Abstract
According to the classical ABC model, B-function genes are involved in determining petal and stamen development. Most core eudicot species have B class genes belonging to three different lineages: the PI, euAP3, and TM6 lineages, although both Arabidopsis and Antirrhinum appear to have lost their TM6-like gene. Functional studies were performed for three gerbera (Gerbera hybrida) B class MADS-box genes—PI/GLO-like GGLO1, euAP3 class GDEF2, and TM6-like GDEF1—and data are shown for a second euAP3-like gene, GDEF3. In phylogenetic analysis, GDEF3 is a closely related paralogue of GDEF2, and apparently stems from a duplication common to all Asteraceae. Expression analysis and transgenic phenotypes confirm that GGLO1 and GDEF2 mediate the classical B-function since they determine petal and stamen identities. However, based on assays in yeast, three B class heterodimer combinations are possible in gerbera. In addition to the interaction of GGLO1 and GDEF2 proteins, GGLO1 also pairs with GDEF1 and GDEF3. This analysis of GDEF1 represents the first functional characterization of a TM6-like gene in a core eudicot species outside Solanaceae. Similarly to its relatives in petunia and tomato, the expression pattern and transgenic phenotypes indicate that GDEF1 is not involved in determination of petal identity, but has a redundant role in regulating stamen development.
Keywords: Asteraceae, evo–devo, flower development, organ identity
Introduction
In core eudicots, the identity of petals and stamens is known to be specified by B class MADS-box genes. The best studied model species, Arabidopsis (Arabidopsis thaliana) and Antirrhinum (Antirrhinum majus), have two B genes: PISTILLATA (PI) and APETALA3 (AP3), and GLOBOSA (GLO) and DEFICIENS (DEF), respectively. The two proteins interact to form a heterodimer that is required for their function in DNA binding (Schwarz-Sommer et al., 1992; Tröbner et al., 1992; Goto and Meyerowitz, 1994). Phylogenetic reconstructions show that PI/GLO- and AP3/DEF-like genes form separate gene lineages that arose from a duplication event that occurred before the rise of modern angiosperms (Kim et al., 2004; Hernandez-Hernandez et al., 2007). The AP3/DEF lineage has undergone another duplication event at the base of core eudicots, resulting into two paralogous lineages, euAP3 and TOMATO MADS BOX GENE6 (TM6). The euAP3 and TM6 lineage genes encode specific C-terminal motifs, called the euAP3 and paleoAP3 motif (Kramer et al., 1998, 2004; Kramer and Irish, 2000). The novel euAP3 motif seems to have originated from the ancestral paleoAP3 motif via a simple frameshift mutation close to the gene duplication event (Vandenbussche et al., 2003; Kramer et al., 2006). Though the C-terminal motifs are highly conserved within the gene lineages, suggesting a functional importance, their exact function remains unclear (Piwarzyk et al., 2007; Su et al., 2008).
Members of the TM6 lineage are the most poorly studied B class genes as neither Arabidopsis nor Antirrhinum have TM6 gene copies (Lamb and Irish, 2003). The most thorough functional studies on TM6 lineage genes so far have been performed in Solanaceae, on Petunia hybrida TM6 (PhTM6) (Rijpkema et al., 2006) and tomato (Solanum lycopersicum) TM6 (de Martino et al., 2006). The expression patterns of these TM6 lineage genes differ from those of PI/GLO and euAP3 lineage genes in that they have a broader expression domain. Their transcripts are detected not only in petals and stamens, but also in carpels and ovules (Vandenbussche et al., 2004; de Martino et al., 2006). The petunia and tomato TM6 genes are involved in stamen development and not in petal development, while the euAP3 genes play a role in both petal and stamen development. The petunia PhTM6 and PhDEF act completely redundantly in stamen development, while the tomato TAP3 and TM6 are only partly redundant: the TAP3 gene has unique functions in stamen development that cannot be fulfilled by TM6 (de Martino et al., 2006; Rijpkema et al., 2006).
To obtain a better understanding of the functional differentiation of B class MADS-box genes, PI/GLO- and AP3/DEF-like genes have been analysed in gerbera. Gerbera belongs to the large sunflower (Asteraceae) family, which lies in a lineage of asterids distinct from the position of Solanaceae (Angiosperm Phylogeny Group II, 2003). Typical of Asteraceae, the inflorescence comprises hundreds of tightly packed flowers with specialized structures and functions. Three distinct flower types can be distinguished: the outermost ray and trans flowers have showy petals and non-functional rudimental stamens, whereas the disk flowers in the centre are smaller and hermaphroditic with functional stamens. Three gerbera B class genes that belong to different subfamilies have previously been identified. GERBERA GLOBOSA-LIKE1 (GGLO1) is a PI/GLO lineage gene, GERBERA DEFICIENS-LIKE2 (GDEF2) is a euAP3 gene, and GDEF1 is a TM6-like gene (Yu et al., 1999). Here functional analyses for these genes are presented and, furthermore, data are shown for a fourth gerbera B gene, GDEF3, which appears to be the result of an Asteraceae-specific gene duplication in the euAP3 subfamily. The data indicate that the PI/GLO and euAP3 lineage genes GGLO1 and GDEF2, respectively, function in petal and stamen development and encode the classical B-function in gerbera. An additional novel phenotype, the conversion of petals into ovary wall-like tissues, was also observed when B-function genes were down-regulated. Expression of the TM6 lineage gene GDEF1 differs from that of the other gerbera B class genes in several aspects, such as being absent during the early petal primordia initiation. However, transgenic phenotypes do not show a unique function for GDEF1 but instead suggest it to be mainly redundant with other gerbera B class genes. Functional redundancy is further supported by the yeast two- and three-hybrid assays which show that the gerbera B class proteins form three kinds of heterodimers with parallel multimeric protein interaction capacities.
