Abstract
The aim of this study was to investigate the role of the amino acid permease gene AAP6 in regulating phloem amino acid composition and then to determine the effects of this altered diet on aphid performance. A genotype of Arabidopsis thaliana (L.) was produced in which the function of the amino acid permease gene AAP6 (At5g49630) was abolished. Plants homozygous for the insertionally inactivated AAP6 gene had a significantly larger mean rosette width than the wild type and a greater number of cauline leaves. Seeds from the aap6 mutant were also significantly larger than those from the wild-type plants. Sieve element (SE) sap was collected by aphid stylectomy and the amino acids derivatized, separated, and quantified using Capillary Electrophoresis with Laser Induced Fluorescence (CE-LIF). In spite of the large variation across samples, the total amino acid concentration of SE sap of the aap6 mutant plants was significantly lower than that of the wild-type plants. The concentrations of lysine, phenylalanine, leucine, and aspartic acid were all significantly lower in concentration in the aap6 mutant plants compared with wild-type plants. This is the first direct demonstration of a physiological role for an amino acid transporter in regulating SE composition in vivo. The amino acid availability in sieve element sap is thought to be the major limiting factor for aphid growth and reproduction. Despite the changes in their diet, the aphid Myzus persicae (Sulzer) displayed only small changes in feeding behaviour on mutant plants when measured using the Electronic Penetration Graph (EPG) technique. Salivation by the aphid into the SE (E1 phase) was increased on mutant plants but there was no significant effect on other feeding EPG behaviours, or in the rate of honeydew production. Consistent with the small effect on aphid feeding behaviour, there was only a small effect of reduced sieve element amino acid concentration on aphid reproduction. The data are discussed in relation to the regulation of phloem composition and the role of phloem amino acids in regulating aphid performance.
Keywords: AAP6, amino acid, aphid, Arabidopsis thaliana, capillary electrophoresis, EPG, herbivore, Myzus persicae, phloem, sieve element
Introduction
The translocation of organic nutrients round the plant largely occurs in the phloem. Tissues exporting nutrients (‘sources’, such as photosynthesizing leaves) load them into the elongated sieve tubes that link different parts of the plant. Water follows by osmosis generating a hydraulically driven bulk flow of sap through files of sieve elements until they are unloaded in tissues that are net importers of nutrients (‘sinks’, such as the root system and the growing shoot tips). The pressures exerted in the translocation system are obvious since sap exudes from the severed stylets of phloem-feeding aphids (Pritchard, 1996). The translocation system is of vital importance to plants since the delivery of sugars, amino acids, and other nutrients provides the solutes required to drive cell elongation and the metabolites required both for respiration and for the synthesis of the macromolecules that form new structures. Regulation of the delivery of solutes to different parts of the plant is critical for a fundamental understanding of growth and development and informs attempts to manipulate resource allocation (e.g. during grain filling) in crop plants.
While there is information on the individual solute transporters in some plant species, there is no comprehensive information on the transport processes that regulate concentrations of a wide range of solutes at sources and sinks (Pritchard, 2007). The solutes present at the highest concentrations in the phloem are sucrose, amino acids, and K+ ions. There is an increasing amount of information about the phloem-located SUC2/SUT1 transporter that loads sucrose into sieve tubes (Stadler and Sauer, 1996; Doering Saad et al., 2002; Slewinski et al., 2009). Potassium ions are present at concentrations of between 50–150 mM and play a crucial role in the phloem transport of photoassimilates (Gould et al., 2004). For example, Arabidopsis thaliana mutant plants lacking K+ phloem channels (AKT2/3) were found to contain only half the sucrose content of the wild type. It is believed that they regulate the phloem osmotic potential which, in turn, controls the activity of sucrose transporters (Deeken et al., 2002). Other work has indentified mRNAs for aquaporins, nitrate transporters, ATPases, CNG channels, ABC, and sucrose transporters in the sieve element sap of barley and Ricinus (Doering Saad et al., 2002, 2006; Gaupels et al., 2008).
Amino acids form the second most abundant class of organic compounds found in the phloem sap after sucrose (Rentsch et al., 1998). Two main super-families of amino acid transporters have been identified so far: the amino acid, polyamine, and choline (APC) transporter super-family and the amino acid transporter (ATF) super-family (Fischer et al., 1995). The plant APCs are subdivided into two subgroups: the cationic amino acid transporters (CATs) and proteins most homologous to the yeast γ-aminobutyric acid (GABA) permease-related family (Fischer et al., 1998). The only transporter of the APC superfamily to have been characterized in any detail is AtCAT1 (Frommer et al., 1995). Recently, a bi-directional amino acid transporter, BAT1 has been identified in Arabidopsis and its location in vascular tissue has led to the suggestion that it may be involved in phloem unloading (Dündar and Bush, 2009).
Much more is known about members of the ATF family and, to date, five subclasses have been defined. These include the amino acid permeases (AAPs) of which eight members have been identified so far (Fischer et al., 1995, 1998; Okumoto et al., 2002), the lysine, histidine transporters (LHTs) such as LHT1 (Chen and Bush, 1997), the proline transporters (ProTs) such as ProT1 and ProT2 (Rentsch et al., 1996), the putative auxin transporters (AUXs) with AUX1 identified as an auxin carrier (Bennett et al., 1996), and the aromatic and neutral amino acid transporter (ANT1) which has been identified as a transporter of aromatic and neutral amino acids as well as arginine and auxin (Chen et al., 2001).
The AAPs (amino acid permeases) have been the most heavily studied group in the model species Arabidopsis thaliana. AAP1 was the first of the group to be characterized (Frommer et al., 1993; Hsu et al., 1993) and, subsequently, seven related permeases (AAP2–AAP8) have been reported in this species (Fischer et al., 1995; Kwart et al., 1993; Okumoto et al., 2002). Some have been associated with SE-specific transport of amino acids (Frommer et al., 1993; Kwart et al., 1993; Fischer et al., 1995, 2002; Hirner et al., 1998; Okumoto et al., 2002; Koch et al., 2003; Slewinski et al., 2009; Tedeger et al., 2007; Williams and Miller 2001).
