Abstract
To investigate whether the high expression levels of actin-depolymerizing factor genes are related to pollen development, three GhADF genes (cDNAs) were isolated and characterized in cotton. Among them, GhADF6 and GhADF8 were preferentially expressed in petals, whereas GhADF7 displayed the highest level of expression in anthers, revealing its anther specificity. The GhADF7 transcripts in anthers reached its peak value at flowering, suggesting that its expression is developmentally-regulated in anthers. The GhADF7 gene including the promoter region was isolated from the cotton genome. To demonstrate the specificity of the GhADF7 promoter, the 5′-flanking region, including the promoter and 5′-untranslated region, was fused with the GUS gene. Histochemical assays demonstrated that the GhADF7:GUS gene was specifically expressed in pollen grains. When pollen grains germinated, very strong GUS staining was detected in the elongating pollen tube. Furthermore, overexpression of GhADF7 gene in Arabidopsis thaliana reduced the viable pollen grains and, consequently, transgenic plants were partially male-sterile. Overexpression of GhADF7 in fission yeast (Schizosaccharomyces pombe) altered the balance of actin depolymerization and polymerization, leading to the defective cytokinesis and multinucleate formation in the cells. Given all the above results together, it is proposed that the GhADF7 gene may play an important role in pollen development and germination.
Keywords: ADF, cotton, F-actin, gene expression, pollen development
Introduction
In higher plants, reproductive processes take place within two specialized floral organ systems, the stamen and the pistil (Drews and Goldberg, 1989). The stamen, which is the male reproductive organ, is composed of a filament and an anther. The filament provides the anther with structural support and nutrients. The anther, where pollen grains are produced, also has four lobes which are, from outside to inside, the epidermis, the endothecium, the middle layer, and the tapetum. Anther development in Arabidopsis is divided into 14 stages based on anther histogenesis (Sanders et al., 1999). Subsequent to stage 5, pollen mother cells (PMCs) undergo meiosis to produce haploid microspores, followed by two mitotic divisions to form pollen. During pollen development, the tapetum and middle layer degenerate and eventually undergo dehiscence to allow the release of mature pollen grains.
After release from the anther, pollen begins germinating after hydration. In germinating pollen grains, F-actin assembles around the germination pores just before tube emergence (Gibbon et al., 1999). Subsequently, many long actin cables are oriented axially throughout the shank of the elongating pollen tubes. The streaming of secretory vesicles and other large organelles correlates with the actin in the process of tube elongation. They form a reverse-fountain cytoplasmic streaming in which the acropetal lanes move along the edge of the tube and then reverse direction at the base of the clear zone to move basipetally through the core of the tube (Vidali and Hepler, 2001). Therefore, actin plays a prominent role in maintaining pollen tube growth and reorientation.
In the different cellular processes, multiple actin binding proteins regulate actin dynamics by maintaining the optimum equilibrium between unpolymerized actin molecules (G-actin) and assembled actin filaments (F-actin). Actin binding proteins are the proteins [such as profilins, actin-depolymerizing factors (ADFs)/cofilins, villins, thymosins, DNasel, capping protein, and fimbrims] that affect the cytoskeleton architecture dynamics (McCurdy et al., 2001). Among them, ADF plays a vital role in the regulation of F-actin filament assembly in cells (Bamburg and Bray, 1987). The ADF/cofilin-family of proteins are highly conserved low-molecular-weight (15–20 kDa) actin-modulating proteins in eukaryotic cells (Chen et al., 2000). These proteins bind to actin in both polymerized and monomeric forms and sever F-actin into fragments and promote the dissociation of monomers from the pointed end of the filament (Mabuchi, 1983). It was confirmed that ADF have dual functions in regulating the dynamics of F-actin filaments. ADF not only enhances the depolymerization of microfilaments at their minus (or slow-growing) ends, but also accelerates the polymerization of F-actin under certain conditions (Gibbon, 2001).
ADF proteins are encoded by a multiple gene family in plants, animals, and micro-organisms. ADF genes have been identified in some higher plants, such as Lilium longiflorum, Brassica napus, maize, and Arabidopsis (Kim et al., 1993; Rozycka et al., 1995; Lopez et al., 1996; Dong et al., 2001a). Previous studies revealed that AtADF1 and AtADF6 were mainly expressed in the vascular tissue of all organs, whereas AtADF5 was exclusively expressed in the root tip meristem of Arabidopsis (Dong et al., 2001a). Overexpression of AtADF1 resulted in the disappearance of thick actin cables in different cell types and caused irregular organs. The results demonstrated that AtADF1 is a key regulator of F-actin organization in Arabidopsis (Dong et al., 2001b). Another study indicated that AtADF7, AtADF8, and AtADF10 were expressed specifically in the pollen tube of Arabidopsis (Maciver and Hussey, 2002). Moreover, some identified ADFs were also confirmed to be pollen-specific, such as maize ADF1 and ADF2, lily ADF1, and an ADF of Brassica napus (Kim et al., 1993; Chung et al., 1995; Rozycka et al., 1995; Lopez et al., 1996; Smertenko et al., 2001). In a phylogenetic analysis of all known plant ADFs, the seven pollen ADFs (ZmADF1, ZmADF2, LiADF1, BnADF1, AtADF7, AtADF8, and AtADF10) fall in one clade, while other ADFs (i.e. ZmADF3 and AtADF1) that are known to be expressed in vegetative tissues fall within the other group (Maciver and Hussey, 2002). The different ADF/cofilin proteins in the pollen-specific clade share conserved sequences (approximately 79% similarity). Overexpression of NtADF1 resulted in the reduction of axially oriented actin cables in transformed pollen tubes and in the inhibition of pollen tube growth in a dose-dependent manner (Chen et al., 2002). Thus, the proper regulation of actin turnover by NtADF1 is critical for pollen tube growth. Although pollen-specific ADF genes have been well characterized in some plants, to date, little is known about the role of the cotton ADF genes in flower development, especially in pollen development. The isolation and molecular characterization of three cotton ADF genes are reported here. GhADF6 and GhADF8 were preferentially expressed in petals, whereas GhADF7 was specifically expressed in the anthers of cotton. Overexpression of the GhADF7 gene in Arabidopsis resulted in a reduction in the percentage of viable and germinated pollen grains, and in yeast (Schizosaccharomyces pombe) cells led to the defective cytokinesis and multinucleate formation.
Materials and methods
Plant materials
Cotton (Gossypium hirsutum) seeds were surface-sterilized with 70% ethanol for 30–60 s and 10% H2O2 for 30–60 min, followed by washing with sterile water. The sterilized seeds were germinated on half-strength MS medium under a 12/12 h light/dark cycle at 28 °C. Cotyledons, roots, and hypocotyls were collected from sterile seedlings, and other tissues were derived from cotton plants grown in a greenhouse or field for DNA and RNA isolation.