Materials and methods
Isolation of full-length GDEF3
Based on the expressed sequence tag (EST) sequence of GDEF3 (G0000700006A02), primers for standard 5′ rapid amplification of cDNA ends (RACE) were designed. Ray flower petals (developmental stages 2 and 3; Helariutta et al. 1993) were used as starting material and cDNA was synthesized as described in Laitinen et al. (2008). Full-length cDNA amplification by RT-PCR (with a forward primer 5′-ATCCAAATCAATGGCGAGAG-3′ and a reverse primer 5′-CCGTCATAATCCAAA-TCAGACA-3′) was performed to ensure that the 5′ fragment originated from the same transcript. The cDNA was cloned into ZeroBlunt vector and sequenced in both directions. The full-length sequence for GDEF3 has been deposited in GenBank (http://www.ncbi.nlm.nih.gov) under accession number FJ817421.
Phylogenetic analysis
Phylogenetic analyses were performed on corresponding nucleotide and amino acid alignments for the MADS and K domains of selected class B MADS-box factors. Both parsimony and maximum likelihood methods were used on nucleotide and amino acid alignments. Since the analyses were largely congruent, only the maximum likelihood results from the nucleotide sequence alignment are described and presented. For these analyses, the options used with the PHYML (Guindon and Gascuel, 2003) web interface (Guindon et al., 2005) were: (i) a starting tree constructed with the BIONJ (Gascuel, 1997); (ii) best of NNI- and SPR-swapped trees; (iii) a random tree from five data addition sequences; (iv) the HKY85 evolutionary model (Hasegawa et al., 1985); (v) estimated proportion of invariable sites=0.122; (vi) six substitution rate categories; (vii) estimated gamma distribution parameter=1.170; (viii) transition/transversion ratio: 2.883; (ix) empirical base frequencies; (x) tree topology optimization; and (xi) branch length and rate parameter optimization. A total of 500 boostrap resampling replicates were used to estimate support for clades (Felsenstein, 1985).
RNA blot and in situ hybridizations
RNA blots were done as previously reported by Broholm et al. (2008). The floral organs were collected from ray and disk flowers at different developmental stages (pooled at stages 3, 5, 7, and 9 for ray; and at stages 6 and 8 for disk flowers). The developmental stages of the gerbera inflorescence have been described in Helariutta et al. (1993). For gene-specific probes, the 3′ end of the cDNA was used (for GGLO1, 217 bp). The plasmid pGGLO1as was digested with XhoI and BglII; for GDEF2 (423 bp) pHTT664.3 with EcoRI and BamHI; for GDEF3 (335 bp) G0000700006A02 with SacII and KpnI; and for GDEF1 (283 bp) pHTT661.2 with EcoRI and BamHI. In situ hybridization analyses using gene-specific probes were performed as in Elomaa et al. (2003). Probe concentration for GGLO1, GDEF2, and GDEF3 was 0.4 μg ml−1 kb−1, and for GDEF1 0.5 μg ml−1 kb−1. Detection time was 16 h for GGLO1 and GDEF2, and 40–45 h for GDEF3 and GDEF1.
Plant transformation and analysis of transgenic lines
Agrobacterium-mediated transformation of the full-length gene constructs 35S-GGLO1, 35S-GDEF2, and 35S-GDEF1 into gerbera was performed as previously described (Elomaa and Teeri, 2001). Integration of the transgene was verified using standard DNA hybridization. Scanning electron microscopy (SEM) analysis of the transgenic flower organs was performed as described by Uimari et al. (2004).
Yeast assays
Full-length gerbera B class MADS-box cDNAs (GGLO1, GDEF1, GDEF2, and GDEF3) were introduced into the Gateway system using PCR (PCR Cloning System with Gateway Technology with pDONR221, Invitrogen). Primers flanking the first methionine of the gene and the stop codon were designed according to Invitrogen's instructions. Two nucleotides were added between the attB1 sequence and the start codon. The Gateway primer sequences are shown in Supplementary Table S2 available at JXB online. The PCRs were run according to the guidelines of the Phusion DNA polymerase (Finnzymes). The PCR products were polyethylene glycol (PEG) purified and recombined to pDONR221 plasmid to create the Gateway entry clones, according to Invitrogen's instructions.
The entry clones were recombined into the activation domain- and binding domain-containing plasmids pDEST22 and pDEST32 (Invitrogen). The pDEST22 and pDEST32 derivatives carrying the gerbera B class MADS-box genes were transformed into both yeast strains PJ69-4A and PJ69-4α (James et al., 1996). The pDEST32 clones containing N-terminal binding domain fusions were tested for autoactivation by plating them on the yeast medium SD lacking adenine (SD –Ade) and histidine (SD –His), and supplemented with 1, 5, 10, 25, and 50 mM 3-amino-1,2,4-triazole (3-AT) (Sigma A8056). To obtain yeast double transformants, the A and α types of yeast strains were mated by pipetting them on top of each other on SD Complete plates containing all the essential amino acids. Yeast double transformants were plated on selection plates SD –Leu –Trp, and these colonies were replica plated on YPAD, SD –Leu –Trp –Ade, and SD –Leu –Trp –His + 25 mM 3-AT essentially as described in the ProQuest Two-Hybrid System Manual (Invitrogen PQ10001-01). X-gal assay was performed for yeast cells grown on YPAD plates according to Invitrogen's instructions.
