Abstract
Oryza meridionalis Ng. is a wild relative of Oryza sativa L. found throughout northern Australia where temperatures regularly exceed 35 °C in the monsoon growing season. Heat tolerance in O. meridionalis was established by comparing leaf elongation and photosynthetic rates at 45 °C with plants maintained at 27 °C. By comparison with O. sativa ssp. japonica cv. Amaroo, O. meridionalis was heat tolerant. Elongation rates of the third leaf of O. meridionalis declined by 47% over 24 h at 45 °C compared with a 91% decrease for O. sativa. Net photosynthesis was significantly higher in O. sativa at 27 °C whereas the two species had the same assimilation rates at 45 °C. The leaf proteome and expression levels of individual heat-responsive genes provided insight into the heat response of O. meridionalis. After 24 h of heat exposure, many enzymes involved in the Calvin Cycle were more abundant, while mRNA of their genes generally decreased. Ferredoxin-NADP(H) oxidoreductase, a key enzyme in photosynthetic electron transport had both reduced abundance and gene expression, suggesting light reactions were highly susceptible to heat stress. Rubisco activase was strongly up-regulated after 24 h of heat, with the large isoform having the largest relative increase in protein abundance and a significant increase in gene expression. The protective proteins Cpn60, Hsp90, and Hsp70 all increased in both protein abundance and gene expression. A thiamine biosynthesis protein (THI1), previously shown to act protectively against stress, increased in abundance during heat, even as thiamine levels fell in O. meridionalis.
Keywords: Calvin Cycle, dark reaction, ferredoxin-NADP(H) oxidoreductase, heat shock protein, heat stress, leaf elongation, O. meridionalis, Rubisco activase, thiamine biosynthesis protein (THI1)
Introduction
The Intergovernmental Panel on Climate Change is predicting a likelihood of more intense, more frequent, and longer lasting heat waves (IPCC, 2007). Although rice is a pan-tropical grass and therefore relatively well adapted to high temperatures in comparison with other cereals such as wheat (Triticum aestivum L.), peak temperatures will increase over this century, providing an abiotic stress to which cultivated rice might not be adapted. The ability of modern rice (Oryza sativa L.) to be cultivated in hotter climatic regimes may be limited by its narrow gene pool as domesticated rice has only about 10–20% of the genetic diversity found in wild progenitors (Zhu et al., 2007).
The pre-eminent method currently practised for improving abiotic stress tolerance in rice cultivars is to source germplasm for desirable traits. Recent attempts to do so have been successful, with backcrossing of O. sativa ssp. japonica and indica leading to substantial improvements in resistance to many abiotic stresses (Ali et al., 2006; Lafitte et al., 2006; Cheng et al., 2007). Although this approach has been productive, it has been limited to O. sativa. With more than 20 known species within the Oryza genus, a large source of genetic material remains to be exploited (Brar and Khush, 1997).
Oryza meridionalis Ng. is a wild rice species likely to have abiotic stress tolerance, as it shows high levels of genetic diversity between geographically isolated accessions, probably as a result of selective pressure on isolated gene pools (Juliano et al., 2005). Oryza meridionalis was first recognized as a species in 1981, found in northern Australia (Ng et al., 1981) and subsequently in West Papua, Indonesia (Lu and Silitonga, 1999). Phylogenetic analysis of Adh1 and Adh2 genes, as well as mitochondrial and chloroplast microsatellites support O. meridionalis as a divergent lineage within the rice genome (Ge et al., 1999; Nishikawa et al., 2005), similar to that of O. sativa. The extent of phenotypic diversity in O. meridionalis has not been described: however, it shares the AA genome with O. sativa, making O. meridionalis a good candidate for genetic improvement of cultivated rice through introgression of new genetic material. For example, the introgression of two genes, yld1.1 and yld2.1 from the progenitor of modern rice, O. rufipogon, improved grain yield by 18% and 17%, respectively, in an elite hybrid breed of O. sativa (Xiao et al., 1998). This improvement occurred without the detection of any deleterious impacts on other desirable traits. Previous transgenic manipulations targeting osmotic solute production, transcription factors, oxidative stress detoxification and ion transport have provided increased resistance in many species to stress such as heat (Wang et al., 2003b). The genetic bottleneck in domesticated rice, in conjunction with the probability that its wild relatives have a diverse array of stress-related genes, opens the possibility of rapid genetic improvement through breeding and transgenics.