AAP6 has been characterized as a high affinity transporter, efficiently transporting neutral amino acids and other acidic amino acids (Rentsch et al., 1996). AAP6 was expressed mainly in sink tissues such as roots, sink leaves, and cauline leaves, which depend on the supply of nitrogen from other organs. GUS expression studies revealed that AAP6 was also expressed in the xylem parenchyma cells of Arabidopsis (Okumoto et al., 2002). The xylem contains concentrations of amino acids that are typically orders of magnitude below that of phloem sap (Bi et al., 2007), thus movement of amino acids into the SE necessitates a high affinity transporter (Rentsch et al., 1996) leading to the hypothesis that a function of AAP6 is the transfer of amino acids from the xylem to the phloem (Okumoto et al., 2002).
As well as its central importance in plant growth and development, the phloem is also a key site for the interaction between plants and some sap-feeding insects. Aphids cause direct damage by withdrawing nutrients from the SEs; they also transmit viruses and the physical lesions and honeydew produced can encourage secondary infections. Aphid growth and reproduction are thought to be limited by the availability of nitrogen in the form of amino acids; a subset of nine amino acids are thought to be essential for aphid growth since they cannot be synthesized by the herbivore (Douglas, 2003; Wilkinson and Douglas, 2003).
In some studies, the nutritional quality of the sieve elements correlates positively with the performance and behaviour of the aphid herbivore. In potato, aphid performance increased at higher phloem amino acid levels (Karley et al., 2002). Similarly, aphids feeding on barley growing with additional nitrogen had a higher reproductive rate than those feeding on nutritionally deprived plants (Ponder et al., 2000). However, other studies have failed to demonstrate a positive correlation between SE amino acid concentrations and aphid performance. In a study on a range of native British grasses, SE amino acids increased following drought but aphid reproductive performance was reduced (Hale et al., 2003).
In this study, experiments utilizing a mutation at the AAP6 locus in A. thaliana are described. Having demonstrated that a T-DNA insertion results in the inactivation of the gene, the effects of this loss of function have been examined first at a general phenotypic level and then on the concentrations of amino acids within sieve elements. The latter experiments make use of the collection of sap from individual sieve elements followed by the quantification of individual amino acids in these small samples using a capillary electrophoresis-laser induced florescence (CE-LIF) protocol (Zhu et al., 2005). The effects of changes in sieve element amino acid levels have then been followed in the aphid Myzus persicae with regard to its detailed feeding behaviour, rate of ingestion of sieve element sap, and reproduction. The aims of this study are 2-fold: to determine the role of AAP6 in regulating SE amino acid composition and to test the relationship between the amino acid composition of aphid diet and performance in vivo.
Materials and methods
Plant material
An Arabidopsis thaliana genotype putatively carrying a copy of T-DNA within gene At5g49630 (the AAP6 gene) was identified in the Syngenta Arabidopsis Insertion Library (SAIL) collection (Garlic_1232_C08.b.1a.Lb3Fa) and obtained from the Nottingham Arabidopsis Stock Centre. Plants were grown in compost consisting of two parts John Innes loam-based compost, two parts peat-based compost, one part Silvaperl, and one part Osmocote (a slow release fertilizer) in a growth room maintained at 20–22 °C with an 18/6 h light/dark regime.
Genetic analysis of the aap6 mutant
The occurrence of a T-DNA insertion within an AAP6 allele was confirmed using AAP6-specific primers (AAP6F; CGTTGAACAGAGCTTCCCGGAGC, AAP6R; GGAGCCTGGAAAGGCTTGAAATCC) and a T-DNA-specific primer (SAIL LB3; CTGAATTTCATAACCAATCTCGATACAC). The exact T-DNA insertion site was determined by sequencing appropriate amplification products. Individuals homozygous for (i) the AAP6 allele carrying the T-DNA insertion, and (ii) the wild-type gene (‘negative segregants’) were initially identified by screening using PCR; tests were carried out on their progeny (following selfing) to confirm their homozygous status.
RT-PCR analysis
mRNA populations were extracted from the rosette leaves of wild-type and aap6 mutant Arabidopsis plants and used to produce single-stranded cDNA using the SuperScript II RNase H– Reverse Transcriptase (Invitrogen) following the manufacturer's protocol. These cDNA populations were used as template DNA in PCRs using (i) primers specific to a positive control transcript (actin2: ACT2For: 5′-TGCTGACCGTATGAGCAAAG; ACT2Rev: 5′-CAGCATCATCACAAGCATCC) to give a product of 400 bp, or (ii) to the AAP6 transcript (AAP6For: 5′-TCCCGGAGCATGAAATTGGC; AAP6Rev: 5′-GGCTTGAAATCCTTGAGACTTTG) to yield a product of 1400 bp. In both cases, the primers were designed to span at least one intron present in the gene so that artefactual amplification from the genomic DNA template would be detected. All PCR products were analysed using gel electrophoresis.
Southern blotting to test for T-DNA insertion number
DNA was extracted from wild-type and homozygous aap6 mutant rosette leaves and digested using HindIII before separation by electrophoresis in a 0.8% agarose gel and blotting onto a nitrocellulose filter. A probe was designed against a ∼350 bp region of the bar (BASTA) resistance gene, which is present in the T-DNA of SAIL lines but not elsewhere in the plant genome. This was amplified using primers BastaF (5′-CATGAGCCCAGAACGACGCC) and BastaR (5′-TCTTGAAGCCCTGTGCCTCC), ligated into pGEM-T, transformed into E. coli strain DH5α and its sequence verified. The bar fragment was excised from the pGEM-T plasmid and labelled with α-32P cytosine using the hexanucleotide labelling method (Feinberg and Vogelstein, 1983) and was used to hybridize to complementary DNA on the filter before subjection to autoradiography using Kodak X-O-Mat film (Amersham).
Developmental screening of the aap6 mutant
Wild-type and homozygous aap6 mutant plants were grown as described above in a random block design with a row of ‘guard’ plants around them. The stages of plant development were catalogued using the ‘principal growth stages’ of development indices developed by Boyes et al. (2001).
Measurements of seed volume were carried out as an indication of genotype fitness. Plants were bagged at 3–4-weeks-old (when flower buds became visible; growth stage 5.10) and not watered after 6–7 weeks (when the first siliques started to burst; growth stage 8.00). When drying was complete (growth stage 9.70), seed was collected, 100 seeds randomly selected from each individual plant, and the length and width of each seed measured under a microscope. The equation for the volume of an ellipse (4/3×width×height×length) was used to calculate the volume of the seeds.