Results
Phylogenetic analysis of the gerbera B class MADS-box genes
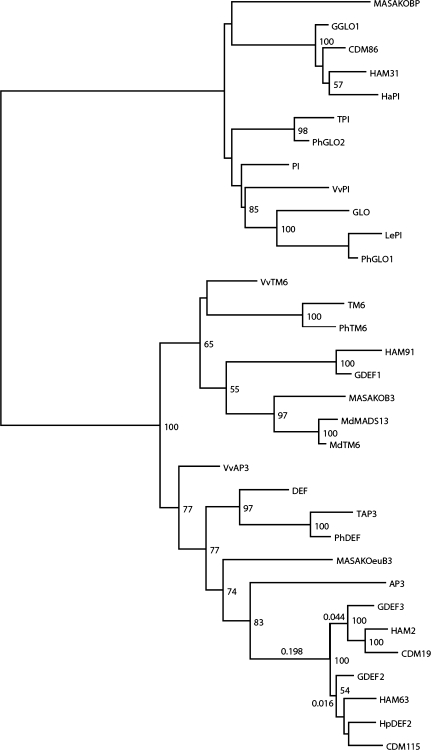
In addition to the three previously isolated gerbera B class genes (GDEF1, GDEF2, and GGLO1), a fourth gene, named GDEF3, was identified by EST sequencing from a gerbera cDNA library that was constructed from early stages of petal development (Laitinen et al., 2005). Maximum likelihood phylogenetic analyses demonstrated that, in general, the B class genes from Asteraceae seem to form specific subgroups within the three B class clades (PI/GLO, euAP3, and TM6) found in core eudicots (Fig. 1).
Fig. 1.
Phylogenetic analysis of selected B class MADS-box genes based on a nucleotide sequence alignment (∼550 bp from the start codon) and the maximum likelihood method. The four gerbera B class genes are located on distinct branches of the tree: GDEF2 and GDEF3 in the euAP3 clade, GDEF1 in the TM6-like clade, and GGLO1 in the PI/GLO clade. Genes from Asteraceae form two paralogous subgroups within the euAP3 clade, the GDEF3-like and GDEF2-like clades. Bootstrap values at nodes represent percentages of times clades appeared in 500 resampling replicates. Lengths for three key branches (in subsititutions per site) among the Asteraceae genes are also indicated for scale. In addition to gerbera, genes were selected from the following species: Antirrhinum majus (GLO, DEF); Arabidopsis thaliana (PI, AP3); Dendranthema grandiflorum (CDM86, CDM115, CDM19); Helianthus annuus (HaPI, HAM31, HAM91, HAM2, HAM63); Hieracium piloselloides (HpDEF2); Malus domestica (MdTM6, MdMADS13); Petunia hybrida (PhGLO1, PhGLO2, PhTM6, PhDEF); Rosa rugosa (MASAKOBP, MASAKOB3, MASAKOeuB3); Solanum lycopersicum (LePI, TPI, TM6, TAP3); and Vitis vinifera (VvPI, VvTM6, VvAP3). GenBank accession numbers are presented in Supplementary Table S1M at JXB online.
In the euAP3 lineage the Asteraceae-specific subgroup is further divided into two paralogous groups, in which GDEF2 and GDEF3 have putative orthologues from both sunflower (Helianthus annuus) and chrysanthemum (Dendranthema grandiflorum) (Fig. 1). This indicates that the euAP3 lineage has undergone a gene duplication event early in the Asteraceae lineage. GDEF2 and GDEF3 both encode the typical euAP3 C-terminal motifs. Alignment of the inferred amino acid sequences of the selected euAP3 lineage members (Supplementary Fig. S1 at JXB online) showed several amino acid changes that were specific to the GDEF2/HAM63/CDM115 and GDEF3/HAM2/CDM19 groups: three were observed in the K-domain and three in the C-terminal region.
Interestingly, the gerbera TM6 lineage gene GDEF1 encodes a paleoAP3 C-terminal motif (YELHDHQHTN) that is highly diverged from the consensus sequence (YGxHDLRLA) (alignment shown, for example, in Kim et al., 2004). The paleoAP3 motif of its putative sunflower orthologue HAM91 (YEPHGLRLD) is much more similar to the consensus paleoAP3 motif than is that of GDEF1.
Expression of B class genes in gerbera tissues
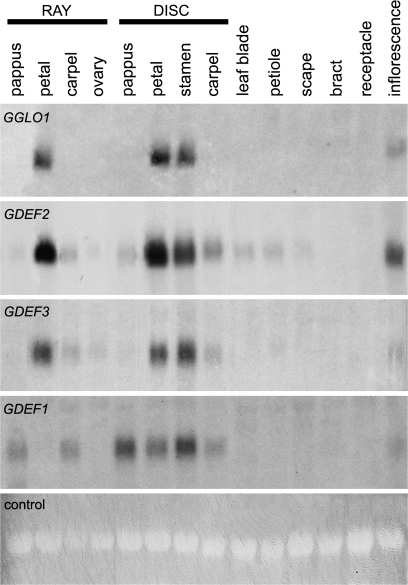
Expression of the gerbera B class genes was studied by RNA blotting and in situ hybridization. Both GGLO1 and GDEF1 were found to be expressed exclusively in floral tissues, whereas GDEF2 and GDEF3 transcripts were also detected (although only weakly) in vegetative leaves and petioles (Fig. 2). GGLO1, GDEF2, and GDEF3 all showed the strongest expression in petals and stamens. GDEF2 and GDEF3 also showed a weak signal in pappus bristles (whorl one) and carpel (stigma and style) (Fig. 2). Only GGLO1 expression was sharply restricted to petals and stamens during organ differentiation (Figs 2 and Fig. 3F, K, P, U), whereas GDEF2 and GDEF3 had broader expression domains (Fig. 2).
Fig. 2.
The expression of gerbera B class genes in floral organs of ray and disk flowers and other tissues. The expression of GGLO1 is restricted to petals and stamens. GDEF2 and GDEF3 are expressed, in addition to petals and stamens, in other floral organs and vegetative tissues. GDEF1 is expressed in all four floral organs in disk flowers, but in ray flowers the expression was not detected in petals. The lowest panel shows ethidium bromide-stained rRNA bands to control RNA loading.
