Abstract
The colour of papaya fruit flesh is determined largely by the presence of carotenoid pigments. Red-fleshed papaya fruit contain lycopene, whilst this pigment is absent from yellow-fleshed fruit. The conversion of lycopene (red) to β-carotene (yellow) is catalysed by lycopene β-cyclase. This present study describes the cloning and functional characterization of two different genes encoding lycopene β-cyclases (lcy-β1 and lcy-β2) from red (Tainung) and yellow (Hybrid 1B) papaya cultivars. A mutation in the lcy-β2 gene, which inactivates enzyme activity, controls lycopene production in fruit and is responsible for the difference in carotenoid production between red and yellow-fleshed papaya fruit. The expression level of both lcy-β1 and lcy-β2 genes is similar and low in leaves, but lcy-β2 expression increases markedly in ripe fruit. Isolation of the lcy-β2 gene from papaya, that is preferentially expressed in fruit and is correlated with fruit colour, will facilitate marker-assisted breeding for fruit colour in papaya and should create possibilities for metabolic engineering of carotenoid production in papaya fruit to alter both colour and nutritional properties.
Keywords: Carotenoid, lycopene, lycopene β-cyclase, papaya fruit
Introduction
Papaya (Carica papaya L.) is a fruit crop widely grown in tropical and sub-tropical environments. There has recently been increased interest in the study of the genome of papaya due to its small genome size of 372 Mb and its short life cycle compared with many other tropical fruit tree crops. The two major papaya fruit flesh colours, red and yellow, are controlled by a single genetic locus with yellow being dominant over red (Storey, 1969). The fruit flesh colour of papaya is determined largely by the carotenoid content. Red-fleshed papaya fruit contain high levels of lycopene, whereas yellow-fleshed fruit do not (Yamamoto, 1964; Chandrika et al., 2003). A draft genome sequence of a transgenic (red) papaya has recently been published (Ming et al., 2008).
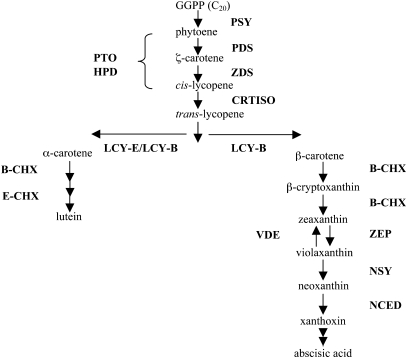
Carotenoid pigments are indispensable for photosynthesis in plants. In many species, carotenoids can also accumulate as secondary metabolites in chromoplasts of flowers, fruits, seeds or roots to provide distinct coloration, ranging from yellow to orange and red. The carotenoid biosynthesis pathway has been extensively studied in many photosynthetic and non-photosynthetic organisms. Carotenoid biosynthesis begins with the formation of phytoene from geranylgeranyl diphosphate (Fig. 1). Lycopene is a precursor of β-carotene and β-cryptoxanthin. In tomato, lycopene accumulation during fruit ripening is due to the down-regulation of lycopene β-cyclase activity (Pecker et al., 1996). Tomato contains two lycopene β-cyclase genes, including one that is expressed in photosynthetic tissues (chloroplast-specific) and one that is expressed predominantly in the fruit and flowers (chromoplast-specific). It was expected that a similar situation may occur in papaya. However, Skelton et al. (2006) were only able to find a single lycopene β-cyclase gene in papaya and its expression profile did not appear to correlate with lycopene production in the fruit. Furthermore, the sequence of this lcy-β gene from both red- and yellow-fleshed papaya cultivars was identical.
Fig. 1.
Carotenoid pathway in plants. PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; HPD, 4-hydroxy phenylpyruvate dioxygenase; PTO, plastid terminal oxidase; CRTISO, carotene cis-trans-isomerase; LCY-E, lycopene-ϵ-cyclase; LCY-B, lycopene β-cyclase; B-CHX, β-carotene hydroxylase; E-CHX, ϵ-carotene hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; NSY, neoxanthin synthase; NCED, 9-cis-epoxycarotenoid dioxygenase.