Construction of a cotton flower cDNA library and isolation of GhADF cDNA clones
Total RNA was extracted from different cotton tissues and a flower cDNA library was constructed as described previously (Wang and Li, 2009). For isolating the full-length clones of the GhADF cDNAs, 2×106 cDNA clones were screened with a α-32P-dCTP-labelled probe (0.4 kb PCR fragment of the GhADF7 gene) generated using a random primer method (Prime-a-Gene Labelling System, Promega). The membrane filters (Hybond N, Amersham Pharmacia Biotech) were hybridized with the labelled GhADF7 probe overnight in the hybridization solution at 68 °C, and were then washed with 0.1× SSC and 0.5% SDS for 30–60 min. The 32P-labelled membranes were exposed to X-ray film at –70 °C. Positive cDNA clones were selected for sequencing.
Northern blot analysis
RNA samples (20 μg lane−1) from different cotton tissues were separated on 1.2% agarose–formaldehyde gels, and transferred onto Hybond-N nylon membrane by capillary blotting. The fragment of the GhADF7 3′-untranslated region was used as a gene-specific probe for Northern hybridization as described previously by Li et al. (2002).
RT-PCR analysis
The expression of the GhADF genes in cotton tissues was analysed by real-time quantitative reverse transcriptase (RT)-PCR using the fluorescent intercalating dye SYBR-Green in a detection system (MJ Research, Opticon 2) described earlier by Li et al. (2005). A cotton polyubiquitin gene (GhUBI1) was used as a standard control in the RT-PCR. A two-step RT-PCR procedure was performed in all experiments. First, total RNA samples (2 μg per reaction) from leaves, stems, cotyledons, roots, anthers, petals, and fibres were reverse transcribed into cDNAs. Then, the cDNAs were used as templates in real-time PCR with gene-specific primers (GhADF6-P1: 5′-AGCTCGAAAAAGATGGCCAACGCT-3′; GhADF6-P2:5′-CTTTATGGCAAAACTTGGAAATTG-3′; GhADF7-P1:5′-CATTGGTGGCAAAATGGCAAACTCA-3′; GhADF7-P2:5′-CCATCTTTGACACCATAACACAT-3′; GhADF8-P1:5′-CGGGAAAAAAAATGGCCAAACGCG-3′; GhADF8-P2:5′-ATCAAAGCAAGTTCCAAGATCCAA-3′). The amplification of the target gene was monitored every cycle by SYBR-Green fluorescence. The Ct (cycle threshold), defined as the PCR cycle at which a statistically significant increase of reporter fluorescence is first detected, is used as a measure for the starting copy numbers of the target gene. Relative quantitation of the target GhADF expression levels was performed using the comparative Ct method. The relative value for the expression level of each GhADF gene was calculated by the equation Y=10ΔCt/3×100% (ΔCt is the difference of Ct between the control GhUBI1 products and the target GhADF products, i.e. ΔCt=CtGhUBI1––CtGhADF). To achieve optimal amplification, PCR conditions for each primer combination were optimized for annealing temperature and Mg2+ concentration. PCR products were confirmed on an agarose gel. The efficiency of each primer pair was detected by using GhADF cDNAs as standard templates, and the RT-PCR data were normalized with the relative efficiency of each primer pair.
Sequence and phylogenetic analysis
Nucleotide and amino acid sequences were analysed using DNAstar (DNAstar Inc). The GhADF peptide sequences were aligned with the ClustalW program (http://www.ebi.ac.uk), and phylogenetic analysis was employed to investigate the evolutionary relationships among the GhADF genes. A Neighbor–Joining tree was generated in MEGA3.1. A bootstrap analysis with 1000 replicates was performed to assess the statistical reliability of the tree topology.
Genome Walker PCR
Genome Walker libraries were constructed using the Universal Genome Walker kit (Clontech) described earlier (Li et al., 2002). Primers for PCR-based DNA walking in Genome Walker Libraries were gene-specific GhADF7-P1 (5′-TGATGTTGTAATGAGGATTAAAAGAAC-3′) and GhADF7-P2 (5′-ACCATTTTGCCACCAATGAGAGAGAAT-3′) in a complementary chain +35 to +9 and +5 to -22 of the GhADF7 gene from the translational initiation site and the adapter sequence AP1 (5′-GTAATACGACTCACTATAGGGC-3′) and AP2 (5′-ACTATAGGGCACGCGTGGT-3′). Genome Walker PCR reactions were carried out twice successively using the Advantage-HF PCR Kit (Clontech), which is a KlenTaq-based system with Pfu-like high fidelity and Taq-like high efficiency in the amplification of the DNA template, by the method described previously (Li et al., 2002).
Construction of the GhADF7p:GUS chimeric gene and plant transformation
A SalI site and an XbaI site were introduced at the 5′-end and the 3′-end of the GhADF7 5′-upstream region (including the putative promoter fragment and the untranslated region before the initiation codon ATG), respectively, by the PCR method. The SalI/XbaI fragment (1.0 kb) was then subcloned into the pGEM-T vector. Plasmid DNA containing the GhADF7 5′-upstream fragments was digested with SalI and XbaI, and subcloned into SalI and an XbaI sites of the pBI101 vector, replacing the CaMV 35S promoter to generate a chimeric GhADF7p:GUS construct.
Tobacco leaves were cut into 2 cm2 pieces, and infected with Agrobacterium tumefaciens harbouring the GhADF7p:GUS construct. After co-cultivation for 2 d, the explants were cultured on MS selective medium containing 100 mg l−1 kanamycin and 1 mg l−1 6-BA (12/12 h light/dark cycle, 25 °C) for shoot regeneration. The transformed shoots were rooted on MS selective medium without phytohormones, and then were transplanted in soil for growing to maturation.
Arabidopsis transformation was performed by the floral dip method (Clough and Bent, 1998).
Overexpression of the GhADF7 gene in Arabidopsis and assay of pollen viability
The coding sequence of the GhADF7 gene, amplified from its cDNA by PCR with the proofreading Pfu DNA polymerase, was cloned into the GhADF7p:GUS construct with XbaI/SacI sites to replace the GUS gene (named pGhADF7 vector). The pGhADF7 vector was then transferred into Agrobacterium tumefaciens for Arabidopsis transformation by the floral dip method (Clough and Bent, 1998).
Pollen grains from wild-type and GhADF7-transgenic Arabidopsis plants were incubated for 5 min in the basic medium with the addition of 5 μg ml−1 fluorescein diacetate (FDA). After washing out FDA with basic medium, the fluorescence of the pollen grains was observed on a Nikon microscope (Japan) for determining the percentage of viable pollen grains.