Aphid material
Experimental aphids were taken from an anholocyclic Myzus persicae culture derived from a single individual and maintained at the University of Birmingham on well-watered A. thaliana (Columbia-0; Col-0) plants, grown in compost in a controlled environment growth room maintained at 20–22 °C with a 18/6 h L/D cycle. Only apterous aphids were used in the experiments. Prior to each experiment, first instar offspring from an individual aphid were taken from the culture and reared for two complete generations in sponge-sealed cylindrical clip cages on either homozygous aap6 mutant or wild-type A. thaliana plants in order to eliminate maternal effects on subsequent analyses.
Sieve element (SE) sap collection and analysis
Plants were grown until approximately 4 weeks old and bolting had occurred (principal growth stage 5.1–6.0). Cylindrical specimen cages (Agar Scientific Ltd) were secured to the stems and left for 2–3 d. Between 5 and 10 M. persicae adults were then placed in each cage and left overnight. Stylectomy was performed on feeding aphids using high-frequency microcautery (Downing and Unwin, 1977), as outlined in Hale et al. (2003). Stylectomy was always performed in daylight h and at 18–20 °C. Two sets of between 8 and 12 samples of exuding SE sap were collected from the severed stylet bundles of aphids feeding on individual homozygous aap6 mutant and wild-type plants: one set for amino acid analysis and the other for osmotic pressure measurements.
For amino acid analyses, SE sample droplets were expelled from microcapillaries into water-saturated paraffin oil (grade BP). Diameters of the suspended sample droplets were measured using a microscope. Sample volume was calculated using the formula for the volume of a sphere; V=(4/3) πr3. Oil-free samples were then collected in further microcapillaries and then expelled into sterile 0.5 ml microcentrifuge tubes. Samples were stored at –20 °C prior to analysis. Samples were air-dried, amino acids derivatized, and then separated and quantified using capillary electrophoresis with laser-induced fluorescence detection (CE-LIF) at the University of Nottingham as described previously (Zhu et al., 2005). Picolitre osmometry was used to measure osmotic pressure by recording the depression of melting temperature of samples compared to pure water (Tomos et al., 1994).
Aphid reproductive performance
Replicate aap6 mutant and wild-type plants were set up in a randomized block design in a controlled growth room (conditions as described above). Single, first instar aphid nymphs, less than 24 h old, were placed in clip cages on the stems of these plants at growth stage 6.0–6.5. The number of days until the onset of aphid reproduction (d) was recorded and the total number of offspring counted (Md) and removed daily for ‘d’ days. Performance was assessed by calculation of the intrinsic rate of increase (rm) (Wyatt and White, 1977) according to the formula rm=0.738×(lnMd)/d.
Aphid feeding behaviour and growth analysis
To study the detailed feeding behaviour of M. persicae, electrical penetration graphs (EPGs) were obtained using an 8-channel Direct Current (DC) system (Tjallingii and Hogen Esch, 1993). Adult, apterous M. persicae aphids were starved in a Petri dish for 1 h prior to experimentation. A 25 μm diameter gold wire was attached to the dorsum of each aphid using silver conductive paint (Ponder et al., 2001; Hale et al., 2003). Each aphid was placed onto the stem of a plant (aap6 mutant or wild type, as above). Stylet 2.5® software (WF Tjallingii, Wageningen, The Netherlands) was used for data acquisition. The following aspects of aphid feeding behaviour were recorded: (i) total duration of E2 periods (ingestion from SE); (ii) mean frequency of G periods (xylem feeding); (iii) duration of G periods; (iv) mean duration of individual E1 periods (salivation into SE followed by E2); (v) length of time to reach E2; (vi) duration of non-probing activities; and (vii) proportion of probes containing E2 periods.
Honeydew production was measured by clamping plants horizontally suspended over rotating Petri dishes for collection (Hale et al., 2003). Honeydew droplets were collected between 20.00 h and 08.00 h, at a temperature of 20–22 °C. The number of honeydew droplets in each dish was counted using a microscope. The mean honeydew droplet volumes for aphids feeding on several individuals of each of the two experimental plant types were calculated by measuring the diameter of 20–25 honeydew droplets expelled directly into silicone oil (200/50 cS, Dow Corning Corporation, USA). The rate of droplet production was combined with mean droplet volumes to obtain the mean hly volume of honeydew produced by aphids. Honey dew amino acid concentration was measured in the same manner as SE sap.
Adult aphid growth rates were assessed by caging preweighed groups of 10 adult aphids on Arabidopsis plants and reweighing them after 12 h. Nine independent replicates were used for aphids feeding on each of the wild-type and aap6 mutant plants. To assess weight loss due to respiration and water loss, nine groups of 10 adult aphids were taken off plants and left for 30 min to expel any residual honeydew. They were then weighed, left for 4 h in a Petri dish, and reweighed after 4 h.
Aphid haemolymph collection and analysis
Aphids were placed under paraffin oil and a leg excised using the stylectomy needle. Exuding haemolymph was collected in glass capillaries and stored and analysed in the same manner as SE sap.
Statistical analyses
The Student t test was used to detect significant differences between aap6 mutant and wild-type plants, or aphids feeding on them, with respect to data relating to whole plant development, silique production, seed volume, osmotic pressures of SE sap and honeydew, aphid reproductive performance, aphid feeding rate, and the concentrations of total, essential, non-essential, and individual amino acids in SE sap, honeydew, and haemolymph. Where necessary, these data were first logarithmically transformed to achieve normalization (assessed by the Ryan–Joiner test for normality).
Pair-wise comparisons of the different feeding parameters measured by EPG were made between aphids on aap6 mutant and wild-type plants using the non-parametric Mann-Whitney U-test (α=0.05) since the data sets were not normally distributed and transforming the data did not make the data sufficiently normal.
Results
Arabidopsis genotype analysis
Leaf DNA was extracted from individuals of Arabidopsis thaliana that putatively contained a mutant allele (with a T-DNA insertion) and from Col-0 (the background genotype of the putative mutant). This was used as the template in PCRs using a combination of primers specific to either the AAP6 locus or to the T-DNA used for mutagenesis. It was confirmed that individuals of the SAIL line used contain a T-DNA insertion (within intron 2). To determine the effect of the T-DNA insertion in the aap6 locus, the abundance of its transcript was assessed using RT-PCR. Although the predicted 1400 bp amplification product of the AAP6 transcript was readily detectable in extracts from the wild-type rosette leaves, no such product was amplified from aap6 mutant samples (Fig. 1). Amplification products representing a part of a positive control transcript (an actin mRNA) were detected in both wild-type and aap6 mutant samples.