Although Skelton et al. (2006) identified only one lycopene β-cyclase gene, it was hypothesized that papaya would contain a second such gene, similar to the situation in tomato. In Australia, yellow papaya (known locally as paw-paws) are grown more commonly than red papaya. However, germplasm used in papaya breeding in Australia includes both yellow and red cultivars. Tainung (red) and Hybrid 1B (yellow) are two papaya cultivars grown commercially in Australia that differ in fruit colour (Fig. 2) and flavour. The objectives of this project were to clone all lycopene β-cyclase genes present in red and yellow papaya cultivars and to identify the genetic basis for lycopene production in papaya fruit.
Fig. 2.

Ripe fruit of the red Tainung (left) and yellow Hybrid 1B (right).
Materials and methods
Plant material
Leaves, flowers, and fruit from various papaya cultivars were obtained from Queensland Primary Industries and Fisheries, Centre for Tropical Agriculture, Southedge, QLD Australia.
DNA isolation
Genomic DNA was isolated from papaya leaves using a DNeasy Plant kit (Qiagen) according to the manufacturer's instructions.
Design of degenerate primers
An alignment of amino acid sequences of lycopene ϵ-cyclase from tomato, wild tomato, and Arabidopsis; lycopene β-cyclase from tomato and capsanthin-capsorubin synthase from bell pepper (Ronen et al., 1999) was used to identify amino acid regions conserved across these enzymes. Two degenerate forward-primers (NNYGVWEDE-F, NNYGVWVDE-F) and two degenerate reverse-primers (PTFLYAMP1-R, PTFLYAMP2-R) were designed to encode the conserved peptide sequences (Table 1).
Table 1.
Primer sequences used for PCR amplification, SNP detection, and measuring gene expression
| Experiment | Primer name | Primer sequence 5′-3′ |
| Lycopene cyclase | NNYGVWEDE-F | AA(T,C)AA(T,C)TA(T,C)GGIGTITGGGA(A,G)GA(T,C)GA |
| degenerate PCR | NNYGVWVDE-F | AA(T,C)AA(T,C)TA(T,C)GGIGTITGGGTIGA(T,C)GA |
| PTFLYAMP1-R | GGCATIGC(A,G)TA(T,C)AA(A,G)AAIGTIGG | |
| PTFLYAMP2-R | GGCATIGC(A,G)TAIAG(A,G)AAIGTIGG | |
| Lycopene cyclase | Cp_Cycb_F2 | AATGGGGTTAAATTCCTCAA |
| 5′ and 3′-RACE | Cp_Cycb_R2 | TCCTCCAATACCCTCTTCAC |
| Lycopene cyclase | Cp_Cycb_F1_5′Racing | AAACAATCGTCAAGCCCTAG |
| RCA-RACE | Cp_Cycb_R2_3′Racing | GTGAAGAGGGTATTGGAGGA |
| Lycopene cyclase | Cp_LCYb2_ORF_F2 | GCTAGTAAGTTCGGTAACTTCCTTGA |
| Direct sequencing | Cp_LCYb2_ORF_R1 | ATATAGTTTATAATTGCAATCCCTCA |
| iPLEX SNP | Lcy-β1 primer 1 | ACGTTGGATGGGGATCCTACTGTTTCTCTC |
| assay | Lcy-β1 primer 2 | ACGTTGGATGGGTCTTTATGGACTGGAGAG |
| Lcy-β1 extension primer | GCATCTTAACAGCAACA | |
| Lcy-β2 primer 1 | ACGTTGGATGTTCTTGCGTCGACCTTATGG | |
| Lcy-β2 primer 2 | ACGTTGGATGCCCCATTGACAACGCAATTC | |
| Lcy-β2 extension primer | CGTCTTCAGTTTCTTCCTAC | |
| qPCR | Lcy-b2_papaya_qPCR_F | CAGATGCGATTGCGGAGTGC |
| Lcy-b2_papaya_qPCR_R | TGGCCTACCCCTGATCATTCTTGT | |
| Lcy-b1_papaya_qPCR_F | TGGCTATATGGTGGCACGAACTCT | |
| Lcy-b1_papaya_qPCR_R | CAAGGAACCGAACAATGGAATCTG | |
| 18S_rRNA_papaya_qPCR_F | CTCCGGCGTTGTTACTTTGAAGAA | |
| 18S_rRNA_papaya_qPCR_R | CCCGAAGGCCAACAGAATAGGA |
PCR conditions
PCR amplifications using degenerate primers were done in 25 μl reactions consisting of 0.2 mM dNTPs, 2 mM MgSO4, 0.4 mM of both forward and reverse primers, 0.5 U of Platinum Taq polymerase (Invitrogen) and 200 ng of genomic DNA as template. Thermocycler conditions consisted of an initial 3 min denaturation at 95 °C, followed by 30 cycles of 95 °C for 20 s, 48 °C for 30 s, and 68 °C for 60 s.