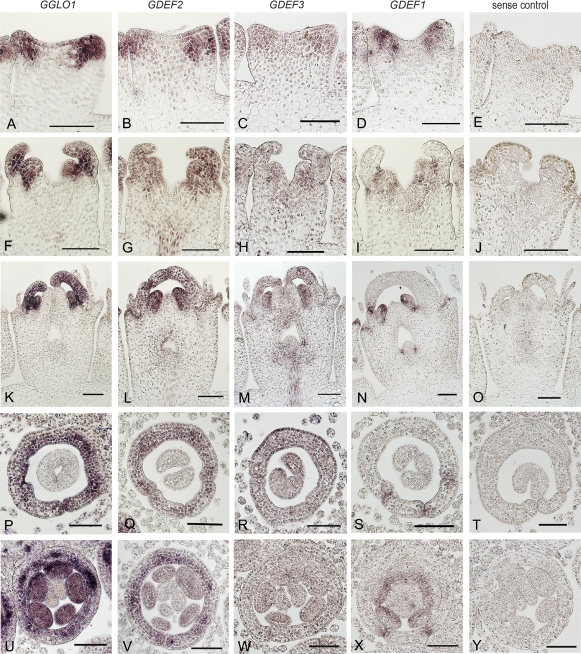
Fig. 3.
In situ analysis of gerbera B class gene expression in floral primordia at stage 2 (A–E), stage 3–4 (F–J), ray flowers (K–O), trans flowers (P–T), and disk flowers (U–Y). GGLO1, GDEF2, and GDEF3 are mainly restricted in petals (pe) and stamens (st). GDEF1 is not detected in early petal primordia (D, I), but later in a spatially restricted pattern in petals (N, S, X), stamens (N, X), and ovule (N). GDEF1 also shows early expression in whorl one (w1) (D, I). Negative controls hybridized with a sense probe are shown in the rightmost panel. A–O represent longitudinal sections and P–Y cross-sections. Scale bars, 100 μm.
While GGLO1, GDEF2, and GDEF3 showed similar expression in both ray and disk flowers (Fig. 2), the expression pattern of GDEF1 varied between the two flower types. In disk flowers, GDEF1 transcripts were detected in all four floral whorls, whereas in ray flowers the expression could not be detected in petals at comparable developmental stages (Fig. 2). The expression of GDEF1 was detected only very weakly at early stages of ray flower petal development (stages 1–3, data not shown), in contrast to the more consistent expression in disk flower petals (Supplementary Fig. S2 at JXB online). All the four gerbera B class genes were expressed in the non-functional stamen rudiments of ray and trans flowers (Fig. 3K, L, M, N for ray flowers, data not shown for trans flower stamens). This suggests that they are not involved in the sex determination of the gerbera flower types.
In situ hybridizations showed further differences in the pattern of GDEF1 expression in comparison with the other gerbera B class genes. GDEF1 transcripts were detected in stamen primordia but not in the early petal primordia (Fig. 3D, I), while the expression of GGLO1, GDEF2, and GDEF3 was activated in both petal and stamen primordia (Fig. 3A, B, C). Later on, GDEF1 was expressed in all four whorls, but in spatially restricted patterns. GDEF1 transcripts were localized in lateral edges of petals in all three flower types (Fig. 3N for ray, S for trans, and X for disk flowers). Thus, the expression of GDEF1 clearly differs from the uniform pattern of GGLO1, GDEF2, and GDEF3 expression in petals.
In disk flower stamens, GDEF1 expression was restricted to the adaxial side (Fig. 3X) and, later in development, to the sterile connective tissue (data not shown). However, RNA blots revealed that GDEF1, together with GGLO1, was expressed in disk flower stamens at later stages than the euAP3 genes GDEF2 and GDEF3 (Supplementary Fig. S2 at JXB online). An additional difference in the GDEF1 expression pattern was strong expression at late stages of carpel development when GDEF2 and GDEF3 were no longer expressed (Supplementary Fig. S2).
Down-regulation of GGLO1 and GDEF2 expression leads to stamen and petal defects
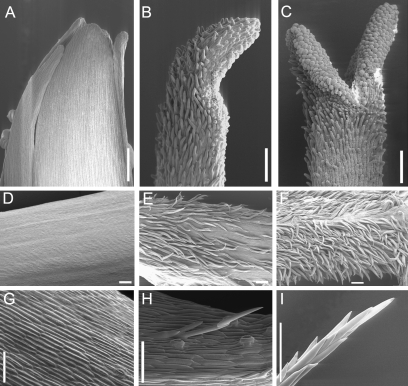
In a previous study, the phenotypic effects of constitutive and reduced expression of GGLO1 were reported (Yu et al. 1999). The lines with reduced GGLO1 expression described in Yu et al. (1999) showed homeotic changes in petals but not in stamens. An additional GGLO1 co-suppression line that shows a stronger phenotype has now been obtained (Tr15, Fig. 4B). In contrast to the previous studies, the identity of both petals and stamens was affected. In accordance with the predicted homeotic changes in B class mutants, stamens were converted into carpeloid structures with carpel-like cells on their surfaces (Fig. 5B). Petals had two different kinds of epidermal trichomes on their abaxial surfaces. On the basal petal tube, trichomes similar to those found in wild-type ovary wall were observed (Fig. 5E). On the distal petal parts (the ligule), there were trichomes that resembled pappus bristles (Fig. 5H). GGLO1 expression was highly reduced in young inflorescences of the Tr15-line. However, the expression of GDEF1, GDEF2, and GDEF3 appeared unaltered (Supplementary Fig. S3 at JXB online).
Fig. 4.
The wild-type gerbera inflorescence (A) and transgenic inflorescence phenotypes caused by co-suppression of GGLO1 expression (B, tr15), co-suppression of GDEF2 (C, tr22), and reduction of GDEF1 expression (D, tr12).
Fig. 5.