Fig. 1.
RT-PCR analysis on cDNA obtained from aap6 mutant and wild-type rosette leaves using: (a) AAP6 gene-specific primers with aap6 cDNA; (b) AAP6 gene-specific primers with WT cDNA; (c) actin2-specific primers with aap6 cDNA; (d) actin2-specific primers with WT cDNA. The four lanes show PCR amplification after 20, 22, 24, and 26 cycles.
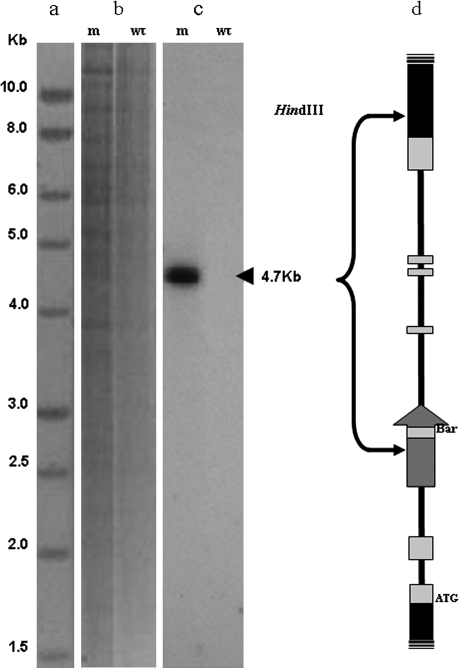
Southern blot analysis was carried out using genomic DNA extracts from mutant aap6 leaves in order to determine the number of T-DNA inserts within the genome of this line. The HindIII-digested DNA samples were probed with a portion of the bar gene, which forms part of the T-DNA construct used to produce the SAIL insertion library. The results (Fig. 2) are consistent with the occurrence of a single T-DNA insertion into the genome and the size of the hybridizing restriction fragment is the same as that predicted by the allele model presented (Fig. 2d) where 4.7 kbp separate HindIII restriction sites within the T-DNA and the AAP6 gene.
Fig. 2.
Southern blot analysis of Arabidopsis genomic DNA using a probe to the BASTA (Bar) gene on the T-DNA insertion: (a) 1 kb ladder. (b) Image of ethidium bromide-stained agarose gel showing genomic DNA digested with HindIII. (c) Autoradiograph revealing hybridization to a single 4.7 kb fragment in the mutant (m) but not the wild type (wt) line. (d) A model of the mutant aap6 allele showing exons (light grey boxes), introns (solid black lines), the position and orientation of the T-DNA insertion including the bar gene, and the positions of two HindIII sites which result in the production of a 4.7 kb DNA fragment to which the bar probe hybridizes.
Effects of abolition of the function of AAP6 on plant phenotype
Whole plant
There were no obvious developmental differences between wild-type and homozygous aap6 mutant plants. However, measurement of quantitative characters revealed some small but significant changes (Table 1). For example, at the time of flowering, the mean rosette width of the aap6 mutant was significantly larger than the wild type and it had a greater number of cauline leaves. Seeds from the aap6 mutant were significantly larger (0.061 mm3) than those from the wild-type plants (0.056 mm3) (P=0.01).
Table 1.
Randomised plot trial of quantitative traits of homozygous aap6 mutant (n=18) and wild type (n=18) plants. Mean values (± standard errors) are given for each genotype; p values show the results of t-tests and are marked ‘*’ if significant.
| aap6 loss of function mutant | Wild type (Col-0) | P-value | |
| Germination in days | 4.7 (0.2) | 4.3 (0.1) | 0.20 |
| Rosette leaf number on day 20 | 8.8±0.3 | 9.1±0.2 | 0.46 |
| Rosette width on day 20 (mm) | 31.0±2.2 | 29.3±1.6 | 0.54 |
| Height on day 30 (mm) | 206.5±19 | 219.7±10 | 0.55 |
| Height on day 35 (mm) | 339.4±15 | 336.9±7.3 | 0.88 |
| Days to flowering | 21.4±0.4 | 21.1±0.4 | 0.50 |
| Height at flowering (mm) | 57.7±3.3 | 52.7±2.5 | 0.24 |
| Rosette leaf number at flowering | 11.39±0.29 | 10.89±0.23 | 0.19 |
| Number of cauline leaves at flowering | 2.94±0.10 | 2.44±0.15 | 0.008* |
| Rosette width at flowering (mm) | 66.1±1.6 | 60.6±0. | 0.005* |
| Number of siliques on day 35 | 54.7±8.7 | 57.1±7.5 | 0.84 |
| Seed Volume (mm3) | 0.061±0.0012 | 0.056±0.0011 | 0.01* |
Sieve element
The total amino acid concentration of SE sap of aap6 plants was significantly lower than that of the wild-type plants (56.04±9.30 mM and 184.71±60.45 mM, respectively; P=0.032, Fig. 3). Similarly, the mean concentrations of essential and non-essential amino acid concentrations in SE sap of aap6 mutant plants were significantly lower than those of the wild types.
Fig. 3.
Mean concentrations (mM ±SE) of amino acids in sieve element sap of wild-type and aap6 mutant plants collected by aphid stylectomy and measured using CE-LIF. Insert indicates the total concentration of amino acids. Asterisks denote significant differences at P <0.05 in the concentration (mM) of amino acids between wild-type and aap6 mutant plants.
Sixteen amino acids were individually identified and quantified; histidine and valine co-eluted and are reported together (Fig. 3). Lysine, phenylalanine, leucine, and aspartic acid were all significantly lower in concentration in aap6 mutant plants compared with wild-type plants, being reduced to 27%, 29%, 32%, and 20% of the wild-type values respectively. The large variances resulted in non-significance of the differences between the mean values of the remaining amino acids.
The mean osmotic pressures of SE sap of the homozygous aap6 mutant and wild-type plants were not significantly different (0.80±0.04 MPa and 0.73±0.04 MPa, respectively).
Effects of the plant mutation on aphid phenotype
Aphid reproductive performance
Despite the large reduction in SE amino acids, aphid reproductive performance, assessed as rm, was only slightly reduced when feeding on aap6 mutant plants (0.268±0.008 compared with 0.243±0.008, respectively; P=0.039).
