Abstract
A quantitative screening method was developed to enable isolation and affinity maturation of peptide ligands specific for a given target from peptide libraries displayed on the outer surface of Escherichia coli using multi-parameter flow cytometry. From a large, random 15-mer peptide library, screening identified a core motif of W-E/D-W-E/D that conferred binding to vascular endothelial growth factor (VEGF). One cycle of affinity maturation resulted in the identification of several families of VEGF-binding peptides having distinct consensus sequences, from which a preferred disulfide constraint emerged. In the second affinity maturation cycle, high affinity peptides were favored by the addition of a decoy protein that bound an adjacent epitope on the display scaffold. The decoy apparently reduced rebinding or avidity effects, and the resulting peptides exhibited consensus at 12 of 19 amino acid positions. Peptides identified and affinity matured using bacterial display were remarkably similar to the best affinity matured using phage display and exhibited comparable dissociation constants (within 2-fold; KD = 4.7 × 10−7 M). Screening of bacterial-displayed peptide libraries using cytometry enabled optimization of screening conditions to favor affinity and specificity and rapid clonal characterization. Bacterial display thus provides a new quantitative tool for the discovery and evolutionary optimization of protein-specific peptide ligands.
Keywords: affinity maturation, bacterial display, peptide, VEGF
Introduction
Peptides are among the largest and most rapidly growing classes of biological diagnostic and therapeutic agents (Lien and Lowman, 2003; Falciani et al., 2005). Peptide display technologies have proven to be essential tools for peptide discovery and subsequent affinity, specificity and stability optimization (Falciani et al., 2005). Although several different display technologies have been used for peptide discovery, including bacteriophage (Sidhu et al., 2000), bacteria (Bessette et al., 2004), yeast (Boder and Wittrup, 1997), ribosome (Mattheakis et al., 1994), RNA and DNA display (Yonezawa et al., 2003), display on the surface of bacteriophage has been applied in the overwhelming majority of cases (Sidhu et al., 2000). The commercial availability of phage display systems and well-established protocols has led to their widespread use to discover protein ligands and enzyme substrates and for affinity and specificity optimization (Aina et al., 2007).
The affinity and specificity properties of peptides identified from random libraries are often insufficient for in vivo applications (Bracci et al., 2003), thus necessitating an affinity maturation process (Levin and Weiss, 2006). Although affinity maturation is routinely utilized to improve the properties of proteins and antibodies, relatively few examples of peptide affinity maturation using display technologies have been reported (Fairbrother et al., 1998; Dwyer et al., 2001; Fleming et al., 2005; Shrivastava et al., 2005). Typically, conserved motifs identified from random libraries are used to design biased libraries, which are then screened using more stringent criteria. Alternatively, when conserved motifs are not present, partial or ‘soft’ randomization can be used wherein a given position is biased toward the parental amino acid, but allowed to vary (Fairbrother et al., 1998). Using this strategy, a family of vascular endothelial growth factor (VEGF)-binding peptides was identified after screening and affinity maturation of cysteine-constrained peptides from a pool of eight phage libraries, with inter-cysteine spacing ranging from 4 to 10 amino acids. The peptide exhibiting the highest affinity (v114; VEPNCDIHVMWEWECFERL) bound the VEGF receptor-binding site with high affinity (KD = 110 nM) and inhibited HUVEC proliferation in vitro (Fairbrother et al., 1998).
Display of heterologous peptides on the surface of bacterial cells was reported as early as 1986, but the use of bacterial display for protein library screening has been slow to mature owing to technical challenges (Daugherty, 2007). Bacterial display combines the advantages of phage and yeast display since large libraries approaching 1011 members can be constructed efficiently in Escherichia coli (Bessette et al., 2004), and analogous to yeast display, libraries can be screened quantitatively using sequential magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). Several display vectors have been described that enable presentation of peptides in outer membrane proteins (Bessette et al., 2004), fimbria (Klemm and Schembri, 2000) and flagella (Lu et al., 2003), yet thus far, the engineering of high-affinity protein-binding peptides via affinity maturation with bacterial display has not been reported. In particular, an important challenge for bacterial display is the accurate assessment of clonal affinity during screening since apparent affinity can be influenced by several factors, including expression level, avidity and rebinding effects and scaffold dependence. High-level peptide display on the cell surface is beneficial for achieving large fluorescence signals for screening using FACS but can skew measurements of apparent affinity through avidity and rebinding effects. The affinity of a cell-surface displayed ligand for a target can differ by orders of magnitude from that measured in solution due to the close proximity of available binding partners on the cell surface (Löfblom et al., 2007) and the mobility of display scaffolds within the membrane (Gibbs et al., 2004). Rebinding effects can slow apparent dissociation rates, or in the case of homomultimeric target proteins, a single target may bind multiple displayed peptides simultaneously leading to underestimates in both dissociation rate and equilibrium dissociation constants.
To investigate the utility of bacterial display for peptide affinity maturation, a large random peptide library was displayed on E.coli using the biterminal display scaffold known as circularly permuted OmpX (CPX) (Rice et al., 2006; Rice and Daugherty, 2008). This bacterial display peptide library was screened to identify peptide ligands specific for VEGF, and two cycles of diversification and quantitative screening via FACS were used to affinity mature the initial leads. Our results compare favorably with those obtained using state-of-the-art phage display systems and demonstrate for the first time that bacterial display provides a highly effective, complementary alternative to phage and yeast systems that is well suited for peptide and mini-protein engineering.
Materials and methods
Bacterial strains and growth conditions, plasmids and reagents
All bacterial display experiments were performed with E.coli strain MC1061 [F-araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (StrR) hsdR2 (rK-mK+) mcrA mcrB1] (Casadaban and Cohen, 1980). All libraries and expression constructs were constructed using pBAD33 (Cmr) (Guzman et al., 1995). For cell-free affinity measurements, peptides were expressed as C-terminal fusions to maltose-binding protein (MBP) using the pMAL-c4X vector (New England Biolabs) expressed in E.coli strain K12 TB1 (F-ara Δ(lac-proAB) [Φ80dlac Δ(lacZ)M15] rpsL(StrR) thi hsdR) (Yanisch-Perron et al., 1985).
