermal titration calorimetry that the full-length intracellular region of plexin-B1 binds R-Ras (Fig. 4 and Table 2). No interaction is detected with H-Ras, confirming the Ras selectivity of plexin-B1 seen in previous cell-based assays (5).
DISCUSSION
Structural Basis of Conformational Changes of the Plexin-B1 Intracellular Region
The molecular mechanisms underlying plexin activation in general and plexin-B1 in particular are not well understood. The insertion of a domain, such as the RBD, into an enzyme, such as plexin-B1, could set up an allosteric mechanism for the regulation of catalytic activity. Domain insertions are not common in the universe of protein structures (e.g. see Ref. 35), but they are being explored in new protein designs (36). Notably, in vitro biochemical studies support this concept in three of four plexins investigated. Specifically, the binding of certain Rho GTPases to the cytoplasmic domain of a number of plexins appeared to disrupt an interaction between the N- and C-terminal GAP region when the segments were supplied as protein fragments (5–8, 13). The observations suggested a large scale reorganization of the GAP-homologous fold upon GTPase binding, although presumably contacts between the segments have to reform in order to obtain a functional protein. Interestingly, the RBD makes contacts with the N-terminal and (less so) with the C-terminal segment of the GAP domain in the structure of the intracellular region of plexin-B1 presented here (Fig. 1a). An activation mechanism based on the transmission of an allosteric signal to both GAP segments and the destabilization of contacts between them is not implausible, although it cannot be verified at present, due to lack of the structure of a dimeric or Rho GTPase-bound entire intracellular plexin region. The data reported in this study, however, support a different mechanistic explanation as follows.
The structure of the plexin-B1 RBD has been determined under a number of different conditions (i.e. as an isolated dimer (21) and reported here as a dimer bound to Rnd1 GTPase) and in the context of the surrounding GAP domain. Comparison of these structures shows that the conformational differences are very limited and occur only in loops known to be flexible in solution (33). There is no indication from these structures that GTPase binding could disrupt RBD-GAP interactions either directly or indirectly through destabilization of contacts involving the coupling loop. Overall then, the structural data do not support a model (above) that involves dramatic internal reorganization of the segments of the plexin intracellular region upon GTPase binding.
A recent mechanistic model for plexin-B1 activation proposed that a change in the configuration of domains, if not an altered oligomerization state, of the intracellular region of the receptor occurs upon Rho GTPase binding (21). The RBD·Rnd1 complex structure presented here crystallized in the dimeric form. A detailed thermodynamic and NMR study, however, confirmed that, in solution, binding of Rnd1 and Rac1 to the RBD opposes its dimerization. Association of GTPases with the dimeric form of the RBD is also possible, albeit with a ∼2–3-fold reduced binding affinity (37). These findings imply that the monomer-dimer versus GTPase binding equilibria are finely balanced. The new data presented here support this model and provide further mechanistic insights. Specifically, by characterizing the structure of the intracellular region of plexin-B1, we discovered that, in a monomeric molecule, the interaction between RBD dimerization loops is replaced by contacts with an adjacent peptide segment that we call the “coupling loop” (Fig. 2a). The competition of this peptide with RBD dimerization rationalizes the observation that the entire intracellular domain crystallized in a monomeric state (and is monomeric in solution) (supplemental Fig. S4). In fact, the localization of the coupling loop in the complete structure is sterically incompatible with dimerization of the RBD via the dimerization loop. Whether the association of the RBD with Rac1 or Rnd1 would stabilize or destabilize binding of this coupling loop is not clear. Nevertheless, the functional data are compatible with a simple mechanism, involving a monomer-dimer equilibrium, that is affected by both coupling loop and GTPase binding.