In vitro pollen germination
Pollen grains were collected from flowers of both wild-type and transgenic tobacco and Arabidopsis plants, and then germinated on BK medium (0.01% H3BO4, 0.02% Ca(NO3)2, 0.01% KNO3, 0.02% MgSO4.7H2O, and 15% sucrose, pH 5.9) (Brewbaker and Kwack, 1963) at 28 °C under dark condition for 2–12 h.
Histochemical assay of GUS gene expression
Histochemical assays for GUS activity in the transgenic tobacco plants were conducted according to a modified protocol (Li et al., 2002). Fresh plant tissues were incubated in 5-bromo-4-chloro-3-indolylglucuronide (X–gluc) solution at 37 °C for 4–6 h, and then cleaned and fixed by rinsing with 100% and 70% ethanol successively. The samples were examined and photographed directly or under a Nikon microscope (Japan).
Overexpression of the GhADF7 gene in fission yeast
The coding sequence of the GhADF7 gene, amplified from its cDNA by PCR with the proofreading Pfu DNA polymerase, was cloned into the yeast vector REP5N with NotI/BamHI sites. Afterwards, the construct was transferred into yeast (S. pombe) cells by electroporation (Bio-Rad MicroPulser Electroporation Apparatus) according to the manufacturer's instructions. Overexpression of the GhADF7 gene in the transformed yeast cells was induced by the method described previously (Li et al., 2007). The yeast cells were observed and photographed under a Nikon microscope (Japan), using empty pREP5N transformants and untransformed cells as controls.
Observation of F-actin structures
Wild-type and transformed yeast cells were fixed in 4% formaldehyde freshly prepared in PBS for 1 h. After washing three times in PBS without formaldehyde, yeast cells were collected and incubated with 0.66 μM TRITC-conjugated phalloidin (P1951; Molecular Probes, Sigma) for 3 h at room temperature. The cells were then washed several times with PBS to remove unbound phalloidin and also incubated in PBS containing 0.6 μg ml−1 4′-6 diamidino-2-phenylindole (DAPI) as a counter stain for nuclei. The specimens were mounted on a glass slide with the same buffer, and observed immediately with a SP5 Meta confocal laser microscope (Leica, Germany).
Results
Isolation and characterization of GhADF cDNAs
By screening the cotton flower cDNA library, three cDNAs encoding ADF proteins were isolated and designated as GhADF6, GhADF7, and GhADF8 (accession numbers in GenBank: DQ402083, DQ402078, and DQ402079, respectively). The isolated GhADF cDNAs contain an encoding region, 5′- and 3′-untranslation region (5′-/3′-UTR). At nucleotide acid level, three GhADFs display relatively low homology. GhADF6 shares 36% identity with GhADF7 and 50% similarity with the GhADF8, while GhADF7 shares 44% homology with GhADF8. Sequence analysis predicted that each of the three cDNAs encodes an ADF protein containing 139 amino acid residues. At the amino acid level, GhADF6 shares 87% identity with GhADF8 and 72% identity with GhADF7, while there is 79% homology between GhADF7 and GhADF8. Comparing the amino acid sequences of the three GhADF proteins, there are 99 conserved sites in which no amino acid substitution occurred, suggesting that these sites may play a key role in maintaining ADF structure and its basic function. On the other hand, a total of 40 substitution sites are presented in the GhADF proteins, indicating that the mutations of ADF proteins occur during evolution (Fig. 1).
Fig. 1.
Comparison of structure and sequence homology among the isolated three GhADF proteins. The black areas indicate amino acids substitutions. The asterisk marks the position of the putative phosphorylation site at the N-terminus. The underlining represents the PIP2/actin-binding site.
Phylogenetic relationships of GhADFs
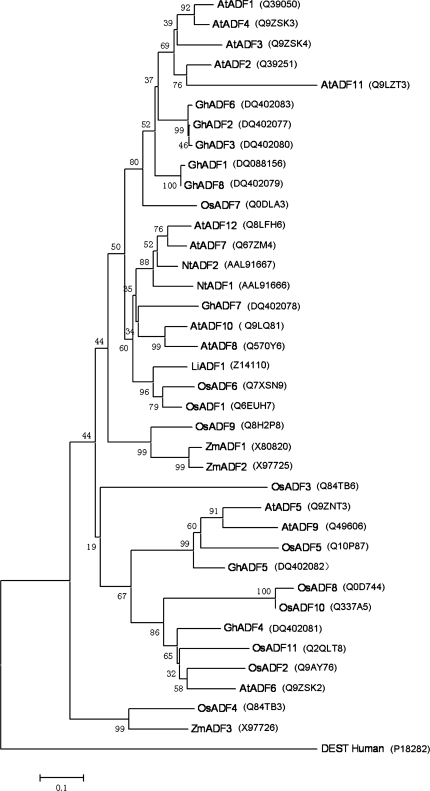
To study the phylogeny of ADF proteins, the complete protein sequences were aligned, and the tree was generated by the maximum parsimony method without any manual adjustment (Fig. 2). As the Arabidopsis thaliana genome sequencing project has been completed, it is possible to analyse the full complement of ADF/cofilin genes from this plant species. The phylogenetic analysis predicts that AtADF7 and perhaps AtADF8 and AtADF10 are pollen-specific proteins, like maize, tobacco, and lily pollen-specific ADFs which fall in the same group with these three AtADFs (Maciver and Hussey, 2002). As shown in Fig. 2, GhADF7 occupied a branch that is located with AtADF8 and AtADF10, whereas the rest of the GhADFs resided on the other subgroups of the tree. This result suggests that GhADF7 may be pollen-specific.
Fig. 2.
Phylogenetic relationships of cotton ADFs and other plant ADFs. The Neighbor–Joining tree was constructed in MEGA3.1 from 1000 bootstrap replicates. The sequences accession numbers were added following. DEST_Human is used as an outgroup.