For cell-surface binding analysis and library sorting, bacteria were subcultured 1:50 from an overnight culture or frozen stock to LB supplemented with chloramphenicol (CM) at 34 µg/ml, and the culture was allowed to grow for 2 h at 37°C with shaking at 250 rpm. Expression of the outer membrane protein scaffold was induced with l(+)-arabinose at a final concentration of 0.02% or 0.04% w/v. Cells expressing peptides in the CPX scaffold (Rice et al., 2006) were allowed to grow for 3 h at room temperature after induction; cells expressing peptides in the eCPX scaffold (Rice and Daugherty, 2008) were allowed to grow for 1 h at 37°C after induction. The selection and recovery of cells during sorting was performed as described (Kenrick et al., 2007). To analyze single clones, 5 µl of culture (corresponding to ∼5 × 106 cells) was used for incubation with protein and subsequent characterization.
Recombinant human VEGF-A (both the 165- and the 121-amino acid isoforms) was purchased from MBL International or Invitrogen. Biotinylation of VEGF was performed using the FluoReporter mini-biotin-XX protein labeling kit (Invitrogen). VEGF was conjugated to Alexa-488 using the Alexa Fluor 488 protein labeling kit. Human serum was purchased from Sigma. For screening in serum, human serum was diluted ∼1/50 in PBS and labeled with the Alexa Fluor 633 protein labeling kit (Invitrogen). For screening where higher serum concentrations were desired, serum was first pre-incubated with bacteria over-expressing the library scaffold at a concentration of 109 cells/ml serum for 30–60 min at 4°C. Centrifugation at 20 800g was used to remove cells, bacterial-binding proteins and other aggregates, and the clarified supernatant was used for competition. Other reagents and their suppliers were as follows: primers (Operon), restriction enzymes (New England Biolabs), streptavidin R-phycoerythrin (SAPE) (Invitrogen), MyOne streptavidin-coated magnetic microbeads (Invitrogen), B-PER II bacterial protein extraction reagent (Pierce Biotechnology), anti-biotin mAB R-phycoeryrthrin (ABPE) (Miltenyi Biotec), neutravidin R-phycoerythrin (NAPE) (Invitrogen), Ni-NTA agarose (Qiagen), synthetic peptides (Anaspec), amylose resin (New England Biolabs). FACS was performed using a FACSAria (BD Biosciences). Surface plasmon resonance (SPR) analysis was performed with a Biacore 3000 (GE Healthcare).
Protein expression and purification
YPet-Mona (YM) was expressed and purified for biterminal library screening and clone characterization using the plasmid pB33YM, which encodes for YPet (Nguyen and Daugherty, 2005) fused to the C-terminal SH3 domain of Mona (You et al., 2006), with a C-terminal 6His tag. For cytoplasmic expression, cells were subcultured 1:50 from an overnight culture to LB–CM and grown for 2 h at 37°C. Expression was induced with 0.2% w/v l(+)-arabinose for 6 h to overnight. Cells were lysed with B-PerII, and protein was purified using Ni-NTA agarose according to the manufacturer's instructions.
A modified pMAL-c4X (pMAL-TEV) vector (a generous gift from CytomX Therapeutics) was used for fusion protein expression wherein the factor Xa cleavage site was replaced by a TEV cleavage site (ENLYFQG) followed by an additional four-amino acid linker sequence of QSGQ. The gene sequence coding the final Gly in the cleavage site and subsequent linker corresponded with the SfiI site present upstream of N-terminal peptides displayed on the pB33eCPX scaffold. Peptides were amplified from pB33eCPX using primers TCGCAACTCTCTACTGTTTC and CCTAGCTCGAGCCCTACCCAGACTGCCCTCC. PCR products were digested with SfiI and XhoI and ligated into similarly digested pMAL-TEV. For protein expression, cells from an overnight culture were diluted 1:100 in rich media (10 g tryptone, 5 g yeast extract, 5 g NaCl, 2 g glucose per liter) with 100 µg/ml carbenicillin and allowed to grow at 37°C to an OD600 of 0.5. Protein expression was induced with IPTG (0.3 mM) for 2 h at 37°C. Cells were then pelleted by centrifugation at 4000g for 20 min and lysed with B-Per II.
For batch purification, the lysate was diluted 1:2 with column buffer (20 mM Tris–HCl at pH 7.4, 200 mM NaCl, 1 mM EDTA) and incubated with amylose resin for 3 h at 4°C with shaking at a ratio of ∼15 ml resin for every liter of culture. The protein–resin mixture was loaded on a column, washed twice with 5 column volumes of column buffer. For continuous purification, the lysate was diluted 1:5 with column buffer and loaded onto an amylose column at 1 ml/min. After loading, the column was washed with 12 volumes of column buffer at 1 ml/min. The MBP fusion proteins were eluted with column buffer supplemented with 10 mM maltose. The eluted proteins were dialyzed to HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20).
Library construction and screening
A library of peptides with 15 randomized amino acid positions (X15) was displayed in N-terminal fusions to CPX (Rice et al., 2006) and screened for binding specifically to VEGF-A165. Magnetic selection (MACS) was first performed with 25 nM biotinylated VEGF to reduce the library size from 9 × 109 to 4.8 × 106 members, as previously described (Bessette et al., 2004). The enriched library population was incubated with 100 nM biotinylated VEGF for 45 min at 4°C, followed by incubation with 3.3 nM SAPE for 45 min at 4°C. Alexa-labeled serum at ∼1% dilution was added to both labeling steps. Two rounds of FACS were performed, sorting for high SAPE fluorescence and low Alexa-633 fluorescence. Two additional rounds of sorting were performed with either 1% labeled serum or 10% unlabeled serum. The VEGF concentration in the final round of sorting was reduced to 50 nM with 1% or 10% unlabeled serum, and the VEGF was labeled with Alexa-488. Arbitrarily selected clones were sequenced and their binding to VEGF in the presence or absence of serum was measured by flow cytometry.
Construction and screening of focused library
A second-generation library was constructed of the form X6-W-E/D-W-E/D-X9 on the N-terminus of enhanced CPX (eCPX). PCR was performed using pB33eCPX-SAPep (described in Rice and Daugherty, 2008) as a template with CGTAGCTGGCCAGTCTGGCCAGNNSNNSNNSNNSNNSNNSTGGGANTGGGANNNS NNSNNSNNSNNSNNSNNSNNSNNSGGAGGGCAGTCTGGGCAGTC as the forward primer and GGCTGAAAATCTTCTCTC (PD180) as the reverse primer. The PCR product was digested with SfiI and ligated into a similarly digested pB33CPX vector. After transformation, the library size was 2 × 108 members.