Aphid feeding behaviour
The results from the EPG experiments analysing aphid feeding behaviour on wild-type and aap6 mutant plants are summarized in Table 2. The mean duration of individual ‘successful’ E1 periods (the salivation period which precedes SE sap ingestion by the aphid) was significantly greater for aphids feeding on aap6 mutant plants than on wild-type plants (P=0.01). There were no other differences in any of the parameters measured for aphids feeding on the two genotypes (P >0.05).
Table 2.
Summary of EPG parameters measured
| Wild type | aap6 mutant | |
| (n=15) | (n=13) | |
| Mean frequency of | ||
| Potential drops (pds) | 76.2±10.7 | 74.5±12.6 |
| Non-probing behaviour | 4.7±1.2 | 4.4±1.0 |
| E1 (total) | 1.9±0.4 | 2.0±0.5 |
| E1 (followed by E2 >10 min) | 1.5±0.3 | 1.3±0.3 |
| E2 periods | 1.7±0.3 | 1.5±0.4 |
| E2 periods (>10 min) | 1.5±0.3 | 1.3±0.3 |
| Xylem feeding (G) | 0.4±0.3 | 0.5±0.1 |
| Mean total duration (s) of | ||
| Non-probing behaviour | 1905.9±742.0 | 832.2±265.3 |
| Pathway activities | 4727.6±849.8 | 5390.2±900.9 |
| E1 (total) | 226.0±139.9 | 571.7±283.2 |
| E1 (followed by E2) | 57.5±11.5 | 224.8±114.7 |
| E2 periods | 14361.6±1675.5 | 12485.0±1941.3 |
| E2 periods (>10 min) | 15381.6±1373.6 | 14693.9±1328.1 |
| Xylem feeding (G) | 386.6±302.7 | 2372.5±1201.1 |
| Mean duration (s) of individual | ||
| E1 (followed by E2) | 34.7±3.5 | 171.9±79.2 |
| Mean time (s) from start of experiment to | ||
| First E1 | 5328.1±1342.8 | 5642.2±1729.7 |
| First E2 (>10 min) | 6458.9±1681.8 | 7949.2±1991.9 |
| Proportion of probes containing | ||
| E2 periods (>10 min) | 0.49±0.12 | 0.37±0.09 |
The mean duration of individual ‘successful’ E1 periods (the salivation period which precedes SE sap ingestion by the aphid) was significantly greater for aphids feeding on aap6 mutant plants than on wild-type plants (P=0.01).
Honeydew production
The absence of large differences in the EPG feeding behaviour between aphids feeding on wild type and aap6 mutants was paralleled by parity in honeydew production. Aphids feeding on wild-type plants produced 2.5±0.2 droplets h−1, compared with 2.5±0.3 droplets h−1 for aphids feeding on aap6 mutant plants. Similarly, there was no difference in the mean volumes of individual honeydew droplets produced by aphids feeding on wild-type plants (10.3±2.7 nl) and aap6 mutants (11.5±3.1 nl). The rates of droplet production and volumes were combined to compare the rate of honeydew volume production (Fig. 4). Overall, there was no difference between the volume of honeydew produced h−1 for aphids feeding on wild-type and aap6 mutant plants.
Fig. 4.
Rate of honeydew production by aphids feeding on wild type or aap6 mutants over a 12 h period commencing at 08.00 h. Each point is the mean of 14 determinations of droplet production rate ±SE. ANOVA indicated no significant difference in honeydew production over time or between genotypes.
Honeydew amino acids
The mean total concentration of amino acids in the honeydew from aphids feeding on aap6 mutant plants was 17.1±10.5 mM compared to 2.2±0.8 mM for those feeding on wild-type plants (Fig. 5); these differences were not significant (P=0.157). There were also no differences in the mean concentrations of essential, non-essential or individual amino acids in the honeydew produced by aphids feeding on each plant type (P >0.05). The mean osmotic pressures of honeydew droplets produced by aphids feeding on wild-type and aap6 plants were similar at 0.84±0.03 MPa and 0.78±0.03 MPa, respectively. In addition, there were no differences in the concentrations of total, essential and non-essential amino acids in the haemolymph of aphids feeding on the two plant genotypes (data not shown, P >0.05) nor in the concentrations of individual amino acids.
Fig. 5.
Mean concentrations (mM ±SE) of amino acids in honeydew collected from aphids feeding on wild-type (n=11) and aap6 (n=10) plants. Insert indicates the total concentration of amino acids. No differences were significant at P <0.05.
Discussion
This study demonstrated a role of the AAP6 transporter in the regulation of sieve element amino acid composition. Despite the change in their diet, there was no large change in the metabolism, behaviour or reproduction of phloem-feeding aphids. The direct measurement of the concentrations of amino acids in sieve element sap has, until recently, been difficult. Aphid stylectomy provides pure sap from single sieve elements, but only provides nanolitre sample volumes. However, the development of the CE-LIF protocol for amino acid quantification has provided a method for measuring concentrations within such small samples (Zhu et al., 2005). The technique involves derivatization of the amino acids, separation by capillary electrophoresis, and detection of the fluorescent analytes by laser induced fluorescence. Using this technique with wheat, it has recently been shown that there is great variation in the concentrations of amino acids in individual sieve elements of the same plant (Gattolin et al., 2008) and this necessitates the collection of large numbers of samples and careful statistical analysis when two genotypes are to be compared.
In the present study, the abolition of AAP6 function reduced the mean total amino acid concentration of SE sap to around 30% of the concentration of the wild type. Despite care taken to sample SE from the same site on the bolting stem from plants of the same age there was large variation in SE amino acid concentration. The previous work in wheat had indicated that such variation had a biological, not an experimental, origin and it was hypothesized that the basis of this variation lies at the regulation of the individual SE-CCC or domain (Gattolin et al., 2008). However, in the present study, despite similar variation in Arabidopsis SE amino acid concentrations, significant reductions were detected in the levels of lysine, phenylalanine, leucine, and aspartic acid. The reduction in the total amino acid concentration of SE sap is consistent with the proposal that the AAP6 transporter moves amino acids from the xylem to the SE (Okumoto et al., 2002). Okumoto et al. (2002) located AAP6 to xylem parenchyma of both roots and leaves of plants prior to bolting. In the present study, sap collection was performed on bolting stems. The two studies are consistent since reduced loading of amino acids in rosette leaf xylem parenchyma in the AAP6 knockout will result in a decrease in amino acids in the SE of the stem supplying the developing seed.