SEM analysis comparing the epidermal cell structure of gerbera floral organs between the wild type (wt) and transgenic line (tr15) with reduced GGLO1 expression. Wt stamen (A) and tr stamen (B), which resembles wt carpel (C). Wt petal tube (D) and tr petal tube (E) with trichomes similar to trichomes in the wt ovary wall (F). Wt petal ligule (G) and tr petal ligule (H) with trichomes that resemble wt whorl one pappus bristles (I). Scale bars, 100 μm.
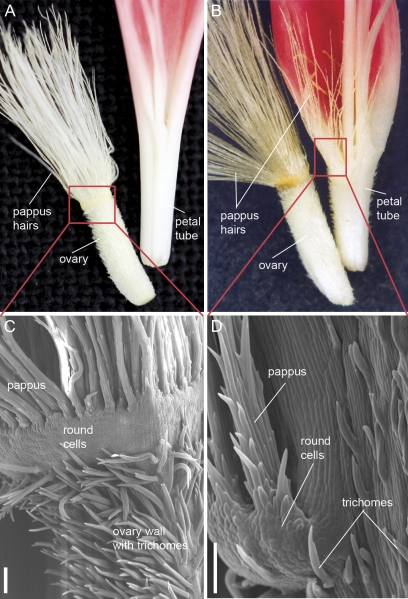
The transgenic lines in which GDEF2 expression was suppressed also showed phenotypic changes in whorls two and three (Supplementary Fig. S4 at JXB online), though their phenotypes were different from those of the GGLO1 down-regulation lines. Conversion of the basal petal tube of ray flowers into an ovary-like structure was even more striking in the GDEF2 down-regulation lines, and three new cell types were observed on the petal epidermis (Fig. 6). The basalmost tubular region of the petals had converted to a tissue covered by similar trichomes to those in wild-type ovary wall. Inside this ovary-like structure there was no ovule but instead what appeared to be normal petal tissue. Above the ovary wall-like tissue, a second round of pappus hairs developed. These emerged from round-shaped cells that were similar to cells from which pappus hairs emerge in the wild type. In addition to the alterations in petals, homeotic conversion of stamens towards carpeloid structures was observed (Supplementary Fig. S4). The homeotic conversion of stamens was seen in only one of the lines (Tr22, Fig. 4C), which showed strongest reduction in GDEF2 expression, and also showed reduction in the expression of GGLO1 (Supplementary Fig. S3). In the other lines, there were no obvious homeotic changes in stamens. However, disk flower stamens showed earlier senescence by turning brown and dry already at anthesis. The stamens produced some pollen but did not release them readily.
Fig. 6.
Comparison of the wild type (A, C) and transgenic line (tr22) with reduced GDEF2 expression (B, D). Transgenic petals had obtained three new cell types. The petal tube was covered by similar trichomes to those in ovary walls (B, D). In addition, a second round of pappus hairs was observed (B, D). SEM analysis of the transgenic petals (D) revealed that at the base of the extra pappus there are round-shaped cells similar to cells observed in the wild type (C) in a ring between the ovary and pappus hairs. Scale bars, 100 μm.
Down-regulation of GDEF1 expression leads to mild phenotypic effects
In both of the GDEF1 antisense lines in which GDEF1 expression was reduced (Fig. 4D, Supplementary Fig. S3 at JXB online), developmental changes in petals and stamens were observed. These changes resembled the mildest phenotypes of the transgenic lines with reduced GDEF2 or GGLO1 expression. However, expression of GDEF2, GDEF3, or GGLO1 in the young inflorescences of these lines was not altered compared with the wild type (Supplementary Fig. S3). Both stereomicroscopic and SEM analysis showed the presence of few ovary wall-like cells in the abaxial side of ray flower petal tubes (data not shown). No pappus bristle-like hairs were observed. Disk flower stamens were similarly brownish at anthesis and unable to release pollen, as described above for the stamens of the mild anti-GDEF2-lines. No obvious homeotic conversions were observed in these stamens (data not shown).
Overexpression of the gerbera B class genes
It was demonstrated previously that transgenic lines expressing GGLO1 ectopically have clear homeotic conversions (Yu et al., 1999). Ectopic expression of GGLO1 caused whorl one pappus bristles to gain petal-like characters, whereas in whorl four, carpels obtained stamen-like traits (Yu et al. 1999). In contrast, the six lines expressing GDEF2 ectopically and five lines expressing GDEF1 ectopically (data not shown) did not show any phenotypic changes compared with the wild type. The expression of GDEF2, GDEF3, GDEF1, and endogenous GGLO1 in the 35S promoter-driven GGLO1 overexpression lines was studied (Supplementary Fig. S5 at JXB online). The expression of GDEF2 was highly up-regulated in whorls one and four as well as in ovaries and leaves. GDEF3 expression was most clearly up-regulated in stamens and whorl four stamenoid carpels, whereas in petals the expression was unaltered. GDEF1 expression was enhanced only in whorl four, and, interestingly, its expression was reduced in the whorl one organs that had petal-like characteristics. The endogenous GGLO1 expression was also up-regulated in these lines and independent of the floral context (Supplementary Fig. S5). In general, overexpression of GGLO1 led to up-regulation of GDEF2, GDEF3, and GDEF1 expression and caused homeotic conversions. In contrast, overexpression of GDEF2 did not alter the expression of GGLO1 (data not shown). The inability of GDEF2 to induce ectopic GGLO1 expression might relate to the strictly restricted expression of GGLO1 in wild-type whorls two and three, and this in turn may explain the lack of phenotypic changes in the GDEF2 overexpression lines.
Protein interactions of the gerbera B class MADS domain proteins
Using GAL4 yeast two-hybrid assays, it could be shown that the GGLO1 protein can form a heterodimer with all the other three gerbera B class proteins GDEF1, GDEF2, and GDEF3 (Supplementary Fig. S6 at JXB online). The interaction capacity of the B class proteins (except GDEF3) was tested with 11 other gerbera AGAMOUS-, SQUAMOSA-, or SEPALLATA3-like MADS domain proteins (S Ruokolainen et al., unpublished results). The B class proteins did not interact with any other class of MADS domain proteins in the pairwise interaction assays. Further, no homodimers or interactions between the euAP3 or TM6 clade members were observed.