GhADF7 is specifically expressed in anthers, while GhADF6 and GhADF8 are predominantly expressed in petals
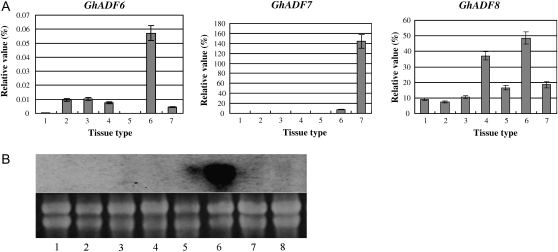
To investigate expression patterns of the isolated GhADF genes, total RNA was extracted from different cotton tissues. The expression patterns of three GhADF cDNA clones were analysed by real-time quantitative SYBR-Green RT-PCR using gene-specific primers as described in the Materials and methods. The relative transcript levels of the isolated GhADF genes are shown in Fig. 3A, using the cotton polyubiquitin gene (GhUBI1) as a standard control to normalize differences in RNA template concentrations (Li et al., 2005). The result revealed that the GhADF7 gene was specifically expressed in anthers, and its expression level reached a relative value of 144 in anthers compared to about 0–7.3 in other tissue types. On the other hand, GhADF6 transcripts were detected mainly in petals, but at very low levels in other tissues. Similarly, GhADF8 mRNAs were predominantly accumulated in petals, and at relatively high levels in cotyledons, but at relatively low levels in other tissues (Fig. 3A). Furthermore, Northern blot analysis, using the 3′-untranslated region (3′-UTRs) of the GhADF7 gene as a probe, demonstrated that the accumulation of the GhADF7 transcripts was specific to anthers. The GhADF7 expression in anthers reached its highest level, but no or little signals were visible in ovules, cotyledons, hypocotyls, fibres, petals, leaves, and roots (Fig. 3B). This result was consistent with the real-time quantitative RT-PCR result and further confirmed that the GhADF7 gene expression is anther-specific.
Fig. 3.
Analysis of expression of the isolated GhADF genes in cotton tissues. (A) Real-time quantitative RT-PCR analysis of the expression of GhADF genes. Total RNA was isolated from various cotton tissues. 1, Leaves; 2, stems; 3, roots; 4, cotyledons; 5, 10 DPA fibres; 6, petals; 7, anthers. (B) RNA gel blot analysis of GhADF7 expression. Upper panel: Autoradiograph of RNA hybridization; Lower panel: RNA gel before being transferred to membrane showing equal loading of RNAs. 1, Cotyledons; 2, leaves; 3, roots; 4, hypocotyls; 5, petals; 6, anthers; 7, fibres; 8, ovules.
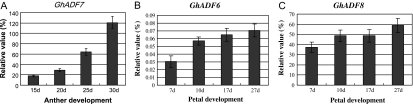
To investigate whether the expression of the GhADF genes is developmentally regulated in flowers, the expression of GhADF6/8 genes was analysed during petal development and GhADF7 expression during anther development. As shown in Fig. 4, GhADF7 activity was gradually increased when the anthers developed, and reached its highest levels in 30-d-old anthers just before flowering. The GhADF6 and GhADF8 genes were expressed in a similar manner during petal development. At the early stages of development, relatively low levels of the gene products were detected in petals. As petals developed further, the expression activities of both genes were gradually enhanced to relatively high levels (Fig. 4). Collectively, GhADF6 and GhADF8 were preferentially expressed in petals, whereas GhADF7 displayed the highest level of expression in anthers, revealing its anther specificity. We are interested in the anther/pollen-specific gene, so our further study focuses on the GhADF7 gene.
Fig. 4.
Analysis of expression patterns of the GhADF genes during cotton flower development. (A) Real-time quantitative RT-PCR analysis of GhADF7 expression in anther development. (B, C) Real-time quantitative RT-PCR analysis of GhADF6 and GhADF8 expression in petal development. d, days.
Isolation and characterization of the GhADF7 gene
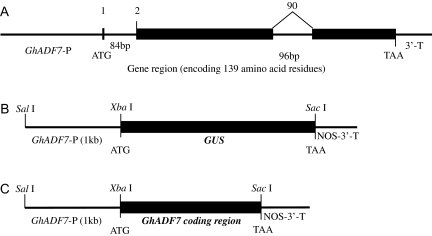
To investigate cotton ADF gene structure, the GhADF7 gene was isolated from cotton genomic DNA by PCR, using Pfu DNA polymerase and gene-specific primers. Based on the sequence of the GhADF7 gene, a 1030 bp fragment of the 5′-upstream region from the translation start site was then isolated by Genome Walker PCR for characterization of the promoter. The complete GhADF7 gene is 1904 bp in length (including a 1030 bp 5′-upstream region, a 600 bp transcribed region, and a 274 bp 3′-downstream sequence). Sequence comparison between its cDNA and genomic clone revealed that the GhADF7 contains two introns splitting its open reading frame into three exons (Fig. 5A). The first intron resides just behind the start codon (ATG) and is 84 bp in length, while the second intron splits codon 90 (Trp90) and is 96 bp in length. The intron positions of the cotton ADF7 gene are exactly identical to those of the Arabidopsis and rice ADF genes, suggesting that intron positions are relatively conserved in plants. Furthermore, GhADF7 showed a remarkable tendency, i.e. the first amino acid (the initiating methionine) to be encoded by a separate exon, like the other plant ADF genes (Maciver and Hussey, 2002).
Fig. 5.
The structure of the GhADF7 gene and construction of GhADF7p:GUS and pGhADF7 expression vectors. (A) GhADF7 gene structure. Exons are denoted by black boxes. Introns, 5′-flanking region and 3′-UTR are denoted by lines. The length of the introns in base pairs are indicated. The number at the boundaries of each exon indicates the codon at which the intron is located. The translation initiation and termination codons are shown. (B) Construction of the chimeric gene between the GhADF7 5′-flanking region and the GUS gene. The length of the 5′-flanking region and cloning sites used for fusion constructs are shown. (C) Construction of the pGhADF7 expression vector containing the GhADF7 coding sequence downstream of its promoter.