Genomic analysis through the use of expression sequence tags and cDNA libraries from stressed plant tissue has been used effectively in identifying the response of plants to stress and discovering the identity of genes involved (Sreenivasulu et al., 2007). An alternative technique previously used for determining the molecular responses of rice to stress is proteomic analysis (Salekdeh et al., 2002; Komatsu et al., 2003; Komatsu and Tanaka, 2004; Lee et al., 2007). The proteomic approach is important to the understanding of the abiotic stress response because many key proteins are regulated translationally and post-translationally (Greenbaum et al., 2003; Lee et al., 2004).
Heat tolerance in the wild rice O. meridionalis was established by comparison of seedling growth and photosynthetic rates at optimal and high temperatures, using O. sativa. ssp. japonica (cv. Amaroo) as a domesticated control cultivar. Based on these findings, proteomic analysis using two-dimensional gel electrophoresis coupled with nanoLC-MS/MS established which proteins might play key roles in the thermotolerance of O. meridionalis. Finally, semi-quantitative RT-PCR on the genes of interest was used to test whether increased protein abundance was transcriptionally regulated.
Materials and methods
Plant material
Seeds of Oryza meridionalis Ng. were collected from a wild accession located in the Cape York Peninsula of Australia (15°41′57′′ S, 145°02′48′′ E). Seedlings were grown in a mixture (1:1:1 by vol.) of silty loam, clay, and organic potting mix. This soil mixture was used throughout all the experiments. A 2 cm layer of vermiculite was placed across the soil surfaces to reduce evaporation and to maintain soil moisture. All experiments were carried out on 22-d-old seedlings that were grown in 500 ml polyvinyl pots in growth chambers (Thermoline Scientific Equipment, Australia) with an illumination of approximately 500 μmol m−2 s−1 and a temperature of 27/22 °C (day/night) with a 12 h photoperiod. Heat-treated plants were held at 45 °C continuously for 24 h, commencing 2 h into a light period. A 12 h dark period followed 10–22 h into the heat treatment, and leaf data (excepting continuous growth measurements) were collected 2 h into the subsequent light period. Seedlings were well watered and fertilized weekly with a commercial liquid fertilizer.
Growth measurements
Seedling growth was determined by measuring elongation of the third leaf blade, using a HR4000 Linear Variable Displacement Transducer (LVDT) with data logged every 6 min by the software program VuGrowth ver. 1.0 (Applied Measurement, Oakleigh, Vic). Seedlings grown in pots were transferred to a growth chamber containing the LVDT unit one day after the emergence of the third leaf blade. For control experiments, seedlings were grown at a constant 27 °C for 46 h with four of the eight measuring stations randomly assigned to each of the two species. Heat experiments were conducted as above with an increase to 45 °C in the period 22–46 h. Both control and heat treatments were repeated in four independent experiments.
Gas exchange and water measurements
Net photosynthetic rates (NPR), respiration rates, stomatal conductance (gs), intercellular CO2 (Ci), transpiration rates (T), and leaf temperatures (Tleaf) were determined using a LI-6400 (Li-Cor, NE, USA) portable gas exchange system. All gas exchange measurements were made within growth chambers on seedlings subjected to previously mentioned temperature and light regimes and a CO2 concentration of 380 μmol mol−1. Cuvette temperatures were adjusted to 27 °C or 45 °C to match growth chamber temperatures for the two treatments. Net photosynthetic rates were measured in the first 3 h of the light period with gas chamber illumination at saturating levels of 1500 μmol m−2 s−1. To minimize the possibility of dry air reducing stomatal conductance, 10 ml of H2O was added to the soda lime canister prior to use and, subsequently, sample chamber relative humidity was maintained above 30% at 45 °C. Respiration rates of seedlings subjected to greater than a 9 h dark period were measured in the absence of light.