In chrysanthemum, yeast three-hybrid analyses have shown that the two euAP3-type proteins CDM115 and CDM19, corresponding to GDEF2 and GDEF3, respectively, act differentially in ternary complex formation (Shchennikova et al., 2004). They both form heterodimers with the PI/GLO-type protein CDM86, but only the CDM86–CDM115 heterodimer forms a ternary complex with AP1/FUL-type proteins and with a class C protein, whereas the CDM86–CDM19 heterodimer does not. To investigate if this apparent diversification in functions is conserved, a yeast three-hybrid experiment was performed for the gerbera GDEF2 and GDEF3 proteins. It was found, however, that gerbera GGLO1–GDEF2 and GGLO1–GDEF3 heterodimers were both capable of forming ternary complexes with the AP1/FUL-type and the C class proteins (Supplementary Fig. S7 at JXB online).
Discussion
Asteraceae-specific B gene duplications and conservation of function
The transgenic phenotypes show that the PI/GLO and euAP3 lineage genes GGLO1 and GDEF2 encode the classical B-function in gerbera. Reduction in the expression of GGLO1 and GDEF2 results in homeotic conversion of stamens towards carpels and of petals towards pappus bristles (modified sepals). Moreover, the expression of GGLO1, as well as both the euAP3 lineage genes GDEF2 and GDEF3, is ubiquitous throughout the development of petals and stamens.
The presence of a paralogous pair of euAP3 lineage genes is shared by gerbera, chrysanthemum, and sunflower. This reflects a common origin, possibly from the genome-wide duplication at the base of the Asteraceae lineage (Barker et al., 2008). It is unlikely that duplicated genes would have avoided non-functionalization or loss in all three lines during the evolutionary history of Asteraceae [∼40–49 million years ago (Mya); Kim et al., 2005; Barker et al., 2008] if they were completely functionally redundant. The average half-life for redundant gene duplicates is only 4 million years (Lynch and Conery, 2003). This implies that the pair of euAP3 lineage genes has probably undergone functional diversification (e.g. sub- or neofunctionalization). However, the GDEF2 and GDEF3 sequences and expression patterns are very similar and their protein interaction capacities seem not to differ, suggesting that they are at least partially redundant. In support of functional diversification, however, in transgenic lines that express GGLO1 ectopically, it was observed that the expression of GDEF2 and GDEF3 is induced differentially.
Sunflower shows the presence of a second PI/GLO-like gene (Dezar et al., 2003; Shulga et al., 2008). An orthologue has not been reported in chrysanthemum and, despite extensive efforts taken to find a second PI/GLO-like gene, gerbera seems to have only GGLO1. An analysis of the gerbera floral transcriptome using EST sequencing (Laitinen et al., 2005, and unpublished sequences obtained by 454 pyrosequencing) has yielded >100 tags similar to GGLO1, but none for a second GGLO gene. The lack of a second GLO-like gene in gerbera and chrysanthemum suggests that the sunflower branch of Asteraceae has had a recent PI/GLO duplication.
B gene expression and autoregulation
The pattern of gerbera B class gene expression in flowers is comparable with that of Antirrhinum, petunia, and tobacco, where PI/GLO-like genes show strictly restricted expression and euAP3 genes a more widespread expression in all four floral organs (Sommer et al., 1990; Davies et al., 1996; Hileman and Irish, 2009). This pattern deviates from that of Arabidopsis, where, in addition to petals and stamens, PI transcripts are detectable in the inner (destined to carpels) and AP3 in the outer (destined to sepals) parts of the flower primordia early in development (Weigel and Meyerowitz, 1993; Goto and Meyerowitz, 1994). This initial expression pattern is evoked independently, but at later developmental stages the expression is regulated through an auto- and cross-regulatory loop, in which the formation of a heterodimer between PI/GLO and euAP3 members is required to maintain expression of these genes specifically in petals and stamens (Schwarz-Sommer et al., 1992; Halfter et al., 1994; Jack et al., 1994).
In gerbera, it was found that ectopic expression of GGLO1 leads to an up-regulation of the expression of GDEF1, GDEF2, and GDEF3 (albeit to different levels; see Supplementary Fig. S5 at JXB online). Remarkably, however, in the transgenic line with reduced GGLO1 expression, homeotic alterations were detected but GDEF2 (as well as GDEF1 or GDEF3) transcription was not diminished. Apparently, even though GGLO1 is capable of regulating GDEF expression, GGLO1 expression is not necessary for the maintenance of GDEF expression. Alternatively, a very low level of GGLO1 is sufficient for autoregulation. The vegetative expression of GDEF2 in tissues where GGLO1 is not expressed further supports the presence of a regulatory mechanism independent from autoregulation.
Transgenic phenotypes suggest a sepaloid origin for the gerbera ovary wall
The homeotic conversion of petals caused by reduced B class gene expression has characteristics unique to gerbera. The abaxial epidermis of basal petal tubes takes on the identity of ovary wall (and not whorl one, which in gerbera is occupied by pappus bristles). The resemblance of petal tubes to ovary wall is especially strong in transgenic lines with reduced GDEF2 expression. Gerbera flowers are epigynous, as they have an inferior ovary located below the floral organs. There are two major morphological interpretations about the origin of the inferior ovary wall; it may be part of the floral axis (and thus be receptacular) or may result from fusion of floral organs with the ovary (and therefore be appendicular) (Gustafsson and Albert, 1999). The present transgenic loss-of-B-function phenotypes showing ovary wall-like tissues in petals suggest the latter alternative, i.e. that the ovary wall might have sepaloid origin, having evolved through fusion of the perianth organs with the ovary.