The library was sorted using one round of MACS and four rounds of FACS. MACS was performed with 10 nM biotinylated VEGF-A165 and a bead:cell ratio of 1:2. Two rounds of FACS were performed using 10 nM biotinylated VEGF followed by secondary labeling with 1 nM ABPE. The VEGF concentration was decreased to 1 nM for the third round of FACS, and secondary labeling was performed with SAPE. For the fourth round of FACS, induced cells were incubated with 100 nM biotinylated VEGF. After 1 h incubation at 4°C, the cells were pelleted by centrifugation and resuspended in cold PBS. An aliquot of this suspension was diluted 1/100 in PBS at room temperature and incubated for 15 min to allow for VEGF dissociation. SAPE was added to a final concentration of 6.6 nM, and the cells were incubated on ice for 30 min for secondary labeling and to stop the dissociation. The cells were pelleted by centrifugation at 3800g for 5 min at 4°C and resuspended in cold PBS prior to analysis by flow cytometry. Individual clones from the third and fourth rounds of sorting were isolated by plating for further characterization.
Construction and screening of biterminal library
For further affinity maturation, a third library was constructed based on the consensus from the second generation of the form X4-C-X4-M/I-W-E/D-W-E/D-C-I/L/M/F-X3. The consensus motif that arose after sorting included only M or F at position 16; however, the degenerate codon that was required at that position (WTK) also encoded I and L. The library template pB33eCPX-P2X was constructed by digesting pBV99 (a generous gift from CytomX Therapeutics) with PstI and HindIII. The resulting fragment, which encoded the 94 C-terminal amino acids of eCPX, a GGS linker and the P2X peptide (HISQWKPKVPNREDKYKK), was ligated into similarly digested pB33eCPX. PCR was performed with pB33eCPX-P2X as the template, CGTAGCTGGCCAGTCTGGCCAGNNSNNSNNSNNSTGCNNSNNSNNSNNS ATKTGGGAKTGGGAKTGCWTKNNSNNSNNSGGAGGGCAGTCTGGGCAGTC as the forward primer and PD180 as the reverse primer. The PCR product was digested with SfiI and ligated into a similarly digested pB33eCPX vector. Electroporation yielded a library of 2 × 108 individual transformants.
The biterminal library was sorted with one round of MACS and four rounds of FACS. MACS was performed using 100 nM VEGF-A165 and a bead:cell ratio of 1:2. For all rounds of FACS, the library population was first incubated with biotinylated VEGF for 45 min at 4°C. The cells were then centrifuged at 3800g for 5 min at 4°C, and the cells were resuspended in 10 nM NAPE with 200 nM YM. Prior to analysis and sorting, the cells were centrifuged at 3800g for 5 min and resuspended in PBS. The VEGF incubation conditions for each of the four FACS rounds were as follows: (1) 10 nM VEGF, (2) 1 nM VEGF, (3) 1 nM VEGF and (4) 10 nM VEGF with 40 µM IgG and 3.5% w/v BSA. Individual clones from rounds 3 and 4 were isolated for further characterization.
Cell surface characterizations
The library population resulting after each round of sorting was assayed for binding to secondary reagents (SAPE, NAPE or ABPE, as appropriate). Where possible, secondary reagents were changed prior to sorting to avoid the enrichment of streptavidin-, neutravidin- and antibiotin-binding peptides. Individual clones were also assayed for binding to secondary reagents before affinity measurements with VEGF were performed. The apparent dissociation rates of peptides from the second-generation library were measured by cytometry. Peptides from each consensus group were labeled with biotinylated VEGF, pelleted by centrifugation and the unbound VEGF removed. The cells were incubated in PBS at room temperature for 15 min, and dissociation was stopped by placing the cells on ice and adding SAPE after 5, 10 and 15 min of incubation. The fluorescence versus time data were used to calculate apparent dissociation rate. Measurements for the highest ranking clones were repeated in triplicate.
The mode of binding for selected peptides from the second-generation library was assayed in comparison to v114 (Fairbrother et al., 1998). Clones expressing second-round peptides were incubated with biotinylated VEGF-A121, an isoform that is composed of an identical receptor-binding domain but lacks a heparin-binding domain, for 45 min at 4°C. Cells were centrifuged at 3800g for 5 min at 4°C and resuspended in 10 nM SAPE for secondary labeling; prior to analysis by cytometry, cells were centrifuged again and resuspended in PBS. In order to determine if the peptides bound in a similar manner to v114, cells expressing second-round peptides were incubated with 10 nM biotinylated VEGF-A165 in the presence of synthetic v114 peptide at serial dilutions of 50–0.016 µM. Secondary labeling was performed with SAPE, as before, and binding inhibition was measured by cytometry.
The apparent affinity of cell-surface-displayed peptides from the biterminal library was ranked using a flow cytometry assay. Individual clones were incubated with 10 nM biotinylated VEGF-A165 for 45 min at 4°C. Cells were pelleted by centrifugation and resuspended in 10 nM NAPE with YMona at 0, 5, 50, 500 and 5000 nM. Relative binding affinity was estimated by measuring the percent decrease in red fluorescence upon incubation with 5 µM YM (F5000) when compared with fluorescence in the absence of YM (F0): (F0–F5000)/(F0–Fbg). The term Fbg corresponds to background cell autofluorescence; thus, this quotient is zero when no change in red fluorescence is observed (F0 = F5000) and 100% when all red fluorescence is lost upon the addition of YM (F5000 = Fbg). As controls, biterminal versions of v114 and second-round clones 2.04 and 2.08 were constructed and assayed. Plasmids encoding eCPX-v114, 2.04 and 2.08 were digested with PstI and HindIII as described above; the resulting fragment containing the VEGF-binding peptide and N-terminal portion of eCPX was ligated into a similarly digested pB33eCPX-P2X vector.