It should be noted that plexin-B1 is known to dimerize through the extracellular region and semaphorin ligand binding is thought to rearrange this structure (22) as part of the receptor activation mechanism. However, ligand- or antibody-mediated clustering of the extracellular portion of a receptor does not necessarily lead to the dimerization of the intracellular region (38). Clearly, conformational changes in the extracellular domain are likely to be complex and have a major regulatory function for the monomer-dimer equilibrium of the intracellular plexin region. For example, cell-based studies have shown that the extracellular domains of plexin-A1 and -B1 have an autoinhibitory function in the absence of the ligand and that their partial removal leads to a constitutive activation of the receptor (8, 39).
The Structure and Functional Data Support a Model Involving Changes in Domain Association for Receptor Activation
We have shown here that the intracellular mutation W1830S, which completely disrupts RBD dimerization in solution (19), has no significant effect on the function of the full-size plexin-B1 receptor, consistent with the proposal that the RBD-mediated dimerization of the intracellular region of plexins is not required for receptor signaling, including GAP function. On the other hand, our data indicate that the W1884S/H1885S mutations in the coupling loop substantially impair plexin activity. This could indicate that (a) the coupling loop can no longer form contacts between the RBD and GAP domains that may be required for function, and/or (b) the coupling loop can no longer disrupt RBD dimerization. The observation that an additional W1830S mutation (preventing dimerization) substantially restores functionality in the W1884S/H1885S mutant can be simply interpreted by saying that the primary function of the coupling loop is to help in disrupting intracellular dimerization (i.e. case b). Thus, when a dimeric state is prevented by the W1830S mutation, the competitive effect of the coupling loop is no longer necessary, and the intracellular moiety basally assumes a monomeric state, which is, again, compatible with activation. Since any interactions between intracellular domains mediated by the coupling loop are likely to be considerably weakened in the presence of the above mutations, this could imply that contacts between the RBD and GAP domain may not be necessary for receptor activity.
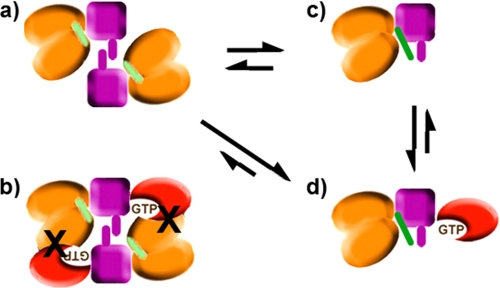
Provided that the orientation of the RBD domains in the dimerized state is maintained in the context of the GAP structure, a model for the full-length intracellular dimeric form can be constructed. This model reveals that the dimeric structure is incompatible with Rnd1/Rac1 GTPase binding to the RBD region, due to significant clashes between the Rho GTPase and GAP segments of the opposing structure (Fig. 6). Given an essentially rigid domain arrangement, the RBD domains are predicted to dissociate upon Rho GTPase binding, emphasizing the mechanistic relevance of the monomer-dimer equilibrium in this model. Fig. 7 schematically shows the dimeric structure, which is unable to bind Rho GTPase due to clashes that would result with the GAP domain. When the coupling loop binds to the dimerization loop, it also destabilizes the dimeric form of the intracellular region of plexin. The monomeric form may then be further stabilized by binding Rho GTPase (21, 37) and is likely to provide a greater accessibility to the catalytic site of the GAP region for Ras GTPases. The combination of the structural and functional results leads to the conclusion that the state of the intracellular plexin-B1 as crystallized in the monomeric form may actually correspond to the active state of the GAP protein or to a GAP “primed” for activity.
FIGURE 7.
Schematic mechanism for monomer formation (as a step toward activation) of the plexin intracellular region. The model shows the putative dimeric form of the intracellular region as free protein (a), with Rho GTPase bound (b) (from Fig. 6), the x-ray structure presented in this paper (c), and a putative Rho GTPase-bound state of the monomeric form (d). The dimerization and coupling loops are shown in violet and green, respectively. The arrows indicate likely populations in equilibrium.