GhADF7p:GUS is specifically expressed in pollen grains
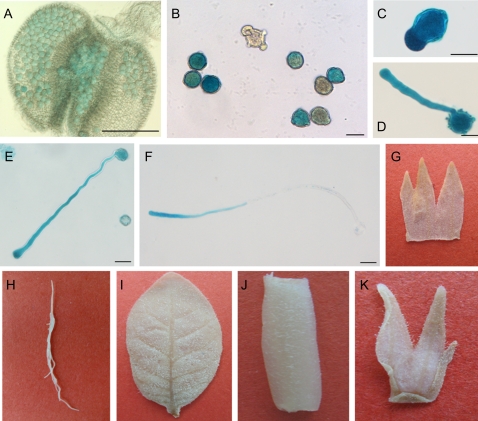
To determine the precise expression pattern of the GhADF7 gene in cotton, the 1 kb GhADF7 5′-flanking region (the putative promoter fragment and untranslated region) before the translational initiation codon ATG was subcloned upstream of the GUS reporter gene in the pBI101 vector, giving rise to the GhADF7p:GUS chimeric gene (Fig. 5B). The GhADF7p:GUS construct was introduced into tobacco and Arabidopsis by Agrobacterium tumefaciens-mediated transformation. The transgenic tobacco and Arabidopsis plants for each independent line were examined with a histochemical assay for GUS expression, using non-transformed plants as negative controls. A total of 41 transgenic tobacco plants and 35 transgenic Arabidopsis plants were obtained. Histochemical assay revealed that the 1 kb GhADF7 promoter showed specific activity in anthers. Strong GUS activity was only observed in pollen grains, whereas no signal was detected in other cell types (including the epidermis, endothecium, vascular bundle, etc) of the transgenic anthers (Fig. 6A, B), and no or very little GUS staining was found in other tissues such as roots, stems, leaves, petals, sepals, ovaries, and ovules (Fig. 6G–K), indicating that the GhADF7 gene produces identical and pollen-specific localization pattern. Furthermore, when the transgenic pollen grains were germinated in vitro, the histochemical assay of GUS activity revealed the localized expression of the GhADF7 gene in germinating pollen grains. A majority of germination-competent pollen grains usually developed a polar protrusion within 30 min after cultivation in vitro. Strong GUS signals were detected in the geminating transgenic pollen grains, especially in the polar protrusion (Fig. 6C). Pollen tubes elongated relatively normally for at least 4–12 h after germination in vitro and showed a detectable level of GUS activity. A high level of GUS activity was found at the apical regions of the pollen tubes, while the shank of these tubes showed weak to moderate GUS signals (Fig. 6D–F). The results suggest that the GhADF7 gene may be involved in pollen development and germination, especially in pollen tube elongation.
Fig. 6.
Histochemical assay of the expression of GUS gene driven by GhADF7 promoter in transgenic tobacco and Arabidopsis plants. (A) Transgenic Arabidopsis anther. Strong GUS activity was detected in pollen grains, but no or only weak GUS staining was found in the other cell types of the transgenic anther. (B) Pollen grains of the transgenic tobacco plants. High levels of GUS expression were observed in transgenic pollen grains. (C–F) Transgenic tobacco pollen grains after germinating for 1, 2, 6, and 12 h, respectively. Very strong GUS expression was detected in the germinating pollen grains, especially at the tips of pollen tubes. (G–K) Other tissues of the transgenic tobacco plants. (G) Petal; (H) root; (I) leaf; (J) stem; (K) sepal. No GUS staining was found in the other tissues of the transgenic tobacco plants. Bar=25 μm.
Overexpression of GhADF7 in Arabidopsis thaliana reduces pollen fertility
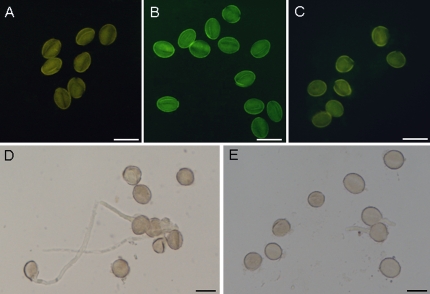
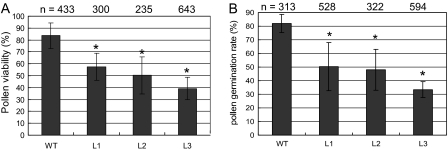
To investigate the role of the GhADF7 gene in plant anthers, the GhADF7 coding sequence amplified from its cDNA was subcloned downstream of its own 5′-flanking region (the putative promoter fragment and untranslated region) in the pBI101 vector, giving rise to the overexpression construct of the GhADF7 gene (Fig. 5C). The construct containing the GhADF7 gene was introduced into Arabidopsis by the floral dip method. The transgenic Arabidopsis seeds were germinated on a selective MS medium containing 100 mg l−1 kanamycin. Twenty-seven transgenic Arabidopsis plants were obtained. RT-PCR analysis revealed that the cotton ADF7 gene, driven by its own promoter, was specifically expressed in anthers of the transgenic Arabidopsis plants (data not shown). To determine whether overexpression of the GhADF7 gene affects pollen cell viability, the percentage of viable transgenic pollen grains and their germination was assayed, using wild-type pollen grains as controls. After staining by fluorescein diacetate (FDA), strong fluorescence of the living cells was observed in the most wild-type pollen grains, but relatively weak or no fluorescence was detected in the most transgenic pollen grains, a large portion of which could not germinate normally in vitro (Fig. 7). Statistical analysis showed that only 39.2–57.3% transgenic pollen grains were positive for FDA staining, whereas 83.7% of control pollen grains were viable (Fig. 8A). The germination rate of transgenic pollen grains in vitro was only 33.4–50.1% which was much lower than the 81.8% of the wild type (Fig. 8B). The results indicated that overexpression of the GhADF7 gene significantly reduced pollen fertility and, consequently, transgenic plants are partially male-sterile.
Fig. 7.
Overexpression of GhADF7 in Arabidopsis thaliana reduces pollen viability. (A) Negative control. Unstained wild-type pollen grains. (B, C) Stained pollen grains. (B) Wild type; (C) GhADF7-transgenic pollen grains. Pollen grains were stained with FDA (fluorescein diacetate) to determine cell viability. Strong fluorescence of living cells was observed in wild-type pollen grains, but only relatively weak or no fluorescence was detected in the most of transgenic pollen grains, indicating that overexpression of the GhADF7 gene in Arabidopsis resulted in a reduction in pollen fertility of the transgenic plants. (D, E) Germinated pollen grains after incubation in BK solution for 6 h. (D) Wild-type pollen grains (controls); (E) GhADF7-transgenic pollen grains. The transgenic pollen grains were fewer and germinated slowly compared with the controls. Bar=20 μm.
Fig. 8.
Statistical analyses of pollen viability and germination of transgenic Arabidopsis plants. (A) Assay of pollen viability. (B) Assay of pollen germination. WT, wild type (control); L1–L3, different transgenic Arabidopsis lines. The asterisk represents significant differences between the transgenic line and control at P <0.05 by the Tukey HSD test. The experiments were repeated three times, bars show standard errors.
Overexpression of GhADF7 in fission yeast hinders cell division due to the defective actin cytoskeleton
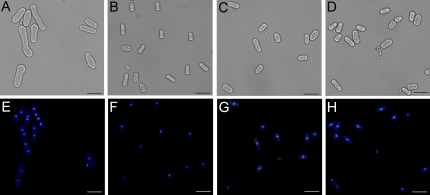
To investigate whether the GhADF7 gene plays a role in cell morphology, the coding sequence of the GhADF7 gene was cloned into a vector pREP5N, and transferred into yeast (S. pombe) cells. Expression of the GhADF7 gene in yeast cells was induced by removing thiamine from the medium. After inducing for 20 h, the transformed cells with the expressing GhADF7 gene grown in the induction medium (Fig. 9A) were remarkably longer than those transformed cells grown in uninduced conditions (Fig. 9B). By contrast, the transformed cell harbouring the empty pREP5N vector displayed a normal length in the induction medium (Fig. 9C), as non-transformed cells did (Fig. 9D). In addition, it was observed that as much as 75% of the cells with the expressing GhADF7 gene were binucleate, even four- and eight-nucleate, at 20 h after induction (Fig. 9E), whereas approximately 1% of the uninduced and control cells were binucleate (Fig. 9F, G, H), when stained with a nucleus-specific fluorescent dye, DAPI. Meanwhile, other abnormal phenotypic cells were observed, such as spherical tips and bifurcate cells.