Measurement of peptide affinities using SPR
The affinity of each peptide for VEGF-A165 was measured using SPR on a Biacore3000 using kinetic analysis of surface-immobilized VEGF to synthetically prepared peptides and MBP-fused peptides. CM5 biosensor chips (GE Healthcare) were activated with EDC (N-ethyl-N'-(3-dimethylaminopropyl)-carbodiimide hydrochloride) and NHS (N-hydroxysuccinimide) according to the manufacturer's instructions. VEGF at a concentration of 2–10 µg/ml in 10 mM sodium acetate, pH 4.5, was injected over the activated surface until 1500 RU were covalently coupled to the chip. Unreacted groups were blocked with ethanolamine according to the manufacturer's protocol. Equilibrium binding data were collected with 2-fold serial dilutions of all peptide constructs in HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20), pH 7.4, at 25°C. Analyte proteins were injected for 1–3 min followed by 3–5 min of dissociation in buffer. MBP fusions were injected at 30 µl/min and the surface regenerated by a 30 s injection of glycine, 10 mM, pH 1.5. For synthetic peptides, the running buffer was modified to include 0.05% Tween 20 and 0.5% DMSO; peptides were injected at 50 µl/min and the surface regenerated with a 1 min injection of running buffer at 100 µl/min. For each MBP analyte, a single concentration series was performed with duplicate measurements of several concentrations. For each synthetic peptide analyte, two concentration series were performed sequentially. The equilibrium binding level was plotted versus fusion concentration to calculate a steady state KD.
Results
Identification of VEGF ligands from a fully random peptide library
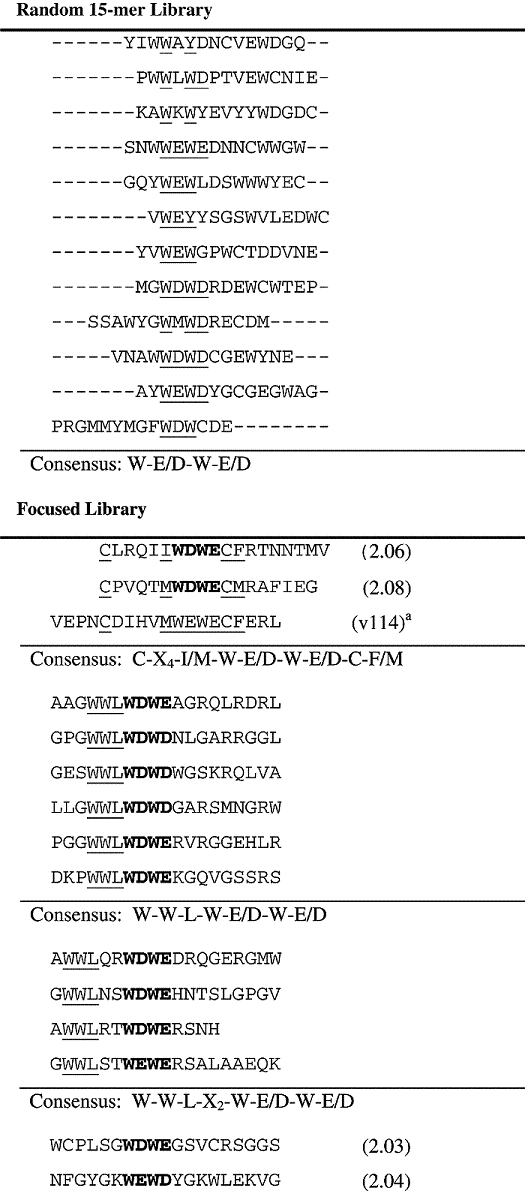
VEGF was selected as a target for peptide screening since the selection and affinity maturation of VEGF-binding peptides using phage display has been described in detail (Fairbrother et al., 1998). VEGF exists as a homodimer whose receptor-binding domain contains two identical receptor-binding sites, to which these phage-derived peptides were found to bind (Pan et al., 2002). To identify sequences that bind specifically to VEGF from a fully random 15-mer peptide library (X15), cells from the library were incubated with VEGF in the presence of competing serum proteins, yielding clones that retained binding in serum. To enable the use of serum as a competitor, it was necessary to deplete the serum of antibodies and other proteins that bind E.coli using an excess of cells expressing the display scaffold without a peptide. The sorting stringency was increased with each successive screening cycle by increasing the serum concentration, up to 10% serum, or decreasing the concentration of VEGF. Peptides binding to VEGF identified from the random library exhibited a consensus of W-E/D-W-E/D, a motif also present in the best of three lead peptides identified by phage display that bind to the receptor-binding site of VEGF (Table I). This motif exhibited specificity for VEGF since 10% serum only partially reduced VEGF binding for most isolated clones, and had no effect on one-third of clones (data not shown).
Table I.
Peptide sequences and consensus motifs isolated from the fully random library and after one round of affinity maturation
 |
Residues contributing to the consensus are underlined; residues constrained by library design are bolded.
VEGF-binding peptide affinity maturation by screening a focused library
To affinity mature the initial VEGF-binding peptide sequences, a focused library was designed and constructed, in which 15 amino acids were randomized surrounding the core consensus motif observed in the first round (X6-W-E/D-W-E/D-X9). This library was displayed on E.coli using the eCPX scaffold (Rice and Daugherty, 2008). The increased display efficiency of this scaffold enabled the use of a shorter induction time and lower VEGF concentrations (1–10 nM) for screening. Sequencing of 30 arbitrarily selected clones from the final cycles of sorting revealed at least four unique consensus groups (Table I). The first and largest group consisted of linear peptides containing the tripeptide WWL upstream of the fixed W-E/D-W-E/D core. The WWL tripeptide was located either immediately adjacent to the core (W-W-L-W-E/D-W-E/D) or separated by two amino acids (W-W-L-X2-W-E/D-W-E/D). The second group included two members with a nine-member cysteine-constrained loop of the form C-X5-W-E/D-W-E/D-C. Two additional novel motifs were represented a single time: an unconstrained sequence that presented with the highest frequency among the selected clones (2.04) and another sequence capable of forming a 13-residue disulfide constrained ring (2.03). Interestingly, two cysteine-constrained sequences (2.06 and 2.08) exhibited an identical loop size and relative placement of the core-binding motif within the cysteine loop as the highest affinity peptide arising from a focused phage display library with built-in disulfide constraints (v114) (Fairbrother et al., 1998). Bacterial display-derived peptides apparently bound VEGF in a manner similar to that of optimized phage-derived peptides v107 and v114 (Fairbrother et al., 1998) since 2.08 and 2.04 both bind the VEGF-A121 isoform, which contains only the receptor-binding domain. Furthermore, the binding of surface-displayed peptide 2.04 was inhibited by synthetically prepared v114 peptide (VEPNCDIHVMWEWECFERL) in a dose-dependent manner with an apparent IC50 of 0.14 µM, as measured by cytometry (data not shown).