GDEF1 may act redundantly in stamen development
Expression of the TM6 lineage gene GDEF1 differs from that of the other gerbera B class genes in several aspects. Instead, it shows similarities with the pattern of PhTM6 expression in petunia (Vandenbussche et al., 2004). Both genes are expressed in all four floral organs and most strongly in stamens and carpels. In petals, their expression becomes similarly weaker at later stages of development. However, there are differences in the initial onset of the expression. In gerbera, GDEF1 transcripts are not detected in the region of emerging petal primordia, whereas in petunia and also in tomato (de Martino et al., 2006), the expression in petals begins already at the primordium stage. The pattern of GDEF1 expression in petals is spatially restricted to lateral edges. Similarly, the tomato TM6 gene shows restricted expression in the lateral margins of petals (de Martino et al., 2006). Adding to the complexity of the GDEF1 expression pattern, stronger and more consistent expression of GDEF1 was detected in disk flower petals than in ray flower petals.
Unlike other B class genes, TM6-like genes are prevalently expressed in carpels. Similarly to gerbera, petunia, and tomato, the grapevine VvTM6 gene shows expression in carpels and ovules in addition to petals and stamens (Poupin et al., 2007). Both in gerbera and in petunia (Vandenbussche et al., 2004), GDEF1 and PhTM6 expression levels remain high in carpels until very late stages of flower development. Whether this expression in carpels is functionally relevant is currently unknown.
Despite the clearly distinct expression pattern, transgenic plants in which GDEF1 expression was suppressed did not reveal any unique phenotypes but only very mild phenotypic changes. Similarly, in petunia, down-regulation of PhTM6 expression alone did not cause any phenotypic changes, and only analysis of the double mutant phtm6 phdef uncovered the role of PhTM6 in stamens (Rijpkema et al., 2006). Although there are no data from simultaneous down-regulation of GDEF1 and GDEF2 (or GDEF3), the present results indicate that GDEF1 is not involved in determining petal identity. In addition to the lack of consistent expression of GDEF1 in petals, further evidence comes from studying GDEF1 expression in GGLO1 overexpression lines, where the identity of pappus bristles has changed into petals. Although GDEF1 is expressed in wild-type pappus bristles, in these homeotic petal-like organs GDEF1 expression is clearly diminished, suggesting that homeotic conversion to petals may in fact require reduction in GDEF1 expression.
The mildness of stamen phenotype due to reduced GDEF1 expression indicates a redundant role for this gene. Suppressed GDEF1 expression caused earlier senescence and diminished release of pollen in disk flower stamens. GDEF1 expression persists longer in wild-type stamens than that of GDEF2 or GDEF3, suggesting that GDEF1 may have an independent regulatory role in late stages of stamen development. However, GDEF1 cannot compensate for the lack of GDEF2 expression in stamens, since similar and stronger changes are detected in the GDEF2 transgenic lines. To define the distribution of regulatory roles between the three AP3-type genes in gerbera, production of GDEF3 down-regulation lines is required. Furthermore, transgenic lines in which two or all of the three genes are down-regulated simultaneously would be informative concerning the overlapping roles that the present work suggests.
Conclusions
The present analysis of B-function genes in gerbera has uncovered many commonalities with the function of orthologues in other systems, particularly other asterids such as Solanaceae. Importantly, gerbera represents an entirely distinct lineage of the asterids that diverged from the lineage containing Solanaceae >100 Mya (Wikström et al., 2001). As such, the general details of euAP3/TM6-like gene function described here can be parsimoniously interpreted as the basal state among the asterids. Although drastic modification has occurred at least once, in Antirrhinum (Lamiales), which has lost its TM6 copy (as has Arabidopsis independently), it can be inferred that even though B-function expression details can be flexible in asterids (Hileman and Irish, 2009), their function has persisted across vast expanses of time with only few modifications.
Supplementary data
Supplementary data are available at JXB online and comprise the following figures and tables.
Figure S1. Amino acid alignment for a selected set of euAP3-clade MADS-box genes.
Figure S2. Expression of the B class genes in gerbera disc flowers during development of petals, stamens, and carpels.
Figure S3. Expression of the B class genes in young inflorescences of the transgenic gerbera lines.
Figure S4. SEM analysis of the transgenic gerbera line (Tr22) with reduced GDEF2 expression.
Figure S5. Expression of the gerbera B class genes in the transgenic GGLO1 overexpression line.
Figure S6. Yeast two-hybrid analysis of the gerbera B class MADS domain proteins.
Figure S7. Yeast three-hybrid analysis of the gerbera B class MADS domain proteins.
Table S1. The B class genes used in the phylogenetic analysis.
Table S2. Primers used for Gateway conversion of the gerbera B class MADS-box genes.
Supplementary Material
Acknowledgments
We thank the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki, for providing laboratory facilities; Eija Takala and Anu Rokkanen for excellent technical assistance; Sanna Peltola for taking care of the plants in the greenhouse; and Anneke Rijpkema for critical reading and helping to improve the manuscript. The Centre of Excellence program of the Academy of Finland supported this work. SKB was supported by the Viikki Graduate School in Biosciences.