Computational Modeling Suggests That R-Ras Can Be Bound to Plexin-B1 in a p120GAP·H-Ras-like Configuration and GTPase Selectivity is Confirmed by Binding Experiments
The suggestion that the monomeric intracellular plexin-B1 structure could represent the active GAP enzyme conformation is supported by placement of H- and R-Ras into the plexin-B1 GAP domain using p120GAP·H-Ras (40–42) as a homology model. Molecular dynamics simulations were used to equilibrate the models, confirming the absence of significant steric clashes. Intriguingly, in contrast to R-Ras, H-Ras begins to drift out of the binding pocket. The biological function of R- and H-Ras GTPases is largely different in cells (43), and thus increased substrate specificity of the GAP appears to be important in plexin signaling. Although the Ras core structures are well conserved, several regions, including the N and C termini of the GTPases, differ significantly and are likely to be used in allowing the plexin-B1 GAP to discriminate between different Ras family members. The molecular dynamics and perturbative docking calculations (not shown) make predictions as to the origin of this behavior and possibly GTPase selectivity. These are being tested by mutagenesis and will be reported in the future.
Experimentally, we have shown here that R-Ras binds to the isolated plexin-B1 intracellular receptor construct, whereas H-Ras does not, confirming the GTPase selectivity that has been observed in cells (5). It should be noted that GAP activity is difficult to measure in vitro for plexins (8, 9), as well as for other GAP proteins (e.g. 44). We have been unable thus far to establish conditions to measure appreciable GAP activity using purified recombinant intracellular plexin-B1, either alone or in the presence of Rnd1, with or without antibody clustering. The protein is primed for function, but an important regulatory aspect could still be missing in the model, such as a small conformational change due to posttranslational modification or localization of the proteins at the plasma membrane.
Biological Implications
A recent model for plexin-B1 receptor activation proposes that a key step toward receptor activation involves the regulation of an equilibrium between a monomeric and a dimeric state of the intracellular region (21). The new model, presented here, has been critically extended by the structural characterization of the entire intracellular region of the receptor and by the discovery of the coupling loop as an additional regulatory element. First, steric clashes between bound Rho GTPases and GAP domains are likely to be severe in the RBD-dimerized form of the intracellular region of the plexin-B1 receptor. Second, the coupling loop provides an additional layer of regulation and functions to further stabilize the monomeric state of the intracellular region. This model can thus take into account the recent finding that Rho GTPase binding alone is a relatively weak input to shift the RBD monomer-dimer equilibrium (37). Such multilevel level regulatory features are becoming well established in cell signaling mechanisms, where often multiple inputs are required to shift an equilibrium to yield full activation (e.g. 45).
Recently, Negishi and co-workers suggested that plexin-C1 and plexin-D1 may be regulated by different mechanisms, compared with plexins of the A and B family (7). Intriguingly, the RBDs of the former proteins are monomeric in solution (data not shown) and do not bind Rho GTPases.8 In fact, plexin-C1 appears to function independently of Rnd1 binding (7). Therefore, a mechanism for RBD dimer destabilization would not be required for plexin-C1 and -D1. This is consistent with the idea that the requirements for Rho GTPase binding, the coupling and the dimerization loops, have been built in as regulatory steps for plexin-A and -B family members. Accordingly, the protein segments that give rise to these features are highly conserved in the latter, but the sequences differ in plexin-C1 and -D1. The disruption of plexin-A and -B family intracellular region dimers or at least a mechanism involving such interdomain rearrangements would represent a step toward receptor activation, similar to the regulatory mechanism known for integrins or receptor tyrosine phosphatases (46, 47). Clearly, the regulation of this event also depends on ligand binding to the extracellular portion of the receptor.
In conclusion, the work presented here characterizes, at the structural and functional level, a new element for plexin regulation: the coupling loop. This protein segment appears to control the interactions between RBD and GAP domains, and it can oppose the dimerization of the RBD and possibly of the entire intracellular region of plexins in synergy with Rho GTPase binding. The present data unveil a multilevel regulatory mechanism underlying plexin activation and prompt future studies that will extend to the extracellular domains and to ligand binding as additional levels of regulation for the intracellular events.