In an attempt to rank the relative apparent affinities of identified VEGF ligands, the apparent dissociation rate constants were measured on the cell surface using flow cytometry. In addition to reflecting intrinsic affinity, this assay carried the caveat that dissociation rates were influenced by scaffold dependence, rebinding and avidity effects, which could not be resolved. Although yielding only slightly faster dissociation at room temperature when compared with cysteine-constrained 2.08 or v114, the high frequency of the clone displaying linear peptide 2.04 was apparently a result of a high display level, rather than improved affinity, since its fluorescence was consistently higher (2–3×) than that of the control clone displaying v114, and the peptide did not retain high affinity in solution (Table II). Two clones displaying peptides with a putative nine-residue cysteine-constrained loop (2.08 and 2.06, Table I) exhibited high similarity and had slow dissociation rates as measured by flow cytometry (data not shown). Since disulfide constraints typically improve peptide affinity, the motif shared between these two peptides was used to design a third library for an additional round of affinity maturation.
Table II.
Affinity improvement between focused and biterminal libraries
|
KD (µM)b |
||||
|---|---|---|---|---|
| Peptide | FACSa (%) | MBP fusion | Synthetic peptidec | |
| Peptide control | ||||
| VEPNCDIHVMWEWECFERL | (v114) | 54 ± 3 | 2 | 0.23 ± 0.01 |
| Peptides isolated after round 2 (focused library) | ||||
| CPVQTMWDWECMRAFIEG | (2.08) | 85 | 43 | |
| NFGYGKWEWDYGKWLEKVG | (2.04) | 67 | >50 | >50 |
| Peptides isolated after round 3 (biterminal library) | ||||
| GPGPCSRLVMWEWECFAAL | (3.33) | 54 | 5 | 0.58 ± 0.05 |
| WPVRCSRFVMWEWECFLRA | (3.30) | 42 | 6 | 0.47 ± 0.07 |
| GGSWCPRLVMWEWECFWPR | (3.53) | 22 | 6 | NB |
| SVGPCGRFVMWEWECFGLL | (3.78) | 76 | 12 | |
| LPVRCSRFVMWEWDCFFGA | (3.08) | 62 ± 1 | 17 | |
| RALSCSRFVMWEWECFVRV | (3.18) | 56 | 19 | |
| LAGRCGRAVIWDWECFAAL | (3.11) | 32 | 37 | |
| FLGGCSRFLMWEWECFGFA | (3.04) | 71 | 44 | |
| VGFRCSRFVMWDWECFWPP | (3.03) | 41 | 61 | |
| AFGACSRFLMWEWECFFPS | (3.07) | 62 ± 2 | NB | |
NB, no binding detected.
aPercent fluorescence lost measured by flow cytometry, as described in Materials and methods with average and standard deviation for data collected on two different days (three for v114); bmeasured by SPR; caverage and standard deviation for two (four for v114) sequential concentration series.
Affinity maturation using biterminal library screening
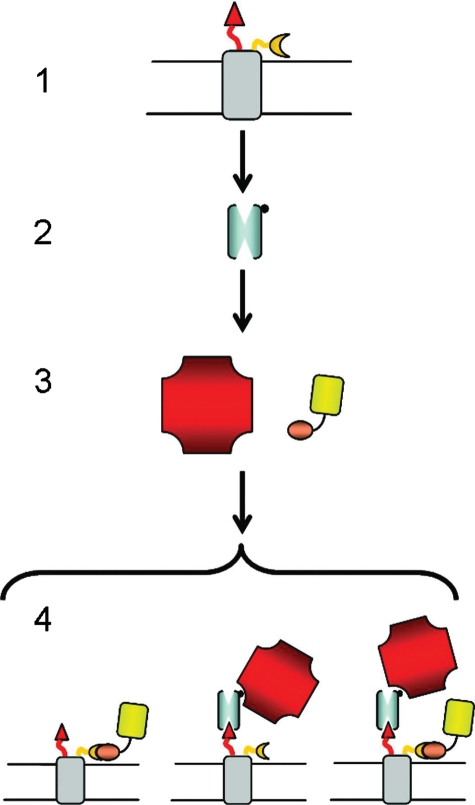
Since cell fluorescence and affinity are not uniformly correlated in cell display systems due to expression level variation (Boder and Wittrup, 1997), a general two-color screening method was applied to favor high affinity. Specifically, a fluorescent protein was used to label a peptide tag introduced at the C-terminus, adjacent to the N-terminally fused peptide (Fig. 1). For this purpose, we used the C-terminal SH3 domain of monocytic adaptor protein (MonaSH3) fused to the C-terminus of a yellow fluorescent protein (YPet) (You et al., 2006) thereby generating a YPet-MonaSH3 fusion (YM). Simultaneous labeling with VEGF and YM resulted in decreased red fluorescence, when compared with the absence of YM, indicating that the secondary reporter was inhibiting VEGF binding to the adjacent N-terminal peptide (Fig. 2). Thus, we reasoned that the fluorescent protein could be used as a decoy to sterically inhibit rebinding and favor high-affinity peptides. The YM decoy binds the peptide ligand P2X (HISQWKPKVPNREDKYKK) with an apparent KD≈1 µM, as measured by cytometry (data not shown). In the presence of excess decoy (>100× the VEGF concentration), only high-affinity VEGF-binding peptides (KD < 1 µM) were expected to remain bound in the presence of the decoy binding (yielding red fluorescence), whereas clones whose apparent affinity was enhanced by rebinding or avidity effects would exhibit reduced labeling (yielding yellow fluorescence). A third-generation focused library was designed based on the consensus between the two cysteine-constrained peptides with four random residues upstream of the first cysteine: X4-C-X4-I/M-W-E/D-W-E/D-C-F/I/L/M-X3. The library was again displayed via fusion to the N-terminus of eCPX with the decoy-binding ligand P2X fused to the eCPX C-terminus. Interestingly, addition of the decoy reduced the frequency of VEGF binders in the initial library by over 10-fold, suggesting that only a small fraction of binders possessed sufficient affinity to remain bound in the presence of the decoy (Fig. 2). Sorting was performed with decreasing concentrations of VEGF in each round, while maintaining a concentration of the decoy yielding at least 5-fold increased yellow fluorescence. Subsequent rounds of sorting with increasingly stringent gates decreased the difference between the fraction of library members bound in the presence and absence of YM (Fig. 2). To further favor affinity and specificity, a final round of sorting was performed in the presence of a mixture of BSA (3.5% w/v) and pooled human IgG (40 µM). Sequencing of 168 randomly selected clones from the final population revealed a consensus remarkably similar to phage peptides v107 and v114, with a strong preference for V and M at positions 9 and 10, respectively, and F occurring exclusively at position 16 (Table III). Unlike phage-derived peptides, positions 6–8 exhibited a strong consensus of S-R-L/F.