Fig. 9.
Overexpression of GhADF7 gene in fission yeast alters cell morphology, leading to the formation of multinucleate cells. (A–D) Cells observed in bright field. (E–H) The same cells observed in dark field. Cells grown in respective conditions were stained with a nucleus-specific fluorescent dye, DAPI. Yeast cells harbouring the GhADF7 gene were induced to undergo longitudinal elongation, resulting in abnormal morphology (A) and the formation of multinucleate cells (E). By contrast, yeast cells harbouring the GhADF7 gene exhibited normal morphology (B) and a single-nucleus (F) in non-induction medium. Likewise, yeast cells harbouring the empty pREP5N vector, grown in induction medium, showed normal morphology (C) and a single-nucleus (G) as those grown under non-induction conditions (D, H). Bar=20 μm. (This figure is available in colour at JXB online.)
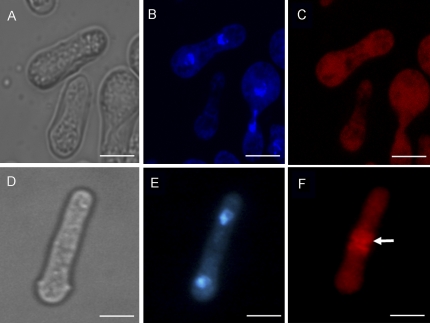
To demonstrate that overexpression of the GhADF7 gene affects the actin dynamics in cells, the actin cytoskeleton was studied in yeast cells using rhodamine–phalloidin staining for F-actin (Fig. 10). The longitudinal F-actin cables were not seen, but replaced by short and random arrays in the cells with expressing GhADF7. Furthermore, the F-actin ring had disappeared in these cells (Fig. 10C). By contrast, almost all the F-actin ring could be observed during metaphase in the dividing control cells (Fig. 10F). The experimental results suggested that the balance of actin depolymerization and polymerization may have been altered, leading to defective cytokinesis and multinucleate formation in the cells, because of the overexpression of the GhADF7 gene in yeast.
Fig. 10.
GhADF7 gene is involved in controlling the organization of the actin cytoskeleton in fission yeast cells. Cells were observed in bright field (A, D), fixed and stained with DAPI (B, E) and TRITC–phalloidin (C, F). (A, B, C) Yeast cells with the GhADF7 gene, grown in induction medium at 30 °C, were induced to lose the F-actin ring necessary for cell division. (D, E, F) Yeast cells harbouring the empty pREP5N vector, grown in induction medium at 30 °C, exhibited normal F-actin ring structure during mitosis. The arrow indicates the F-actin ring. Bar=5 μm.
Discussion
In this work, three additional cotton ADF genes were identified. Our data showed that the GhADF7 gene was specifically expressed in pollen, whereas high levels of GhADF6 and GhADF8 activities were detected in petals (Fig. 3). A previous study indicated that there was a significant delay in the induction of flowering in AtADF1-underexpressed plants, which grew larger than wild-type plants (Dong et al., 2001a,). The F-actin cytoskeleton has been associated with flower induction through signal transport between cells during development of the petals (McLean et al., 1997). Actin binding proteins may be sensitive to changes in the level of the actin cytoskeleton via intercellular signalling processes. Previous studies revealed that GhADF1 and GhADF2 were predominantly expressed in fibres, GhADF5 was mainly expressed in cotyledons, whereas GhADF3 and GhADF4 genes have no tissue specificity (Zhang et al., 2007; Wang et al., 2009). In general, different GhADF genes are expressed in different cotton tissues, suggesting that each ADF isoform may develop the functional difference during evolution.
Eukaryotic ADF/cofilin proteins from organisms as distantly related as yeast and mammals are highly conserved, and are believed to play similar roles in the regulation of actin dynamics and organization (Aizawa et al., 1997). In our study, the structure of GhADF7 has been analysed and compared with the related genes from other organisms. It was found that these gene structures are surprisingly similar to each other. A remarkable tendency for ADF/cofilins is for the first amino acid (or the first few amino acids) to be encoded by a separate exon. Previous studies have revealed that 75% (9/12) Arabidopsis ADF proteins were organized in a similar manner, with the first amino acid to be encoded by a separate exon (Feng et al., 2006). Many ADF/cofilins are known to associate with the phospholipid phosphatidylinositod-4,5-bisphosphate (PIP2), and a short sequence (Trp100–Met115) has been identified to be important for binding to both actin and PIP2 (Yonezawa et al., 1991). In addition, in the conserved N-terminal domain, the serine residue was present in GhADF7, like that in other AtADFs except AtADF5 (Dong et al., 2001b). The conserved serine residue is regulated by phosphorylation to effect the activity of ADF/cofilin proteins. A Ser6-to-Asp conversion abolished the interaction between NtADF1 and F-actin in elongating pollen tubes and reduced its inhibitory effect on pollen tube growth significantly, suggesting that phosphorylation at Ser6 may be a prominent regulatory mechanism for this pollen ADF (Chen et al., 2002).
In germinating transgenic pollen grains, strong GUS signals were found in pollen grains, especially the polar protrusion of the germinating pollen (Fig. 6), indicating that the GhADF7 promoter contained all the cis regulatory elements required for its pollen-specificity, like the other pollen-specific genes in tobacco (Chen et al., 2002) and lily (Allwood et al., 2002). This result is consistent with the GhADF7 expression pattern revealed by real-time quantitative RT-PCR and Northern blot analyses (Fig. 4). Thus, the similarity of the expression of different ADF/cofilin genes indicates that they have a certain conservation in their genomic organization.
Previous studies showed that, both loading onto the stigma and in a hydrated environment, pollen can germinate to give rise to a polar-growing pollen tube in a short time (Edlund et al., 2004). Transcriptomic and proteomic data showed that mature pollen has presynthesized these mRNA and proteins (such as wall remodelling proteins, carbohydrate synthesizing proteins, and energy metabolism proteins) to prepare for quick pollen germination and polar pollen tube growth (Dai et al., 2007). Actomyosin-driven intracellular trafficking and active actin remodelling in the germinating pollen are both important aspects of this rapid germinating process. Rapid actin turnover must also keep pace with the equally rapid pollen tube growth. Therefore, GhADF7 proteins which enhance the depolymerization of actin filaments at their minus or slow-growing ends may be mainly responsible for actin remodelling and pollen polarity growth.