Cloning of PCR products
PCR amplification products were size-separated on a 1.5% agarose/TAE gel containing 0.1 μl ml−1 Gel Red™ fluorescent nucleic acid dye (Biotium), excised and purified using the DNA Clean & Concentrator™ -5 kit (Zymo Research) prior to ligation into the pCR®4-TOPO® vector (Invitrogen).
DNA sequencing
Purified plasmid DNA was sequenced using universal M13 reverse and T7 sequencing primers. Truncated lcy-β2 amplicons were sequenced directly using the primers Cp_LCYb2_ORF_F2 and Cp_LCYb2_ORF_R1 (Table 1). Sequencing was performed by the Australian Genome Research Facility (Brisbane, Australia).
RNA extraction
Total RNA for cDNA synthesis was extracted with TRIzol® (Invitrogen) using a modified protocol described elsewhere (Devitt et al., 2006).
5′- and 3′- rapid amplification of cDNA ends
The gene-specific primers Cp_Cycb_F2 and Cp_Cycb_R2 (Table 1) for 5′- and 3′- rapid amplification of cDNA ends (RACE) were designed based on the degenerate primer-amplified lcy-β2 sequence. Total RNA was isolated from ripe fruit flesh of cultivar Hybrid 1B. The 5′- and 3′-RACE were performed using SMART RACE cDNA Amplification Kit (Clontech) and Platinum Taq DNA polymerase (Invitrogen) in accordance with the manufacturers’ protocols. PCR conditions in the GeneAmp® PCR System 9700 (Applied Biosystems) were as follows: initial denaturation at 94 °C for 3 min followed by 30 cycles with 94 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s, and a final extension at 72 °C for 4 min. The 5′- and 3′-RACE products were cloned into pCR4-TOPO (Invitrogen) and transformed into E. coli TOP10 cells. Cloned RACE products were fully sequenced in both directions.
RCA-RACE
The rolling circle amplification (RCA)-RACE method of Polidoros et al. (2006) was also used to isolate the 5′- and 3′-ends of the lycopene β-cyclase 2 cDNA from papaya. Total RNA from fruit of hybrid 1B and Tainung was used for cDNA synthesis. First-strand cDNA was synthesized using 10 μg of RNA as template and a 5′-phosphorylated oligo(dT)20 as primer. After first-strand cDNA synthesis the RNA strand was removed with RNaseH (Invitrogen) and the cDNA was purified using the DNA Clean & Concentrator™-5 purification kit (Zymo Research). cDNA was eluted and circularized using 150 U of Circligase, 50 μM ATP and 1× reaction buffer (Epicentre Biotechnologies) at 60 °C for 1 h, followed by heat inactivation of the enzyme at 80 °C for 10 min. Linear cDNA was removed by the addition of 20 U of Exonuclease1 and Exonuclease reaction buffer (New England Biolabs) and incubation at 37 °C for 45 min. Circular cDNA was purified using the DNA Clean & Concentrator™-5 purification kit and eluted in 6 μl of nuclease free water. An RCA method using the Illustra™ TempliPhi™ amplification kit was then performed in order to amplify the circular cDNA pool. RCA products were used as template to PCR-amplify the full-length papaya lcy-β2 gene using primers Cp_Cycb_F1_5′Racing and Cp_Cycb_R2_3′Racing (Table 1).
Phylogenetic analysis
Amino acid sequences of various lycopene β- and ϵ-cyclase genes were obtained from GenBank. Multiple alignments of amino acid sequences from papaya LCY-B1 (ABD91578), papaya LCY-B2 (FJ839871), Arabidopsis LCY-B (AAF02819), Arabidopsis LCY-E (AAF82389), tomato LCY-E (CAA74745), tomato LCY-B1 (CAA60170), and tomato LCY-B2 (AAG21133) were generated using ClustalW version 2.0.11 (Larkin et al., 2007) using BLOSUM as the cost matrix and gap penalties of 10 and 0.1 for opening and extending gaps, respectively. A phylogenetic tree was constructed using MEGA 4 (Tamura et al., 2007). The tree was inferred using the Neighbor–Joining methodology and tested by bootstrapping with 1000 replications. Distances were determined using the p-distance option. The values displayed at the tree nodes are percentage consensus support as determined by the bootstrapping.