Fig. 1.
Dual-color labeling scheme for eCPX bacterial display library. (1) E.coli eCPX display scaffold with N-terminal library (triangle) and C-terminal affinity tag (P2X; crescent); (2) the library is incubated with biotinylated target protein (e.g. VEGF); (3) secondary labeling is accomplished with NAPE (red) and a fluorescent probe recognizing the C-terminus (YM; yellow); (4) the library is sorted for clones exhibiting high red and low yellow fluorescence.
Fig. 2.
Cytometric analysis of bacterial display library pools after single (red) or dual color (red and yellow) labeling. Cell populations from unsorted library (A and B), after one cycle of FACS (C and D) and after three cycles of FACS (E and F). A, C and E show the library labeled with VEGF followed by NAPE only; B, D and F show the library labeled with VEGF followed by NAPE and YM. Sorting was performed with VEGF concentrations of 100 nM (A and B) and 1 nM (C–F), with secondary labeling by 10 nM NAPE (all panels) in the presence of 200 nM YM (B, D and F). Gates are drawn to identify the VEGF-binding population, whose frequency is indicated by the given percentage value.
Table III.
Peptide sequences and consensus motifs isolated from the biterminal library
| Clone ID | Peptide sequence |
|---|---|
| 3.33 | GPGPCSRLVMWEWECFAAL |
| 3.30 | WPVRCSRFVMWEWECFLRA |
| 3.53 | GGSWCPRLVMWEWECFWPR |
| 3.78 | SVGPCGRFVMWEWECFGLL |
| 3.08 | LPVRCSRFVMWEWDCFFGA |
| 3.18 | RALSCSRFVMWEWECFVRV |
| 3.11 | LAGRCGRAVIWDWECFAAL |
| 3.04 | FLGGCSRFLMWEWECFGFA |
| 3.02 | VAGRCSRLVMWEWECFLRL |
| 3.03 | VGFRCSRFVMWDWECFWPP |
| 3.07 | AFGACSRFLMWEWECFFPS |
| 3.23 | SVAPCSVLVMWEWECFGMA |
| 3.13 | AGRPCSRFVMWEWECFLAL |
| 3.01 | ALGACSRFVMWEWECFGLS |
| 3.06 | VLGRCSRYVMWEWDCFLLS |
| 3.15 | LGLPCGRLVMWEWECFGFG |
| 3.10 | PRRGVPCSRFVMWEWECFVWP |
| 3.39 | LSGRCSRFVMWEWDCFLQS |
| 3.67 | GLPRCSRFVMWEWECFFGA |
| 3.48 | WLLPCSRFVMWEWECFGLG |
| 3.28 | PFGPCSRLVMWEWECFGLV |
| 3.45 | ARLSCSRFLMWEWECFGLA |
| 3.27 | GPGGCSRLVMWEWECFGLAE |
| Consensus: C-S-R-F/L-V/L-M-W-E-W-E-C-F | |
Residues contributing to the consensus are underlined; residues constrained by library design are bolded.
The apparent affinities of cell-surface-displayed peptides resulting from the second cycle of affinity maturation were ranked based on their ability to bind VEGF in the presence of increasing decoy concentrations (Fig. 3, Table II) before further affinity characterizations were performed. Selected peptides exhibiting a range of affinities in the cell surface assay were expressed as MBP fusions, and their equilibrium dissociation constants were determined using SPR. The apparent affinity for cell-displayed peptides as measured by FACS proved a useful negative predictor of affinity. That is, cell-displayed peptides with >60% red fluorescence loss also exhibited low affinity as MBP fusions (KD > 10 µM), but roughly half of clones with <60% red fluorescence loss also exhibited higher affinity as MBP fusions (KD < 10 µM) (Table II). Equilibrium dissociation constants of all fusion proteins, including MBP-v114, were >1 µM (Table II). Given the reported KD of 0.11 µM (Fairbrother et al., 1998) and our measured value of 0.23 µM (Table II) for v114, this result indicated that the MBP-fusion format underestimated peptide affinity. Given the substantial difference between the apparent affinity of the v114 MBP-fusion (KD = 2 µM) and the reported value for the synthetic peptide (110 nM), three peptides exhibiting the highest relative affinities (3.30, 3.33 and 3.53) as MBP fusions were prepared synthetically for affinity analysis using SPR. Peptides 3.30 and 3.33 exhibited the highest affinities as MBP fusions, with KD's within 2- to 3-fold of MBP-v114. Synthetically prepared peptide 3.53 did not exhibit detectable binding to a VEGF-coated sensor chip. However, peptides 3.30 and 3.33 retained their relative affinity ranking and had dissociation constants of KD = 0.47 and 0.58 µM, respectively, values comparable with that measured for v114 (0.23 µM, Fig. 3, Table II).
Fig. 3.
Characterization of clonal affinity on the cell surface using cytometry and in solution using SPR. (A) Overlaid contour plots showing the red and green fluorescence of three different clones [2.08 (left), 3.30 (center) and v114 (right)] incubated with 10 nM VEGF followed by NAPE and increasing concentrations of YM from 0 (red) to 5 µM (yellow), with intermediate concentrations of 5, 50 and 500 nM shown (from left to right). (B) Langmuir binding isotherm for synthetically prepared peptides measured using SPR. Data for duplicate (3.30 and 3.33) and quadruplicate (v114) concentration series are shown where dashed lines indicate the best-fit isotherm.