It has been reported that F-actin plays an important role in pollen tube growth (Mascarenhas, 1993; Chen et al., 2002), in trichome morphogenesis (Mathur et al., 1999), in root hair tip growth (Miller et al., 1999), in fibre elongation (Li et al., 2005), and in cell elongation of other cell types (Baluska et al., 2000; Waller et al., 2002; Yamamoto and Kiss, 2002). In the tip-growing pollen tube, F-actin might transport secretory vesicles and other large organelles toward the cell periphery, especially near the polar region during tip growth. Similar F-actin arrays were also found for root hair growth (Miller et al., 1999) as well as root tip growth (Blancaflor and Hasenstein, 1997). In addition, phalloidin labelling of fixed pollen tubes from several species has also revealed that disorganized, short actin filaments have been observed in the subapical region and are most concentrated around the base of the clear zone (Chen et al., 2002). Similarly, cotton ADF7 displays its strong activity at the apical region of the pollen tubes. This would result in a higher number of fragmented filaments and generate more barbed ends for actin polymerization in the pollen tube tips where actin filaments are expected to undergo rapid restructuring.
It is believed that a highly dynamic cytoskeleton maintained by significant amounts of actin is essential for the elongation of pollen tubes as well as other cell types. Reduction of the actin level affected pollen tube elongation and resulted in aberrant cytoskeleton structures in these cells (Chen et al., 2003). In a few studies, the effects of experimentally increasing ADF/cofilin protein levels have been investigated in model plants, such as tobacco and Arabidopsis. A previous study indicated that overexpression of Arabidopsis ADF1 resulted in the disappearance of thick actin cables in polar cells (such as cells in etiolated hypocotyls and root hairs), caused irregular cellular morphogenesis, and reduced the growth of cells and organs (Dong et al., 2001b). Overexpression of NtADF1 in pollen tubes reduced fine, axially oriented actin cables and, consequently, inhibited pollen tube growth (Chen et al., 2002). Similarly, our results revealed that overexpression of the cotton ADF7 gene in Arabidopsis remarkably affected pollen viability and, as a result, the transgenic plants were partially male-sterile (Figs 7, 8). In addition, overexpression of plant ADF in yeast (S. pombe) exhibited a diffuse actin staining pattern with some giving dispersed actin dots and a double nucleus (Xia et al., 1996). Likewise, our data indicated that overexpression of the cotton ADF7 in fission yeast displayed the defective actin cytoskeleton in the cells (Fig. 10). Previous studies revealed that, in co-operation with other actin-binding proteins, yeast cells during the metaphase formed a contractile ring arranged in an equatorial plate (Nakano and Mabuchi, 2006). During anaphase, the F-actin ring initiated contraction and the cells shrink to divide the cell into two (Ritsuko and Issei, 2002). By contrast, heterologous expression of the GhADF7 gene in yeast cells may affect the building of the actin ring and arrest cytoplasm division, but seems to have no effect on nucleus division, leading to the formation of multinucleate cells. Additional GhADF7 activity may change the balance of actin depolymerization and polymerization, altering the rate of actin cycling and, ultimately, actin organization in both transgenic Arabidopsis pollen grains and transformed yeast cells.
In conclusion, the GhADF7 gene exhibits its specific expression in cotton pollen grains, especially in the apical regions of elongating pollen tubes. Overexpression of this gene results in reduced pollen fertility in Arabidopsis thaliana as well as defective cytokinesis in fission yeast. These results suggest that GhADF7 proteins may affect the dynamics of actin cycling in cells, particularly active growth points, where actin filaments are expected to undergo rapid restructuring. Thus, the results of this study contribute to the understanding of the role of cotton ADF7 gene in pollen development.
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (Grant nos 30470930, 30871317) and the National Major Project of Breeding for New Transgenic Organisms (Grant nos 2008ZX08009-003, 2009ZX08009-117B).
References
- Aizawa H, Fukui Y, Yahara I. Live dynamics of Dictyostelium cofilin suggests a role in remodelling actin latticework into bundles. Journal of Cell Science. 1997;110:2333–2334. doi: 10.1242/jcs.110.19.2333. [DOI] [PubMed] [Google Scholar]
- Allwood EG, Anthony RG, Smertenko AP, Reichelt S, Drobak BK, Doonan HJ, Weeds AG, Hussey PJ. Regulation of the pollen-specific actin-depolymerizing factor LlADF1. The Plant Cell. 2002;14:2915–2927. doi: 10.1105/tpc.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Salaj J, Mathur J, Braun M, Jasper F, Samaj J, Chua NH, Barlow PW, Wolkmann D. Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Developmental Biology. 2000;227:618–632. doi: 10.1006/dbio.2000.9908. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Bray D. Distribution and cellular localization of actin depolymerizing factor. Journal of Cell Biology. 1987;105:2817–2825. doi: 10.1083/jcb.105.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancaflor EB, Hasenstein KH. The organization of the actin cytoskeleton in vertical and graviresponding primary roots of maize. Plant Physiology. 1997;113:1447–1455. doi: 10.1104/pp.113.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewbaker JL, Kwack BH. The essential role of calcium ion in pollen germination and pollen tube growth. American Journal of Botany. 1963;50:859–865. [Google Scholar]
- Chen CY, Wong EI, Vidali L, Estavillo A, Hepler PK, Wu H, Cheung AY. The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. The Plant Cell. 2002;14:2175–2190. doi: 10.1105/tpc.003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Cheung AY, Wu HM. Actin-depolymerizing factor mediates Rac/Rop GTPase–regulated pollen tube growth. The Plant Cell. 2003;15:237–249. doi: 10.1105/tpc.007153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bernstein BW, Bamburg JR. Regulating actin-filament dynamics in vivo. Trends in Biochemical Sciences. 2000;25:19–23. doi: 10.1016/s0968-0004(99)01511-x. [DOI] [PubMed] [Google Scholar]
- Chung YY, Magnuson NS, An GH. Subcellular localization of actin depolymerizing factor in mature and germinating pollen. Molecular Cell. 1995;5:224–229. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dai SJ, Chen TT, Chong K, Xue YB, Liu SQ, Wang T. Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Molecular and Cellular Proteomics. 2007;6:207–230. doi: 10.1074/mcp.M600146-MCP200. [DOI] [PubMed] [Google Scholar]