Leaf water potential (Ψleaf) was measured using a pressure bomb. The third leaf was cut from plants close to their base and immediately placed in the pressure chamber for measurement of balancing pressure. Relative water content (RWC) was derived from the formula (fresh mass–dry mass)/(turgid mass–dry mass). All gas-exchange and water measurements were made on four plants per pot, with three pots representing each treatment.
Gel electrophoresis
Leaf blades were collected from both control and heat-treated plants (see ‘Plant material’ section) and immediately ground in liquid nitrogen. Leaf powder was washed with TCA and acetone and subjected to phenol/SDS extraction as described by Wang (2003a). The protein extract was resuspended in 200 μl of rehydration buffer [7 M urea, 2 M thiourea, 4% CHAPS (w/v), 30 mM DTT, trace amount of bromophenol blue]. Approximately 250 mg of protein quantified by the Bradford assay (Bio-Rad Protein Assay) was loaded on 11 cm, 4–7 ReadyStrip IPG strips (Bio-Rad) with rehydration following the manufacturer's instructions. Focusing occurred using a PROTEAN IEF Cell (Bio-Rad) at 20 °C with a total of 37 200 focusing hours (200 V for 1 h, 1000 V for 1 h, 4000 V for 3 h, and 8000 V for 3 h). After focusing, IPG strips were placed in re-equilibration buffer (50 mM TRIS pH 8.8, 6 M urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue) for 30 min then run in the second dimension on 8–16% gradient Criterion pre-cast gels (Bio-Rad). All gels were stained with Lava purple (FLUOROtechnics, Sydney, Australia) as directed by the manufacturer's guidelines and fluorescently imaged on a Typhoon 9200 scanner (Amersham Pharmacia Biotech). Harvesting of leaf tissue, sample preparation, and gel electrophoresis experiments were replicated in three independent experiments so that each control and heat-stress sample was biologically and experimentally replicated three times.
Image analysis and protein identification
Spots that were matched across all gels were labelled accordingly and abundances measured as integrated density using ImageJ software (Abramoff et al., 2004). Percentage relative integrated density values were calculated to normalize against loading differences and the values used to determine heat-induced differentially expressed proteins.
Following image analysis, 48 spots that had a heat-induced change in relative integrated density values greater than 1.8-fold (up or down) were selected for further analysis. The fold change values in both Tables 2 and 3 are expressed as abundance (heat)/abundance (control) so in Table 3 smaller values indicate a more significant change. Additional spots of two highly abundant proteins, with a fold change that was consistent across the replicates, but slightly below the 1.8-fold threshold, were also analysed (spots 148 and 132) and included in Table 2.
Table 2.