Functional validation of expression of papaya lycopene β-cyclases
Full-length or truncated versions of the LCY-B2 cDNA clones isolated from Tainung and Hybrid 1B were cloned into pCRII-TOPO (Invitrogen) and used to transform E. coli strain MG1655 PT5-dxs PT5-idi PT5-ispDispF (Yuan et al., 2006) containing pAC-LYC. Plasmid pAC-LYC is a pACYC184 derived vector containing functional carotenoid biosynthesis genes for geranylgeranyl pyrophosphate synthase (crtE), phytoene synthase (crtB), and phytoene desaturase (crtI) from Erwinia herbicola (Cunningham et al., 1994). E. coli colonies containing pAC-LYC accumulate lycopene and appear pink.
The double transformants were plated onto LB agar supplemented with ampicillin (100 μg ml−1) and chloramphenicol (50 μg ml−1). Colonies were incubated overnight at 37 °C followed by 3 d incubation at room temperature in darkness to maximize carotenoid accumulation.
HPLC profiling of carotenoids in E. coli
E. coli containing LYC-B2 clones were grown in 30 ml of LB media in the dark for 24 h at 37 °C. Carotenoid extraction and HPLC analysis methods were adapted from Howe and Tanumihardjo (2006). Cells were sedimented by centrifugation and 6 ml of ethanol (containing 0.1% butylated hydroxyl toluene [BHT]) was added to the E. coli pellet. After vortexing for 20 s, the solution was extracted five times with 3 ml hexane and then centrifugation was used to separate the phases (5000 g for 3 min at 4 °C). Hexane fractions were combined, dried in a centrifugal evaporator at 30 °C, reconstituted in 2 ml methanol/dichloromethane (50/50, v/v), containing 0.1% BHT, filtered through a 0.22 μm syringe filter (Grace) into HPLC vials, and stored under nitrogen at -80 °C prior to HPLC analysis.
The HPLC system consisted of a SIL-10AD VP auto injector (Shimadzu), SCL-10A VP system controller (Shimadzu), LC-10AT VP liquid chromatograph (Shimadzu) and a SPD-M10 A VP diode array detector (Shimadzu). Forty microlitres of each extract was injected onto a YMC C30 Carotenoid Column, 3 μm, 4.6×250 mm (Waters), with a mobile phase consisting of 92% methanol/8% 10 mM ammonium acetate (phase A), and 100% methyl tert butyl ether (phase B). A gradient was used: 0–25 min, 45–5% phase A; 25–27 min, 5% phase A; 27–28 min, 5–45% phase A; 28–33 min, 45% phase A. Lycopene and β-carotene were identified using known standards (Sigma-Aldrich), which showed co-elution and similar absorption spectra.
Quantitative real-time RT-PCR (qPCR)
qPCR was done in a RotorGene RG-3000 instrument (Corbett Life Science) to determine relative lcy-β1 and lcy-β2 gene expression levels in various organs and fruit development stages. PCR primers were designed from available lcy-β1, lcy-β2, and 18s rRNA sequences (see Table 1) using Primer Express software v1.5 (Applied Biosystems). The Power SYBR Green RNA-to-Ct 1-step kit (Applied Biosystems) was used for all qPCR experiments. The reverse-transcriptase reaction was done on total RNA from each sample at 48 °C for 30 min followed by a 10 min denaturation at 95 °C to activate the DNA polymerase. The amplification was carried out using the following cycling parameters: 40 cycles of 95 °C for 15 s, and 60 °C for 60 s. Fluorescence was acquired at 60 °C. A melt curve from 55–99 °C was performed following each experiment to ensure reactions amplified a single product. Each qPCR experiment was repeated three times and the average results of those reactions reported. Two controls were included in each experiment where either no template or no reverse transcriptase was added.