References
- Angiosperm Phylogeny Group II. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society. 2003;141:399–436. [Google Scholar]
- Barker MS, Kane NC, Matvienko M, Kozik A, Michelmore RW, Knapp SJ, Rieseberg LH. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Molecular Biology and Evolution. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RA, Albert VA, Teeri TH, Elomaa P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proceedings of the National Academy of Sciences, USA. 2008;105:9117–9122. doi: 10.1073/pnas.0801359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Di Rosa A, Eneva T, Saedler H, Sommer H. Alteration of tobacco floral organ identity by expression of combinations of Antirrhinum MADS-box genes. The Plant Journal. 1996;10:663–677. doi: 10.1046/j.1365-313x.1996.10040663.x. [DOI] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. The Plant Cell. 2006;18:1833–1845. doi: 10.1105/tpc.106.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezar CA, Tioni MF, Gonzalez DH, Chan RL. Identification of three MADS-box genes expressed in sunflower capitulum. Journal of Experimental Botany. 2003;54:1637–1639. doi: 10.1093/jxb/erg163. [DOI] [PubMed] [Google Scholar]
- Elomaa P, Teeri TH. Transgenic Gerbera. In: Bajaj YPS, editor. Biotechnology in agriculture and forestry 48. Berlin: Springer-Verlag; 2001. pp. 139–154. [Google Scholar]
- Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RAE, Teeri TH. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein–protein and protein–promoter interactions between the anciently diverged monocots and eudicots. Plant Physiology. 2003;133:1831–1842. doi: 10.1104/pp.103.026039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes and Development. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MHG, Albert VA. Inferior ovaries and angiosperm diversification. In: Hollingsworth PM, Bateman RM, Gornall RJ, editors. Molecular systematics and plant evolution, The Systematics Association Special Volume Series edn. New York: Taylor and Francis; 1999. pp. 403–431. [Google Scholar]
- Halfter U, Ali N, Stockhaus J, Ren L, Chua NH. Ectopic expression of a single homeotic gene, the petunia gene green petal, is sufficient to convert sepals to petaloid organs. EMBO Journal. 1994;13:1443–1449. doi: 10.1002/j.1460-2075.1994.tb06398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human–ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Elomaa P, Kotilainen M, Seppänen P, Teeri TH. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae) Plant Molecular Biology. 1993;22:183–193. doi: 10.1007/BF00014927. [DOI] [PubMed] [Google Scholar]
- Hernandez-Hernandez T, Martinez-Castilla LP, Alvarez-Buylla ER. Functional diversification of B MADS-box homeotic regulators of flower development: adaptive evolution in protein–protein interaction domains after major gene duplication events. Molecular Biology and Evolution. 2007;24:465–481. doi: 10.1093/molbev/msl182. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Irish VF. More is better: the uses of developmental genetic data to reconstruct perianth evolution. American Journal of Botany. 2009;96:83–95. doi: 10.3732/ajb.0800066. [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM. Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell. 1994;76:703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Choi KS, Jansen RK. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae) Molecular Biology and Evolution. 2005;22:1783–1792. doi: 10.1093/molbev/msi174. [DOI] [PubMed] [Google Scholar]
- Kim S, Yoo M, Albert VA, Farris JS, Soltis PS, Soltis DE. Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. American Journal of Botany. 2004;91:2102–2118. doi: 10.3732/ajb.91.12.2102. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Irish VF. Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. International Journal of Plant Sciences. 2000;161:S29–S40. [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Su HJ, Wu CC, Hu JM. A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evolutionary Biology. 2006;6:30. doi: 10.1186/1471-2148-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen RA, Ainasoja M, Broholm SK, Teeri TH, Elomaa P. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. Journal of Experimental Botany. 2008;59:3691–3703. doi: 10.1093/jxb/ern216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen RAE, Immanen J, Auvinen P, et al. Analysis of the floral transcriptome uncovers new regulators of organ determination and gene families related to flower organ differentiation in Gerbera hybrida (Asteraceae) Genome Research. 2005;15:475–486. doi: 10.1101/gr.3043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RS, Irish VF. Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proceedings of the National Academy of Sciences, USA. 2003;100:6558–6563. doi: 10.1073/pnas.0631708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary demography of duplicate genes. Journal of Structural and Functional Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- Piwarzyk E, Yang Y, Jack T. Conserved C-terminal motifs of the arabidopsis proteins APETALA3 and PISTILLATA are dispensable for floral organ identity function. Plant Physiology. 2007;145:1495–1505. doi: 10.1104/pp.107.105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupin MJ, Federici F, Medina C, Matus JT, Timmermann T, Arce-Johnson P. Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene. 2007;404:10–24. doi: 10.1016/j.gene.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. The Plant Cell. 2006;18:1819–1832. doi: 10.1105/tpc.106.042937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig WE, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO Journal. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchennikova AV, Shulga OA, Immink R, Skryabin KG, Angenent GC. Identification and characterization of four chrysanthemum MADS-box genes, belonging to the APETALA1/FRUITFULL and SEPALLATA3 subfamilies. Plant Physiology. 2004;134:1632–1641. doi: 10.1104/pp.103.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga OA, Shchennikova AV, Angenent GC, Skryabin KG. MADS-box genes controlling inflorescence morphogenesis in sunflower. Russian Journal of Developmental Biology. 2008;39:2–5. [PubMed] [Google Scholar]
- Sommer H, Beltran JP, Huijser P, Pape H, Lonnig WE, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO Journal. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K, Zhao S, Shan H, Kong H, Lu W, Theissen G, Chen Z, Meng Z. The MIK region rather than the C-terminal domain of AP3-like class B floral homeotic proteins determines functional specificity in the development and evolution of petals. New Phytology. 2008;178:544–558. doi: 10.1111/j.1469-8137.2008.02382.x. [DOI] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lonnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO Journal. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uimari A, Kotilainen M, Elomaa P, Yu D, Albert VA, Teeri TH. Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proceedings of the National Academy of Sciences, USA. 2004;101:15817–15822. doi: 10.1073/pnas.0406844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Theissen G, Van de Peer Y, Gerats T. Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Research. 2003;31:4401–4409. doi: 10.1093/nar/gkg642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. The Plant Cell. 2004;16:741–754. doi: 10.1105/tpc.019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. Activation of floral homeotic genes in arabidopsis. Science. 1993;261:1723–1726. doi: 10.1126/science.261.5129.1723. [DOI] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society B: Biological Sciences. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pöllänen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH. Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae) The Plant Journal. 1999;17:51–62. doi: 10.1046/j.1365-313x.1999.00351.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.