Acknowledgments
Drs. Christina Kiel and Luis Serrano (EMBL-CRG, Barcelona) helped us early in this project with a homology model of plexin-B1 based on p120GAP. Crystallographic data shown in this report were derived from work performed at Argonne National Laboratory, Structural Biology Center, beam line 19ID, at the Advanced Photon Source. Data collection at GM/CA CAT, beam line 23ID-B, has been funded by NCI Grant Y1-CO-1020 and NIGM Grant Y1-GM-1104 from the National Institutes of Health. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Calculations were carried out at the Case Western Reserve University High Performance Cluster, at the Ohio Supercomputer Center (Columbus, OH), and at LoneStar (Austin, TX) via a TeraGrid Award (to M. B.).
Addendum
While this paper was under review (and following the release of the plexin-B1 coordinates in late May 2009), the structure of the plexin-A3 intracellular region also became available. It should be noted that the structures are very similar (Cα root mean square deviation of 0.95 Å for 430 matching residues). However, the interpretation of the structures, drawn in terms of possible molecular mechanisms by He et al. (48) is substantially different from that presented here.
This work was supported in part by National Institutes of Health Grants R01GM73071 and K02HL084384 (to M. B.) and in part by the Structural Genomics Consortium, a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
The atomic coordinates and structure factors (codes 3HM6 and 2REX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
P. K. Hota and M. Buck, unpublished data.
- GAP
- GTPase-activating protein
- RBD
- Rho GTPase binding domain.
REFERENCES
- 1.Tamagnone L., Comoglio P. M. (2000) Trends Cell Biol. 10, 377–383 [DOI] [PubMed] [Google Scholar]
- 2.Kruger R. P., Aurandt J., Guan K. L. (2005) Nat. Rev. Mol. Cell Biol. 6, 789–800 [DOI] [PubMed] [Google Scholar]
- 3.Neufeld G., Shraga-Heled N., Lange T., Guttmann-Raviv N., Herzog Y., Kessler O. (2005) Front. Biosci. 10, 751–760 [DOI] [PubMed] [Google Scholar]
- 4.Pasterkamp R. J., Verhaagen J. (2001) Brain Res. Brain Res. Rev. 35, 36–54 [DOI] [PubMed] [Google Scholar]
- 5.Oinuma I., Ishikawa Y., Katoh H., Negishi M. (2004) Science 305, 862–865 [DOI] [PubMed] [Google Scholar]
- 6.Toyofuku T., Yoshida J., Sugimoto T., Zhang H., Kumanogoh A., Hori M., Kikutani H. (2005) Nat. Neurosci. 8, 1712–1719 [DOI] [PubMed] [Google Scholar]
- 7.Uesugi K., Oinuma I., Katoh H., Negishi M. (2009) J. Biol. Chem. 284, 6743–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oinuma I., Katoh H., Negishi M. (2004) J. Neurosci. 24, 11473–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohba Y., Mochizuki N., Yamashita S., Chan A. M., Schrader J. W., Hattori S., Nagashima K., Matsuda M. (2000) J. Biol. Chem. 275, 20020–20026 [DOI] [PubMed] [Google Scholar]
- 10.Rohm B., Rahim B., Kleiber B., Hovatta I., Püschel A. W. (2000) FEBS Lett. 486, 68–72 [DOI] [PubMed] [Google Scholar]
- 11.Pasterkamp R. J. (2005) Trends Cell Biol. 15, 61–64 [DOI] [PubMed] [Google Scholar]
- 12.Püschel A. W. (2007) Adv. Exp. Med. Biol. 600, 12–23 [DOI] [PubMed] [Google Scholar]
- 13.Turner L. J., Nicholls S., Hall A. (2004) J. Biol. Chem. 279, 33199–33205 [DOI] [PubMed] [Google Scholar]
- 14.Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 15.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 16.Canagarajah B., Leskow F. C., Ho J. Y., Mischak H., Saidi L. F., Kazanietz M. G., Hurley J. H. (2004) Cell 119, 407–418 [DOI] [PubMed] [Google Scholar]