In Arabidopsis, as with other plants, there are multiple AAP transporters with overlapping specificities, so that it is not surprising that not all neutral and acidic amino acids were affected. It has been reported that the AAP6 gene encodes a transporter of aspartic acid as well as other neutral and acidic amino acids (Rentsch et al., 1996; Fischer et al., 2002; Okumoto et al., 2002). In the present study, SE aspartic acid concentration was decreased by 80% in the aap6 mutant. In addition, the neutral amino acids phenylalanine and leucine were also significantly reduced. The significant decrease in lysine implies that the AAP6 transporter is also able to transport some basic amino acids.
In spite of the clear reduction in total amino acid concentration within the aap6 mutants, the morphological phenotype and general development of the plants was little changed when grown under ‘ideal’ conditions. Phloem translocation is achieved by the hydraulically driven flow of sap through sieve tubes (Pritchard, 2007). Since SE osmotic pressure and whole plant phenotype was largely unchanged despite the large reduction in SE amino acid concentration, it is likely that the levels of other SE solutes (e.g. sucrose or cations) were elevated to maintain SE osmotic pressure and therefore translocation efficiency. However, there were some small but significant differences between the two genotypes. The mutant had a larger mean rosette width and a greater number of cauline leaves than the wild type. While seed number was unchanged in mutant plants (data not shown), seed volume was significantly greater in the aap6 mutant plants. This is consistent with a role for AAP6 gene function in the allocation of resources to seeds; however, there was no significant effect on the number of siliques produced (data not shown). A seed-related phenotype is not restricted to aap6 plants; for example, a loss of function mutant in AAP8 significantly reduced seed number in Arabidopsis (Schmidt et al., 2007). It is not obvious how loss of function in the AAP6 transporter would increase allocation to seeds, especially as SE amino acids were reduced. Increased seed volume may represent a secondary effect, due to, for example, a larger rosette diameter increasing capture and allocation of resources.
Sieve element sap is the major component of aphid diet and its composition can affect aphid growth. Generally, SE sap represents a non-ideal diet for aphids, being hyperosmotic and low in amino acids (Douglas, 2006). In addition to such direct effects, SE composition may influence aphid performance by affecting location and acceptance by a probing aphid. Despite the large variation in SE concentration, aphids on the different genotypes would experience altered amino acid concentration in their diet since over the period of the rm measurement they would sample many individual SEs. The significant reduction in the average SE amino acid concentration between the wild type and the mutant was also determined from an average of a number of SEs. Therefore, despite the variation in SE amino acids, aphids on the aap6 knockout mutant would indeed receive a reduced amino acid concentration in their diet. The alteration in diet did not greatly affect aphids feeding on the aap6 mutant; rm was slightly but significantly reduced from 0.268 to 0.243. The aphid Rhopalosiphum padi feeding on barley grown under low nitrogen conditions had its rm reduced from 0.46 to 0.41 (Ponder et al., 2000). By contrast, Arabidopsis with a mutation in the amino acid carrier ANT1 had an altered SE amino acid composition but no significant change in rm (Hunt et al., 2006).
Despite the absence of large effects on reproduction, there were some small changes in aphid feeding behaviour. While there was no difference in the time taken ingesting sap from the SE (E2 phase), Myzus persicae spent an increased length of time salivating into the SEs of aap6 mutant plants (E1 phase), suggesting the reduced acceptance of SE sap on the mutant plants. During E1, aphids inject a watery saliva into the SE which is proposed to condition the sap at the feeding site (Tjallingii and Hogen Esch, 1993). Aphids feeding from plants on which they perform well, salivate into the sieve element for a shorter period of time before initiating sustained ingestion (Van Helden and Tjallingii, 1993; Wilkinson and Douglas, 1998). Aphid saliva has recently been demonstrated to overcome SE blockage by forisomes (Will et al., 2007). In addition to overcoming direct defence, some aphids are able to increase the nutritional quality of their diet, sometimes by altering SE amino acid composition (Prado and Tjallingii, 1997), with a role hypothesized for E1 salivation. Ponder et al. (2000) reported an increase in the time spent salivating (E1) prior to the onset of ingestion (E2) by aphids feeding on nitrogen-deficient barley.
Whilst the electrical penetration graph method system determines the duration of phloem ingestion (E2) feeding, it does not directly measure the rate of sap ingestion; this was achieved by measuring honeydew production. Since the amino acid content of the SE was reduced in the aap6 mutant, a compensatory change in the rate of SE sap ingestion might have been expected. However, this was not observed, since M. persicae feeding on wild-type and aap6 mutant plants produced similar amounts of honeydew. Amino acid analyses of the honeydew confirmed that the efficiency with which aphids are able to remove these compounds from their diet was similar between the two genotypes since there was no significant difference in the amino acid content of honeydew or haemolymph.
Previous studies have shown that nitrogen can positively influence aphid reproductive performance (Van Emden, 1966; Harrewijn, 1970; Ponder et al., 2000, 2001; Sandström, 2000; Karley et al., 2002). It is generally assumed and often empirically demonstrated that increased dietary amino acids correlated with increased performance. It was therefore predicted that aphids feeding on the aap6 knockout would have reduced performance. However, there was no large difference in reproductive performance despite the significant change in SE amino acid composition. Other studies have reported a lack of a clear positive correlation between SE amino acid composition and reproductive rate. In droughted grasses, an increase in SE amino acids was accompanied by a decrease in aphid performance (Hale et al., 2003) while an increase in the proportions of certain SE amino acids in the Arabidopsis mutant ANT1 did not result in any change in aphid fitness (Hunt et al., 2006). It is becoming clear that prediction of aphid performance is more complex that a simple correlation with the nitrogen content of the diet, with many other factors potentially contributing to output. For example, a study using artificial diets concluded that the presence of secondary plant compounds in the diet was more important in determining aphid fitness than amino acid concentration (Tosh et al., 2003). The cost of osmoregulation is receiving more attention (Shakesby et al., 2009), but this is unlikely to be a factor in the present study as there was no significant difference in the osmotic pressures of the sieve element sap between the wild type and the mutant.