PCR efficiency determination and estimation of starting concentration
Non-baseline-corrected PCR data were exported from the RotorGene RG-3000 instrument for use in the analysis. Baseline correction and the average PCR efficiency for each of the three genes was determined using the algorithms developed by Ruijter et al. (2009) contained within version 11.0 of the qPCR analysis program, LinRegPCR (Ramakers et al., 2003). The starting concentration (NO) of mRNA from each gene in each tissue sample was calculated using values provided in the LinRegPCR output file by division of the fluorescence threshold (Nt) by the amplication efficiency (E) raised to the power of the normalized fluorescence cycle threshold (ΔCt):
Cycle thresholds for each gene in each tissue were normalized by deducting the Ct of 18S rRNA from the cycle threshold of the sample of interest. NO is expressed in arbitrary fluorescence units. Relative gene expression is calculated by directly dividing the NO of a sample of interest by the NO of a reference sample.
Single nucleotide polymorphism (SNP) assays
iPLEX assays (Sequenom) were used to identify lcy-β1 and lcy-β2 alleles present in various red- and yellow-fruited papaya cultivars. Primers used for PCR amplification and extension are listed in Table 1.
Results
A DNA fragment of ∼500 bp was PCR-amplified from Hybrid 1B and Tainung genomic DNA using degenerate primers designed from regions conserved across lycopene β- and ϵ-cyclases and capsanthin-capsorubin synthase. Sequencing of 16 cloned DNA fragments from each cultivar revealed two different, but related, gene sequences from both cultivars: lcy-β1 and a second putative lycopene β-cyclase, designated lcy-β2.
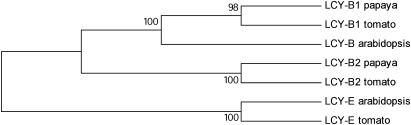
In order to clone a full-length expressed gene sequence of the newly identified lcy-β2, 5′- and 3′-RACE was performed using a combination of two methods: SMART-RACE and RCA-RACE. RCA-RACE amplification using primers designed from sequence discovered using the SMART RACE cDNA amplification method successfully identified full-length lcy-β2 sequences from both Tainung and Hybrid 1B. The full length lcy-β2 sequence from Hybrid 1B (GenBank accession FJ839871) contained an open-reading frame encoding 500 amino acids which shared 52% sequence identity with LCY-B1 and contained a predicted plastid transit peptide of 71 amino acids. Alignment of the papaya lycopene β-cyclase amino acid sequences with tomato and arabidopsis lycopene β- and ϵ-cyclases, and subsequent phylogenetic analysis (Fig. 3), revealed that papaya lcy-β2 was most similar to the chromoplast-specific lycopene β-cyclase gene from tomato.
Fig. 3.
Phylogenetic tree based on amino acid sequences of lycopene cyclase enzymes. Lycopene ϵ-cyclase from tomato (CAA74745) and Arabidopsis (AAF82389) were compared with Arabidopsis lycopene β-cyclase (AAF02819), lycopene β-cyclase 1 and 2 from tomato (CAA60170 and AAG21133), and papaya (ABD91578 and FJ839871).
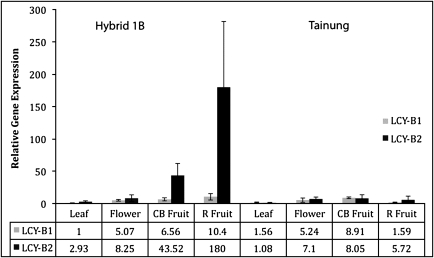
To determine the relative expression levels of lcy-β1 and lcy-β2 genes in red and yellow papaya cultivars the relative mRNA levels of both genes were measured in Tainung and Hybrid 1B leaves, flowers and fruit using qPCR (Fig. 4). The expression level of lcy-β2 was generally higher than that of lcy-β1. However, the major difference between lcy-β1 and lcy-β2 was a large increase in lcy-β2 gene expression in Hybrid 1B fruit relative to the other tissues. The lcy-β2 transcript levels in Hybrid 1B were 15-fold higher in colour-break fruit and 60-fold higher in ripe fruit than in leaves. The lcy-β2 transcript levels were 18-fold higher than lcy-β1 levels in ripe Hybrid 1B fruit. Such a significant difference in lcy-β1 and lcy-β2 transcript levels between fruit and leaves was not found in Tainung, where there was a relatively small increase in lcy-β2 gene expression in fruit compared with leaf (Fig. 4).
Fig. 4.