- 17.Pena V., Hothorn M., Eberth A., Kaschau N., Parret A., Gremer L., Bonneau F., Ahmadian M. R., Scheffzek K. (2008) EMBO Rep. 9, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drugan J. K., Rogers-Graham K., Gilmer T., Campbell S., Clark G. J. (2000) J. Biol. Chem. 275, 35021–35027 [DOI] [PubMed] [Google Scholar]
- 19.Tong Y., Hughes D., Placanica L., Buck M. (2005) Structure 13, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong Y., Hota P. K., Hamaneh M. B., Buck M. (2008) Structure 16, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong Y., Chugha P., Hota P. K., Alviani R. S., Li M., Tempel W., Shen L., Park H. W., Buck M. (2007) J. Biol. Chem. 282, 37215–37224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antipenko A., Himanen J. P., van Leyen K., Nardi-Dei V., Lesniak J., Barton W. A., Rajashankar K. R., Lu M., Hoemme C., Püschel A. W., Nikolov D. B. (2003) Neuron 39, 589–598 [DOI] [PubMed] [Google Scholar]
- 23.Vikis H. G., Li W., Guan K. L. (2002) Genes Dev. 16, 836–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger T. C. (2003) Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 26.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 27.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta. Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P., Cowtan K. (2004) Acta. Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31.Fiser A., Sali A. (2003) Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 32.Barberis D., Artigiani S., Casazza A., Corso S., Giordano S., Love C. A., Jones E. Y., Comoglio P. M., Tamagnone L. (2004) FASEB J. 18, 592–594 [DOI] [PubMed] [Google Scholar]
- 33.Bouguet-Bonnet S., Buck M. (2008) J. Mol. Biol. 377, 1474–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmadian M. R., Kiel C., Stege P., Scheffzek K. (2003) J. Mol. Biol. 329, 699–710 [DOI] [PubMed] [Google Scholar]
- 35.Bernards A., Settleman J. (2004) Trends Cell Biol. 14, 377–385 [DOI] [PubMed] [Google Scholar]
- 36.Radley T. L., Markowska A. I., Bettinger B. T., Ha J. H., Loh S. N. (2003) J. Mol. Biol. 332, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hota P. K., Buck M. (2009) Protein Sci. 18, 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vilar M., Charalampopoulos I., Kenchappa R. S., Simi A., Karaca E., Reversi A., Choi S., Bothwell M., Mingarro I., Friedman W. J., Schiavo G., Bastiaens P. I., Verveer P. J., Carter B. D., Ibáñez C. F. (2009) Neuron 62, 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi T., Strittmatter S. M. (2001) Neuron 29, 429–439 [DOI] [PubMed] [Google Scholar]
- 40.Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 41.Scheffzek K., Lautwein A., Kabsch W., Ahmadian M. R., Wittinghofer A. (1996) Nature 384, 591–596 [DOI] [PubMed] [Google Scholar]
- 42.Mittal R., Ahmadian M. R., Goody R. S., Wittinghofer A. (1996) Science 273, 115–117 [DOI] [PubMed] [Google Scholar]
- 43.Ehrhardt A., Ehrhardt G. R., Guo X., Schrader J. W. (2002) Exp. Hematol. 30, 1089–1106 [DOI] [PubMed] [Google Scholar]
- 44.Venkateswarlu K., Brandom K. G., Lawrence J. L. (2004) J. Biol. Chem. 279, 6205–6208 [DOI] [PubMed] [Google Scholar]
- 45.Buck M., Xu W., Rosen M. K. (2004) J. Mol. Biol. 338, 271–285 [DOI] [PubMed] [Google Scholar]
- 46.Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 47.Barr A. J., Ugochukwu E., Lee W. H., King O. N., Filippakopoulos P., Alfano I., Savitsky P., Burgess-Brown N. A., Müller S., Knapp S. (2009) Cell 136, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He H., Taehong Y., Terman J. R., Zhang X. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15610–15615 [DOI] [PMC free article] [PubMed] [Google Scholar]