Discussion
To our knowledge, this is the first demonstration that bacterial display coupled with screening via FACS is a highly effective tool for peptide ligand discovery and subsequent affinity maturation. Although the pairing of microbial surface display and FACS for quantitative library screening and rapid clone characterization was described over a decade ago (Boder and Wittrup, 1997; Georgiou et al., 1997), several technical problems slowed the emergence of bacterial display/FACS as a simple and quantitative alternative to phage display panning experiments (Daugherty, 2007). Nevertheless, improvements in bacterial display scaffolds, expression systems and screening strategies that allow for expression normalization (Löfblom et al., 2005, 2007; Rice and Daugherty, 2008) have created new opportunities to apply bacterial display/FACS to discover and optimize peptides. Using the Gram-positive host Staphylococcus carnosus, libraries of 3 × 109 members were created and screened via FACS yielding affibodies towards tumor necrosis factor alpha (TNF-α) (Kronqvist et al., 2008); however, in this study, pre-enrichment was performed using phage display rather than MACS in order to reduce the library to a size that can be screened by FACS (<108). Larger peptide libraries of 5 × 1010 members have been previously constructed in E.coli, as constrained insertions into the outer membrane protein OmpA (Bessette et al., 2004). However, not surprisingly, these scaffold-constrained libraries yielded peptides whose affinity was scaffold-dependent (Bessette et al., 2004). In an effort to address this problem, a bacterial display scaffold was developed by CPX wherein both N- and C-termini are localized on the cell surface allowing display of unconstrained peptides (Rice et al., 2006). Peptide libraries constructed using the CPX display scaffold yielded peptide ligands for streptavidin that retained high affinity when removed from the cell surface, with apparent dissociation rates within 2- to 4-fold of their cell-surface measurements (Rice et al., 2006). The CPX display scaffold was subsequently evolved (eCPX) to improve the display of diverse peptides by reducing the impact of the passenger peptide upon the surface localization efficiency (Rice and Daugherty, 2008). Here we applied, for the first time, the eCPX display system to affinity mature peptides specific for a challenging homodimeric target (VEGF) yielding peptides with dissociation constants as low as 470 nM.
Somewhat surprisingly, affinity maturation using bacterial display yielded peptides highly similar, in both sequence and binding affinity, to those previously isolated and affinity matured using phage display (Fairbrother et al., 1998). One of the best peptides identified using bacterial display shared 10 identities of 19 amino acids to the best phage display-generated peptide (v114) and had similar affinity. Even so, there were significant differences between the phage and bacterial display affinity maturation procedures. For phage display, eight cysteine-constrained libraries having loop sizes of 4–10 amino acids were used for the initial selection, while a single large bacterial display library of random 15-mers was used. Surprisingly, panning of the phage display library pool enriched just three distinct binders without significant similarity, necessitating the use of a soft-randomization procedure for the first cycle of affinity maturation. Here, bacterial display yielded a large family of non-constrained peptides with a consensus for a W-E/D-W-E/D motif mediating specific VEGF binding. This high-quality consensus information enabled design and of an ordinary degenerate codon library incorporating the motif. This library, in turn, identified preferred flanking residues and an optimal cysteine-spacing relative to the WEWE core motif. A second cycle of affinity maturation using a biterminal library screen enhanced the degree of sequence convergence among isolated VEGF-binding clones. Equilibrium dissociation constants for the best bacterial display identified peptides were within 2-fold of that of the best known VEGF-binding peptide—with KD values determined using SPR of 0.47 and 0.58 µM for 3.30 and 3.33, respectively, versus 0.23 µM for v114. The extent of sequence and functional convergence for peptides discovered and optimized using two different display technologies has not been reported previously, further validating bacterial display as a powerful tool for identifying functional binding peptides.
Variation in peptide display level between distinct clones in a library can adversely impact accurate affinity screening, since highly expressed peptides can have a higher apparent affinity due to avidity and rebinding effects. The benefits of quantitative measurement of protein display level via a secondary fluorescent reporter have been established for yeast (Boder and Wittrup, 1997) and Staphylococcus display systems (Löfblom et al., 2007; Rockberg et al., 2008), yet in these systems, high display level was considered desirable. However, a high level display of peptide ligands on the cell surface can be expected to give rise to rebinding effects, wherein peptides exhibiting fast association and dissociation rates effectively trap a target protein on a dense array of possible ligands on the cell surface. Furthermore, target proteins with two or more identical binding sites, such as VEGF, can bind multiple neighboring peptides on the cell surface. In such cases, apparent affinity constants measured for peptides on the cell surface can differ by orders of magnitude from those of peptides in free solution (Löfblom et al., 2007). In the present study, we attempted to favor monovalent binding between VEGF and surface-displayed peptides by sorting library members with both low expression (yellow fluorescence) and high VEGF binding (red fluorescence) using a biterminal display scaffold. Addition of the decoy probe (YM) did not enable expression normalization but greatly impacted the ability of peptides to remain bound to VEGF and decreased the number of binding clones in the library by more than 10-fold (Fig. 2), for a given VEGF concentration. We hypothesize that addition of the decoy (YM) acts as a generic competitor by sterically interfering with rebinding or the formation of VEGF-peptide complexes with 1:2 stoichiometry.
In addition to providing sorting criteria, the biterminal display system enabled the apparent affinity ranking of peptides on the cell surface by measuring the fraction of VEGF-binding lost with increasing concentrations of decoy protein YM. This metric effectively captured the affinity improvement of clones isolated from the final library relative to those from the second library, with a fluorescence loss of 85% for parent clone 2.08 when compared with an average of 58% loss for all the isolated third-round peptides, comparable with the v114 control (54%) (Fig. 3, Table II). Using this metric, the peptides least likely to maintain a high affinity when removed from the cell surface could be identified, i.e. peptides exhibiting >60% change in red fluorescence as measured on the cell surface proved lowest affinity when measured as MBP fusions (KD > 10 µM). However, a red fluorescence change <60% did not guarantee high affinity when the peptide was removed from the cell surface, perhaps owing to orientation or solubility changes that occurred when changing display scaffolds. To confirm peptide affinity, MBP fusions of peptides exhibiting a range of apparent affinities by FACS were created and assayed for binding by SPR, and VEGF binders with high affinities as MBP fusions maintained their relative affinity ranking as synthetically prepared peptides (Table III).