It may be that the knockout of AAP6 did not reduce SE amino acids below the critical threshold for aphid performance; this may be possible in plants containing knockouts of two or more amino acid transporters in order to alter dietary nutritional quality to a greater magnitude, but this may also grossly impair plant function. The contribution of essential amino acids to the aphid by the bacterial symbiont Buchnera is critically important in determining the relationship between aphid performance and diet (Douglas, 2003); the response of aphids without these symbionts to dietary changes may reveal more direct relationships than can be observed in their presence (Douglas et al., 2001).
References
- Bennett MJ, Marchant A, Green HG, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Bi JL, Castle SJ, Toscano NC. Amino acid fluctuations in young and old orange trees and their influence on glassy-winged sharpshooter (Homalodisca vitripennis) population densities. Journal of Chemical Ecology. 2007;33:1692–1706. doi: 10.1007/s10886-007-9346-6. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of arabidopsis: a model for high throughput functional genomics in plants. The Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Bush DR. LHT1, lysine- and histidine-specific amino acid transporter in Arabidopsis. Plant Physiology. 1997;115:1127–1134. doi: 10.1104/pp.115.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Ortiz-Lopez A, Jung A, Bush DR. ANT1, an aromatic and neutral amino acid transporter in Arabidopsis. Plant Physiology. 2001;125:1813–1820. doi: 10.1104/pp.125.4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Geiger D, Fromm J, Koroleva O, Ache P, Langenfeld-Heyser R, Sauer N, May ST, Hedrich R. Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta. 2002;216:334–344. doi: 10.1007/s00425-002-0895-1. [DOI] [PubMed] [Google Scholar]
- Douglas AE. The nutritional physiology of aphids. Advances in Insect Physiology. 2003;31:73–140. [Google Scholar]
- Douglas AE. Phloem-sap feeding by animals: problems and solutions. Journal of Experimental Botany. 2006;57:747–754. doi: 10.1093/jxb/erj067. [DOI] [PubMed] [Google Scholar]
- Douglas AE, Minto LB, Wilkinson TL. Quantifying nutrient production by the microbial symbionts in an aphid. Journal of Experimental Biology. 2001;204:349–358. doi: 10.1242/jeb.204.2.349. [DOI] [PubMed] [Google Scholar]
- Doering-Saad C, Newbury HJ, Bale JS, Pritchard J. Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. Journal of Experimental Botany. 2002;53:631–637. doi: 10.1093/jexbot/53.369.631. [DOI] [PubMed] [Google Scholar]
- Doering-Saad C, Newbury HJ, Couldridge CE, Bale JS, Pritchard J. A phloem-enriched cDNA library from Ricinus: insights into phloem function. Journal of Experimental Botany. 2006;57:3183–3193. doi: 10.1093/jxb/erl082. [DOI] [PubMed] [Google Scholar]
- Downing N, Unwin DM. A new method for cutting the mouthparts of feeding aphids, and for collecting plant sap. Physiological Entomology. 1977;2:275–277. [Google Scholar]
- Dündar E, Bush DR. BAT1, a bidirectional amino acid transporter in Arabidopsis. Planta. 2009;229:1047–1056. doi: 10.1007/s00425-009-0892-8. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling dna restriction endonuclease fragments to high specific activity. Analytical Biochemistry. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB. Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. Journal of Biological Chemistry. 1995;270:16315–16320. doi: 10.1074/jbc.270.27.16315. [DOI] [PubMed] [Google Scholar]
- Fischer WN, Andre B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, Frommer WB. Amino acid transport in plants. Trends in Plant Science. 1998;3:188–195. [Google Scholar]
- Fischer WN, Loo DDF, Koch W, Ludewig U, Boorer KJ, Tegeder M, Rentsch D, Wright EM, Frommer WB. Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. The Plant Journal. 2002;29:717–731. doi: 10.1046/j.1365-313x.2002.01248.x. [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Riesmeier JW. Expression cloning in yeast of a cDNA encoding a broad specificity amino acid permease from Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1993;90:5944–5948. doi: 10.1073/pnas.90.13.5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Unseld M, Ninnemann O. Seed and vascular expression of a high-affinity transporter for cationic amino acids in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1995;92:12036–12040. doi: 10.1073/pnas.92.26.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattolin S, Newbury HJ, Bale JS, Tseng HM, Barrett DA, Pritchard J. A diurnal component to the variation in sieve tube amino acid content in wheat. Plant Physiology. 2008;147:912–921. doi: 10.1104/pp.108.116079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupels F, Buhtz A, Knauer T, Deshmukh S, Waller F, van Bel AJE, Kogel KH, Kehr J. Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. Journal of Experimental Botany. 2008;59:3297–3306. doi: 10.1093/jxb/ern181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould N, Thorpe MR, Minchin PEH, Pritchard J, White PJ. Solute is imported to elongating root cells of barley as a pressure driven-flow of solution. Functional Plant Biology. 2004;31:391–397. doi: 10.1071/FP03231. [DOI] [PubMed] [Google Scholar]
- Hale BK, Bale JS, Pritchard J, Masters GJ. Effects of host plant drought stress on the performance of the bird cherry-oat aphid, Rhopalosiphum padi (L.): a mechanistic analysis. Ecological Entomology. 2003;28:666–677. [Google Scholar]
- Harrewijn P. Reproduction of the aphid Myzus persicae related to the mineral nutrition of potato plants. Entomologia Experimentalis et Applicata. 1970;13:307–319. [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB. Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. The Plant Journal. 1998;14:535–544. doi: 10.1046/j.1365-313x.1998.00151.x. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Chiou TJ, Chen L, Bush DR. Cloning a plant amino acid transporter by functional complementation of a yeast amino acid transport mutant. Proceedings of the National Academy of Sciences, USA. 1993;90:7441–7445. doi: 10.1073/pnas.90.16.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt EJ, Pritchard J, Bennett MJ, Zhu X, Barrett DA, Allen T, Bale JS, Newbury HJ. The Arabidopsis thaliana/Myzus persicae model system demonstrates that a single gene can influence the interaction between a plant and a sap-feeding insect. Molecular Ecology. 2006;15:4203–4213. doi: 10.1111/j.1365-294X.2006.03090.x. [DOI] [PubMed] [Google Scholar]