Expression of lcy-β1 and lcy-β2 in various tissues of papaya Hybrid 1B and Tainung. Relative levels of lcy-β1 and lcy-β2 mRNA from leaf, flower, colour-break (CB) fruit, and ripe (R) fruit of cultivars Hybrid 1B and Tainung were measured by qPCR using gene-specific primers. Expression data were normalized to the expression of 18S rRNA. Expression levels in each tissue are relative to the expression of lcy-β1 in leaf tissue of Hybrid 1B.
In order to demonstrate function of lcy-β2 from Tainung and Hybrid 1B, each gene was cloned into pCRII-TOPO and transformed into E. coli harbouring the plasmid pAC-LYC, which contains genes enabling lycopene biosynthesis. Each gene was cloned as either a full-length, or a truncated sequence lacking the predicted chloroplast transit peptide. Lcy-β2 from Hybrid 1B was functional in that truncated clones were able to convert lycopene to β-carotene, but the gene from Tainung was not functional in either full-length or truncated clones (Table 2). β-carotene was undetectable by HPLC in Tainung, while 70% of the available lycopene was converted to β-carotene in Hybrid 1B. These results suggest that a non-functional lcy-β2 may be responsible for the accumulation of lycopene in the red Tainung fruit.
Table 2.
Expression of Papaya lycopene β-cyclase genes in E. coli analysed by HPLC profiling
| Papaya lycopene β-cyclase gene | Lycopene (%) | β-carotene (%) |
| None (pAC-LYC only) | 100 | 0 |
| Hybrid 1B lcy-β2 | 50 | 50 |
| Hybrid 1B lcy-β2 (without transit peptide) | 30 | 70 |
| Tainung lcy-β2 | 100 | 0 |
| Tainung lcy-β2 (without transit peptide) | 100 | 0 |
Comparison of the nucleotide sequences of lcy-β2 from Hybrid 1B and Tainung revealed two polymorphisms, both in the coding region of the gene. At nucleotide 607, Hybrid 1B contained a cytosine (C) that was substituted for an adenosine (A) in Tainung. Starting at position 881 Hybrid 1B has a run of five thymidines (T), whereas Tainung had an insertion of an additional two Ts. The A/C sequence polymorphism leads to a single amino acid change, whereas the TT/- mutation leads to a frame-shift and premature translation termination. To determine whether either of these polymorphisms was correlated with the production of red versus yellow fruit colour, SNP and sequencing assays were done on 12 colour-differentiated cultivars. Genotyping using an iPLEX SNP assay for the A/C polymorphism at position 607 showed that all papaya cultivars with A at this position produced red fruit, except for Kapoho, which is yellow (Table 3). The remaining yellow-fruit cultivars tested contained either C or a heterozygous A/C polymorphism at this position. PCR amplification and sequencing of the lcy-β2 gene from Kapoho confirmed that it contained an adenosine at position 607, but it lacked the TT insertion at position 881 (Table 3). Both full-length and truncated lcy-β2 clones from Kapoho were used to transform a lycopene-producing strain of E. coli, which confirmed that it contained a functional lcy-β2 gene able to convert lycopene to β-carotene (data not shown). Therefore, it could be concluded that the TT insertion at 881 is responsible for the inactivation of lcy-β2 in papaya and not the A/C polymorphism at position 607 in the gene sequence. Sequencing of lcy-β2 from a number of yellow- and red-fleshed papaya cultivars confirmed that all of the red cultivars contained the TT insertion at position 881 (Table 3).
Table 3.
Combined results of genotyping and sequencing of lcy-β2 alleles across various papaya cultivars
| Cultivar | Sequence (607) | SNP (607) | Insertion (881) | Phenotype |
| Hybrid 1B | C | C,A | – | Yellow |
| BB9H | C | N.D. | – | Yellow |
| Kapoho | A | A | – | Yellow |
| Peter Gray | C | C | – | Yellow |
| Tainung | A | A | TT | Red |
| Maradol Roja | A | A | TT | Red |
| Paris | A | A | TT | Red |
| Eksotica | A | A | TT | Red |
| Mission Beach Red | A | A | TT | Red |
| Sunrise Solo | A | A | TT | Red |
| Subang | A | A | TT | Red |
Discussion
Our attempt to isolate all of the lycopene cyclase genes from papaya yielded two different lycopene β-cyclases. One of these genes had previously been cloned and characterized by Skelton et al. (1996), but the second lycopene β-cyclase is new. The newly discovered lycopene β-cyclase gene (lcy-β2) is more closely related to chromoplast-specific β-cyclases from other species than to the papaya lcy-β1 gene, suggesting that it is also a chromoplast-specific gene and may play a significant role in carotenoid production and control of fruit colour in papaya. This was supported by gene expression studies which showed that lcy-β2 is much more highly expressed in ripe fruit than in leaves.