Overall, our results demonstrate that eCPX bacterial display is an effective tool for the identification and affinity maturation of peptides. Bacterial display enables rapid construction and screening of large libraries (1011) similar to those used in phage display. Moreover, like yeast display, bacterial display allows for the use of quantitative multi-parameter screening criteria to identify clones with desired affinity and specificity characteristics (Garcia-Rodriguez et al., 2007). The relative simplicity of working with E.coli streamlines library manipulation and individual clone characterization using cytometry.
Funding
This work was supported by a National Science Foundation CAREER award [BES-0449399] to P.S.D.; the Center of Cancer Nanotechnology Excellence at the National Institutes of Health [5 U54 CA119335-04] and a National Science Foundation graduate fellowship to S.A.K.
Acknowledgements
We thank Jerry Thomas for carefully reading the manuscript.
Footnotes
Edited by Andrew Bradbury
References
- Aina O.H., Liu R., Sutcliffe J.L., Pan C.-X., Lam K.S. Mol. Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- Bessette P.H., Rice J.J., Daugehrty P.S. Protein Eng. Des. Sel. 2004;17:731–739. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Wittrup K.D. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Bracci L., Falciani C., Lelli B., Lozzi L., Runci Y., Pini A., De Montis M.G., Tagliamonte A., Neri P. J. Biol. Chem. 2003;278:46590–46595. doi: 10.1074/jbc.M308615200. [DOI] [PubMed] [Google Scholar]
- Casadaban M.J., Cohen S.N. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Daugherty P.S. Curr. Opin. Struct. Biol. 2007;17:474–480. doi: 10.1016/j.sbi.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Dwyer J.J., Dwyer M.A., Kossiakoff A.A. Biochemistry. 2001;40:13491–13500. doi: 10.1021/bi011703b. [DOI] [PubMed] [Google Scholar]
- Fairbrother W.J., Christinger H.W., Cochran A.G., Fuh G., Keenan C.J., Quan C., Shriver S.K., Tom J.Y., Wells J.A., Cunningham B.C. Biochemistry. 1998;37:17754–17764. doi: 10.1021/bi981931e. [DOI] [PubMed] [Google Scholar]
- Falciani C., Lozzi L., Pini A., Bracci L. Chem. Biol. 2005;12:417–426. doi: 10.1016/j.chembiol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Fleming T.J., et al. J. Mol. Recognit. 2005;18:94–102. doi: 10.1002/jmr.722. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez C., Levy R., Arndt J.W., Forsyth C.M., Razai A., Lou J., Geren I., Stevens R.C., Marks J.D. Nat. Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- Georgiou G., Stathopoulos C., Daugherty P.S., Nayak A.R., Iverson B.L., Curtiss R. Nat. Biotechnol. 1997;15:29–34. doi: 10.1038/nbt0197-29. [DOI] [PubMed] [Google Scholar]
- Gibbs K.A., Isaac D.D., Xu J., Hendrix R.W., Silhavy T.J., Theriot J.A. Mol. Microbiol. 2004;53:1771–1783. doi: 10.1111/j.1365-2958.2004.04242.x. [DOI] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., Beckwith J. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick S., Rice J., Daugherty P. Curr. Protoc. Cytom. 2007;Chapter 4 doi: 10.1002/0471142956.cy0406s42. Unit 4.6. [DOI] [PubMed] [Google Scholar]
- Klemm P., Schembri M.A. Microbiology. 2000;146:3025–3032. doi: 10.1099/00221287-146-12-3025. [DOI] [PubMed] [Google Scholar]
- Kronqvist N., Löfblom J., Jonsson A., Wernérus H., Ståhl S. Protein Eng. Des. Sel. 2008;21:247–255. doi: 10.1093/protein/gzm090. [DOI] [PubMed] [Google Scholar]
- Levin A.M., Weiss G.A. Mol. Biosyst. 2006;2:49–57. doi: 10.1039/b511782h. [DOI] [PubMed] [Google Scholar]
- Lien S., Lowman H.B. Trends Biotechnol. 2003;21:556–562. doi: 10.1016/j.tibtech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Löfblom J., Wernérus H., Ståhl S. FEMS Microbiol. Lett. 2005;248:189–198. doi: 10.1016/j.femsle.2005.05.040. [DOI] [PubMed] [Google Scholar]
- Löfblom J., Sandberg J., Wernérus H., Ståhl S. Appl. Environ. Microbiol. 2007;73:6714–6721. doi: 10.1128/AEM.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., LaVallie E.R., McCoy J.M. Methods Mol. Biol. 2003;205:267–280. doi: 10.1385/1-59259-301-1:267. [DOI] [PubMed] [Google Scholar]
- Mattheakis L.C., Bhatt R.R., Dower W.J. Proc. Natl Acad. Sci. USA. 1994;91:9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.W., Daugherty P.S. Nat. Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- Pan B., Bing L., Russell S.J., Tom J.Y.K., Cochran A.G., Fairbrother W.J. J. Mol. Biol. 2002;316:769–787. doi: 10.1006/jmbi.2001.5370. [DOI] [PubMed] [Google Scholar]
- Rice J.J., Daugherty P.S. Protein Eng. Des. Sel. 2008;21:435–442. doi: 10.1093/protein/gzn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J.J., Schohn A., Bessette P.H., Boulware K.T., Daugherty P.S. Protein Sci. 2006;15:825–836. doi: 10.1110/ps.051897806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockberg J., Löfblom J., Hjelm B., Uhlén M., Ståhl S. Nat. Methods. 2008;5:1039–1045. doi: 10.1038/nmeth.1272. [DOI] [PubMed] [Google Scholar]
- Shrivastava A., et al. Protein Eng. Des. Sel. 2005;18:417–424. doi: 10.1093/protein/gzi049. [DOI] [PubMed] [Google Scholar]
- Sidhu S.S., Lowman H.B., Cunningham B.C., Wells J.A. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Viera J., Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yonezawa M., Doi N., Kawahashi Y., Higashinakagawa T., Yanagawa H. Nucleic Acids Res. 2003;31:e118. doi: 10.1093/nar/gng119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X., Nguyen A.W., Jabaiah A., Sheff M.A., Thorn K.S., Daugherty P.S. Proc. Natl Acad. Sci. USA. 2006;103:18458–18463. doi: 10.1073/pnas.0605422103. [DOI] [PMC free article] [PubMed] [Google Scholar]