- Dong CH, Kost B, Xia G, Chua NH. Molecular identification and characterization of the Arabidopsis AtADF1, AtADF5, and AtADF6 genes. Plant Molecular Biology. 2001a;45:517–527. doi: 10.1023/a:1010687911374. [DOI] [PubMed] [Google Scholar]
- Dong CH, Xia GX, Hong Y, Ramachandran S, Kost B, Chua NH. ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. The Plant Cell. 2001b;13:1333–1346. doi: 10.1105/tpc.13.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Goldberg RB. Genetic control of flower development. Trends in Genetics. 1989;5:256–261. doi: 10.1016/0168-9525(89)90098-x. [DOI] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D. Pollen and stigma structure and function: the role of diversity in pollination. The Plant Cell. 2004;16:S84–S97. doi: 10.1105/tpc.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Liu QP, Xue QZ. Comparative study of rice and Arabidopsis actin-depolymerizing factors gene families. Journal of Plant Physiology. 2006;163:69–79. doi: 10.1016/j.jplph.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Kovar RD, Staiger JC. Latrunculin B has different effects on pollen germination and tube growth. The Plant Cell. 1999;11:2349–2363. doi: 10.1105/tpc.11.12.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon BC. Actin monomer-binding proteins and the regulation of actin: dynamics in plants. Journal of Plant Growth Regulation. 2001;20:103–112. [Google Scholar]
- Kim SR, Kim Y, An G. Molecular cloning and characterization of anther-preferential cDNA encoding a putative actin-depolymerizing factor. Plant Molecular Biology. 1993;21:39–45. doi: 10.1007/BF00039616. [DOI] [PubMed] [Google Scholar]
- Li XB, Cai L, Cheng NH, Liu JW. Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiology. 2002;130:666–674. doi: 10.1104/pp.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Fan XP, Wang XL, Cai L, Yang WC. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. The Plant Cell. 2005;17:859–875. doi: 10.1105/tpc.104.029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang XL, Huang GH, Li XB. Molecular characterization of cotton GhTUA9 gene specifically expressed in fibre and involved in cell elongation. Journal of Experimental Botany. 2007;58:3227–3238. doi: 10.1093/jxb/erm167. [DOI] [PubMed] [Google Scholar]
- Lopez I, Anthony RG, Maciver SK, Jiang CJ, Khan S, Weeds AG, Hussey PJ. Pollen specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proceedings of the National Academy of Sciences, USA. 1996;93:7415–7420. doi: 10.1073/pnas.93.14.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I. An actin-depolymerizing protein (depactin) from starfish oocytes: properties and interaction with actin. Journal of Cell Biology. 1983;97:1612–1621. doi: 10.1083/jcb.97.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Hussey PJ. The ADF/cofilin family: actin-remodelling proteins. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-5-reviews3007. reviews 3007.1-3007.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP. Molecular mechanisms of pollen tube growth and differentiation. The Plant Cell. 1993;5:1303–1314. doi: 10.1105/tpc.5.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Spielhofer P, Kost B, Chua NH. The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development. 1999;126:5559–5568. doi: 10.1242/dev.126.24.5559. [DOI] [PubMed] [Google Scholar]
- McCurdy DW, Kovar DR, Staiger CJ. Actin and actin-binding proteins in higher plants. Protoplasma. 2001;215:89–104. doi: 10.1007/BF01280306. [DOI] [PubMed] [Google Scholar]
- McLean BG, Hempel FD, Zambryski PC. Plant intercellular communication via plasmodesmata. The Plant Cell. 1997;9:1043–1054. doi: 10.1105/tpc.9.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DD, de Ruijter NCA, Bisseling T, Emons AMC. The role of actin in root hair morphogenesis: studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. The Plant Journal. 1999;17:141–154. [Google Scholar]
- Nakano K, Mabuchi I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Molecular Biology of Cell. 2006;17:1933–1945. doi: 10.1091/mbc.E05-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsuko A, Issei M. F-actin ring formation and the role of F-actin cables inthe fission yeast Schizosaccharomyces pombe. Journal of Cell Science. 2002;115:887–898. doi: 10.1242/jcs.115.5.887. [DOI] [PubMed] [Google Scholar]
- Rozycka M, Khan S, Lopez I, Greenland AJ, Hussey PJ. A Zea mays pollen cDNA encoding a putative actindepolymerizing factor. Plant Physiology. 1995;107:1011–1012. doi: 10.1104/pp.107.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther development defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reproduction. 1999;11:297–322. [Google Scholar]
- Smertenko AP, Allwood EG, Khan S, Jiang CJ, Maciver SK, Weeds AG, Hussey PJ. Interaction of pollen-specific actin-depolymerizing factor with actin. The Plant Journal. 2001;25:203–212. doi: 10.1046/j.1365-313x.2001.00954.x. [DOI] [PubMed] [Google Scholar]
- Vidali L, Hepler PK. Actin and pollen tube growth. Protoplasma. 2001;215:64–67. doi: 10.1007/BF01280304. [DOI] [PubMed] [Google Scholar]
- Waller F, Riemann M, Nick P. A role for actin-driven secretion in auxin-induced growth. Protoplasma. 2002;219:72–81. doi: 10.1007/s007090200007. [DOI] [PubMed] [Google Scholar]
- Wang HY, Wang J, Gao P, Jiao GL, Zhao PM, Li Y, Wang GL, Xia GX. Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnology Journal. 2009;7:13–23. doi: 10.1111/j.1467-7652.2008.00367.x. [DOI] [PubMed] [Google Scholar]
- Wang XL, Li XB. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. The Plant Journal. 2009;57:473–486. doi: 10.1111/j.1365-313X.2008.03700.x. [DOI] [PubMed] [Google Scholar]
- Xia GX, Ramachandran S, Hong Y, Chan YS, Simanis V, Chua NH. Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. The Plant Journal. 1996;10:761–769. doi: 10.1046/j.1365-313x.1996.10040761.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Kiss JZ. Disruption of the actin cytoskeleton results in the promotion of gravitropism in inflorescence stems and hypocotyls of Arabidopsis. Plant Physiology. 2002;128:669–681. doi: 10.1104/pp.010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa N, Homma Y, Yahara I, Sakai H, Nishida E. A short sequence responsible for both phosphoinositide binding and actin binding activities of cofilin. Journal of Biological Chemistry. 1991;266:17218–17221. [PubMed] [Google Scholar]
- Zhang CW, Guo LL, Wang XL, Zhang H, Shi HY, Xu WL, Li XB. Molecular characterization of four ADF genes differentially expressed in cotton. Journal of Genetics and Genomics. 2007;34:347–354. doi: 10.1016/S1673-8527(07)60037-X. [DOI] [PubMed] [Google Scholar]