- Karley AJ, Douglas AE, Parker WE. Amino acid composition and nutritional quality of potato leaf phloem sap for aphids. Journal of Experimental Biology. 2002;205:3009–3018. doi: 10.1242/jeb.205.19.3009. [DOI] [PubMed] [Google Scholar]
- Koch W, Kwart M, Laubner M, Heineke D, Stransky H, Frommer WB, Tegeder M. Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. The Plant Journal. 2003;33:211–220. doi: 10.1046/j.1365-313x.2003.01618.x. [DOI] [PubMed] [Google Scholar]
- Kwart M, Hirner B, Hummel S, Frommer WB. Differential expression of two related amino acid transporters with differing substrate specificity in Arabidopsis thaliana. The Plant Journal. 1993;4:993–1002. doi: 10.1046/j.1365-313x.1993.04060993.x. [DOI] [PubMed] [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W. High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. Journal of Biological Chemistry. 2002;277:45338–45346. doi: 10.1074/jbc.M207730200. [DOI] [PubMed] [Google Scholar]
- Ponder KL, Pritchard J, Harrington R, Bale JS. Difficulties in location and acceptance of phloem sap combined with reduced concentration of phloem amino acids explain lowered performance of the aphid Rhopalosiphum padi on nitrogen deficient barley (Hordeum vulgare) seedlings. Entomologia Experimentalis et Applicata. 2000;97:203–210. [Google Scholar]
- Ponder KL, Pritchard J, Harrington R, Bale JS. Feeding behaviour of the aphid Rhopalosiphum padi (Hemiptera: Aphididae) on nitrogen and water-stressed barley (Hordeum vulgare) seedlings. Bulletin of Entomological Research. 2001;91:125–130. [PubMed] [Google Scholar]
- Prado E, Tjallingii WF. Effects of previous plant infestation on sieve element acceptance by two aphids. Entomologia Experimentalis et Applicata. 1997;82:189–200. [Google Scholar]
- Pritchard J. Aphid stylectomy reveals an osmotic step between sieve tube and cortical cells in barley roots. Journal of Experimental Botany. 1996;47:1519–1524. [Google Scholar]
- Pritchard J. Solute transport in the phloem. In: Flowers TJ, Yeo AR, editors. Plant solute transport. Oxford, UK: Blackwell Publishing; 2007. pp. 235–309. [Google Scholar]
- Rentsch D, Boorer KJ, Frommer WB. Structure and function of plasma membrane amino acid, oligopeptide and sucrose transporters from higher plants. Journal of Membrane Biology. 1998;162:177–190. doi: 10.1007/s002329900355. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. The Plant Cell. 1996;8:1437–1446. doi: 10.1105/tpc.8.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström J. Nutritional quality of phloem sap in relation to host plant alternation in the bird cherry-oat aphid. Chemoecology. 2000;10:17–24. [Google Scholar]
- Schmidt R, Stransky H, Koch W. The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta. 2007;12:805–813. doi: 10.1007/s00425-007-0527-x. [DOI] [PubMed] [Google Scholar]
- Shakesby AJ, Wallace IS, Isaacs HV, Pritchard J, Roberts DM, Douglas AE. A water-specific aquaporin involved in aphid osmoregulation. Insect Biochemistry and Molecular Biology. 2009;39:1–10. doi: 10.1016/j.ibmb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Slewinski TL, Meeley R, Braun DM. Sucrose transporter1 functions in phloem loading in maize leaves. Journal of Experimental Botany. 2009;60:881–892. doi: 10.1093/jxb/ern335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N. Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Botanica Acta. 1996;109:299–306. [Google Scholar]
- Tegeder M, Tan Q, Grennan AK, Patrick JW. Amino acid transporter expression and localization studies in pea (Pisum sativum) Functional Plant Biology. 2007;34:1019–1028. doi: 10.1071/FP07107. [DOI] [PubMed] [Google Scholar]
- Tjallingii WF, Hogen Esch T. Fine structure of aphid stylet route in plant tissues in correlation with EPG signals. Physiological Entomology. 1993;18:317–328. [Google Scholar]
- Tomos AD, Hinde P, Richardson P, Pritchard J, Fricke W. Microsampling and measurements of solutes in single cells. In: Harris N, Oparka KJ, editors. Plant cell biology: a practical approach. Oxford University Press; 1994. pp. 297–314. [Google Scholar]
- Tosh CR, Powell G, Holmes ND, Hardie J. Reproductive response of generalist and specialist aphid morphs with the same genotype to plant secondary compounds and amino acids. Journal of Insect Physiology. 2003;49:1173–1182. doi: 10.1016/j.jinsphys.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Van Emden HF Studies on the relations of insect and host plant. III. A comparison of the reproduction of Brevicoryne brassicae and Myzus persicae (Homoptera: Aphididae) on Brussel's sprout plants supplied with different rates of nitrogen and potassium. Entomologia Experimentalis et Applicata. 1966;9:444–460. [Google Scholar]
- Van Helden M, Tjallingii WF. Tissue localization of lettuce resistance to the aphid Nasonovia ribisnigri using electrical penetration graphs. Entomologia Experimentalis et Applicata. 1993;68:269–278. [Google Scholar]
- Will T, Tjallingii WF, Thonnessen A, van Bel AJE. Molecular sabotage of plant defense by aphid saliva. Proceedings of the National Academy of Sciences, USA. 2007;104:10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson TL, Douglas AE. Plant penetration by pea aphids (Acyrthosiphon pisum) of different plant range. Entomologia Experimentalis et Applicata. 1998;87:43–50. [Google Scholar]
- Wilkinson TL, Douglas AE. Phloem amino acids and the host plant range of the polyphagous aphid, Aphis fabae. Entomologia Experimentalis et Applicata. 2003;106:103–113. [Google Scholar]
- Williams LE, Miller AJ. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:659–688. doi: 10.1146/annurev.arplant.52.1.659. [DOI] [PubMed] [Google Scholar]
- Wyatt IJ, White PF. Simple estimation of intrinsic rates from aphids and tertanyxhid mites. Journal of Applied Ecology. 1977;14:757–766. [Google Scholar]
- Zhu X, Shaw PN, Pritchard J, Newbury J, Hunt EJ, Barrett DA. Amino acid analysis by micellar electrokinetic chromatography with laser-induced fluorescence detection: Application to nanolitre-volume biological samples from Arabidopsis thaliana and Myzus persicae. Electrophoresis. 2005;26:911–919. doi: 10.1002/elps.200410259. [DOI] [PubMed] [Google Scholar]