The functional analysis of LCY-B2 activity across a number of papaya genotypes was conducted using E. coli engineered for enhanced carotenoid substrate accumulation. The identification of a functional LCY-B2 in the yellow-fleshed Kapoho variety was particularly helpful in challenging our early hypothesis that the A/C sequence polymorphism at position 607 and resulting amino acid change may have been responsible for the reduced enzyme functionality in red-fleshed varieties.
The position 607 SNP assay identified a correlation between SNP and red flesh colour in all genotypes tested except Kapoho. Complete sequencing of multiple lcy-β2 clones from all varieties across both DNA strands successfully identified the TT insertion at position 881 in all red-fleshed varieties. From the combination of all these data it was concluded that lcy-β2 encodes a chromoplast-specific lycopene β-cyclase gene, which controls papaya fruit colour. A TT insertion in the lcy-β2 gene results in an inactive chromoplast-specific lycopene β-cyclase and leads to the accumulation of lycopene in the fruit of red-fleshed varieties.
Over the the past 10 years, a number of studies of carotenogenic fruit have identified the key importance of lycopene cyclase activity in determining the profile of fruit carotenoids. Studies of carotenoid mutants of tomato by Ronen et al. (2000) led to the map-based cloning of a second, chromoplast-specific, lycopene β-cyclase and a new model of alternative carotenoid regulation in chromoplasts and chloroplasts in tomato.
The presence of two lycopene β-cyclases in papaya is similar in some respects to a number of other species that accumulate lycopene in their fruit. Tomato contains two lycopene β-cyclase genes, CrtL-b and B, with the B gene expressed exclusively in chromoplast-containing tissues of flowers and fruits (Ronen et al., 2000). However, in contrast to the situation in papaya, expression of the tomato B gene in fruits is low and limited to a short period around the breaker stage, leading to mostly lycopene and a little β-carotene accumulation in wild-type tomato fruit. Overexpression of a bacterial lycopene β-cyclase in tomato fruit has been shown to increase β-carotene production (Wurbs et al., 2007).
While citrus species do not typically accumulate lycopene in fruit, lycopene-accumulating mutants in grapefruit and orange do exist, and their carotenoid content and carotenoid pathway gene expression have recently been studied. A chromoplast-specific lycopene β-cyclase (Csβ-LCY2) has been characterized from Citrus sinensis (cv. Navel) and the lycopene-accumulating mutant, Citrus paradise (cv. Star Ruby) (Alquézar et al., 2009). In Navel oranges, Csβ-LCY2 was highly expressed during fruit maturation in contrast to Csβ-LCY1, which displayed a low, unchanging expression pattern during fruit maturation. Two alleles of the LCY2 gene were isolated and 16 amino acid changes were proposed to account for the difference between the functional β-LCY2a allele from Navel orange and the almost inactive allele, β-LCY2b, from Star Ruby grapefruit.
The presence of two lycopene β-cyclase genes can account for the observed carotenoid accumulation in red and yellow papaya fruit. In yellow fruit, both lcy-β1 and lcy-β2 are expressed, although the lcy-β2 gene is expressed at a much higher level and accounts for 95% of all β-cyclase mRNA. The level of β-cyclase enzyme activity resulting from the expression of β1 and β2 is sufficient to convert all of the available lycopene to β-carotene (and subsequently to β-cryptoxanthin). However, in red papaya fruit, the β2 transcript level is reduced and the gene product is non-functional. Therefore, the only β-cyclase enzyme activity would be from the lcy-β1 gene. This lcy-β1 activity is not sufficient to convert all of the lycopene to β-carotene, resulting in accumulation of both lycopene and β-carotene in red-fleshed papaya fruit.
The isolation and characterization of the two lycopene β-cyclase genes from papaya has potential applications to enhance papaya breeding and genetic improvement. The most obvious use of this information is for the early selection of fruit colour in papaya breeding through the development of a molecular marker assay to target the TT insertion in lcy-β2. Further modification and selection of carotenoid composition of papaya may also be possible through the up- or down-regulation of β-cyclases and other genes in the carotenoid pathway.
Acknowledgments
The authors wish to thank Buddy Cunningham (University of Maryland) for the provision of the E. coli colour complementation vector pAC-BETA and Wonchul Suh (DuPont) for providing the E. coli strain MG1655 PT5-dxs PT5-idi PT5-ispDF. Papaya fruit, flowers, and leaves were provided by Mareeba staff of the Department of Employment, Economic Development and Innovation (DEEDI), Primary Industries and Fisheries. The authors wish to thank Roger Mitchell and David Innes for expert technical assistance with RACE for the lycopene cyclase gene and qPCR analysis, respectively. This work was supported by DEEDI under the Tropical Fruit Genomics Initiative and by the University of Queensland.
References
- Alquézar B, Zacarias L, Rodrigo MJ. Molecular and functional characterization of a novel chromoplast-specific lycopene β-cyclase from Citrus and its relation to lycopene accumulation. Journal of Experimental Botany. 2009;60:1783–1797. doi: 10.1093/jxb/erp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika UG, Jansz ER, Wickramasinghe SMDN, Warnasuriya ND. Carotenoids in yellow- and red-fleshed papaya (Carica papaya L.) Journal of the Science of Food and Agriculture. 2003;83:1279–1282. [Google Scholar]
- Cunningham FX, Jr, Sun Z, Chamovitz D, Hirschberg J, Gantt E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp. strain PCC7942. The Plant Cell. 1994;6:1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devitt LC, Sawbridge T, Holton TA, Mitchelson K, Dietzgen RG. Discovery of genes associated with fruit ripening in Carica papaya using expressed sequence tags. Plant Science. 2006;170:356–363. [Google Scholar]
- Howe JA, Tanumihardjo SA. Evaluation of analytical methods for carotenoid extraction from biofortified maize (Zea mays sp.) Journal of Agricultural and Food Chemistry. 2006;54:7992–7997. doi: 10.1021/jf062256f. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Chenna R, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–997. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecker I, Gabbay R, Cunningham FX, Jr, Hirschberg J. Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Molecular Biology. 1996;30:807–819. doi: 10.1007/BF00019013. [DOI] [PubMed] [Google Scholar]
- Polidoros AN, Pasentsis K, Tsaftaris AS. Rolling circle amplification-RACE: a method for simultaneous isolation of 5’ and 3’ cDNA ends from amplified cDNA templates. BioTechniques. 2006;41:35–36. doi: 10.2144/000112205. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. The Plant Journal. 1999;17:341–351. doi: 10.1046/j.1365-313x.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proceedings of the National Academy of Sciences, USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research. 2009 doi: 10.1093/nar/gkp045. 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton RL, Qingyi L, Srinivasan R, Manshardt R, Moore PH, Ming R. Tissue differential expression of lycopene β-cyclase gene in papaya. Cell Research. 2006;16:731–739. doi: 10.1038/sj.cr.7310081. [DOI] [PubMed] [Google Scholar]
- Skelton RL, Yu Q, Srinivasan R, Manshardt R, Moore PH, Ming R. Tissue differential expression of lycopene β-cyclase gene in papaya. Cell Research. 1996;16:731–739. doi: 10.1038/sj.cr.7310081. [DOI] [PubMed] [Google Scholar]
- Storey WB. Papaya. In: Ferwerda FP, Wit F, Veenman H, Zoen NV, editors. Outlines of perennial crop breeding in the tropics. The Netherlands: Wageningen; 1969. pp. 389–408. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. Mega4: molecular evolutionary genetics analysis (Mega) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. The Plant Journal. 2007;49:276–288. doi: 10.1111/j.1365-313X.2006.02960.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto H. Differences in carotenoid composition between red- and yellow-fleshed papaya. Nature. 1964;201:1049–1050. doi: 10.1038/2011049a0. [DOI] [PubMed] [Google Scholar]
- Yuan LZ, Rouviere PE, LaRossa RA, Suh W. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metabolic Engineering. 2006;8:79–90. doi: 10.1016/j.ymben.2005.08.005. [DOI] [PubMed] [Google Scholar]