Protein spots with increased abundance upon heat stress
| Functional classification | Spot no. | Fold-changea | Accession nob. | Putative proteinc | Peptide no./log e score | Theoretical protein mass (kDa) |
| Photosynthetic metabolism | 127 | 7.5 | BAA97583 | Rubisco activase, large isoform | 27/–151 | 51.4 |
| 136 | 4.0 | BAA97584 | Rubisco activase small isoform | 69/–216 | 47.9 | |
| 133 | 2.4 | BAA97584 | Rubisco activase small isoform | 85/–297 | 47.9 | |
| 161 | 2.4 | NP_001056711 | Transketolase, chloroplast | 5/–36 | 73.4 | |
| 57 | 1.8 | NP_001056711 | Transketolase, chloroplast | 68/–295 | 73.4 | |
| 143 | 3.7 | EAY98560 | Phosphoglycerate kinase, chloroplast | 68/–316 | 30.5 | |
| 148 | 1.3 | NP_001047825 | Phosphoribulokinase | 40/–201 | 44.8 | |
| 132 | 1.5 | NP_001052293 | Sedoheptulose-1,7-bisphosphatase, chloroplast | 31/–124 | 42.2 | |
| Protective proteins | 27 | 2.9 | NP_001066567 | Chaperonin-60, chloroplast alpha subunit | 30/–211 | 61.1 |
| 31 | 2.3 | NP_001056601 | Chaperonin-60, chloroplast beta subunit | 44/–260 | 64 | |
| 371 | 1.9 | NP_001056601 | Chaperonin-60, chloroplast beta subunit | 7/–42 | 64 | |
| 6 to 8 | 2.1 | NP_001042206 | Unknown protein | 27/–190 | 84.1 | |
| EAZ45094 | HSP90 | 23/–176 | 103.9 | |||
| NP_001063456 | HSP90 | 19/–137 | 74.8 | |||
| 1 to 5 | 1.8 | EAZ33719 | Chloroplast heat shock protein 70 | 30/–163 | 73.5 | |
| AAA18541 | Triosephosphate isomerase | 18/–84 | 27 | |||
| NP_001060741 | Ascorbate peroxidase | 13/–96 | 27.1 | |||
| NP_001066486 | HSP70 | 12/–66 | 74 | |||
| NP_001068278 | Unknown protein | 12/–57 | 26 | |||
| 35 | 2.0 | AAB63469 | Endosperm lumenal binding protein (HSP70) | 26/–157 | 73.5 | |
| Thiamine biosynthesis | 175 | 3.7 | NP_001059841 | Thiamine biosynthesis protein (THI1) | 16/–66 | 37.2 |
| Energy production | 48 | 2.6 | AAA84588 | ATP synthase CF1 beta subunit | 79/–268 | 53.9 |
| Glycine metabolism | 92 | 2.7 | EAZ01651 | Glycine dehydrogenase | 49/–248 | 111.4 |
| Unknown function | 126 | 1.9 | EAY81919 | Hypothetical protein of the AAA family | 4/–22 | 39.6 |
| 154 | 1.9 | NP_001065830 | Hypothetical protein | 34/–151 | 41.6 |
The fold-change values were derived from heat/control spot volumes.
NCBI listed accession numbers of proteins in downloaded rice database matched by Xtandem algorithm to nanoLC-MS/MS spectra.
MS matched proteins were BLAST searched (Altschul et al., 1997) and high scoring homologues provided annotation for putative O. sativa proteins.
Table 3.
Protein spots with decreased abundance upon heat stress
| Functional classification | Spot no. | Fold-changea | Accession no. | Putative protein | Peptide no./log e score | Theoretical protein mass (kDa) |
| Photosynthetic metabolism | 250 | 0.254 | CAG34174 | Rubisco, large subunit | 16/–38 | 52.8 |
| 254 | 0.255 | CAG34174 | Rubisco, large subunit | 11/–57 | 52.8 | |
| 255 | 0.303 | CAG34174 | Rubisco, large subunit | 19/–102 | 52.8 | |
| 173 | 0.481 | BAA97584 | Rubisco activase small isoform | 22/–108 | 47.9 | |
| 181 | 0.508 | BAA97584 | Rubisco activase small isoform | 56/–201 | 47.9 | |
| 147 | 0.548 | BAA97584 | Rubisco activase small isoform | 29/–160 | 47.9 | |
| 245 | 0.549 | BAA97584 | Rubisco activase small isoform | 11/–54 | 47.9 | |
| CAG34174 | Rubisco, large subunit | 9/–45 | 52.8 | |||
| 131 | 0.395 | NP_001052293 | Sedoheptulose-1,7-bisphosphatase, chloroplast | 60/–213 | 42.2 | |
| 348 | 0.553 | NP_001061210 | Photosystem II 10 kDa polypeptide | 4/–14 | 13.2 | |
| Carbon metabolism | 220 | 0.541 | NP_001048847 | Glyceraldehyde-3-phosphate dehydrogenase | 10/–75 | 47.1 |
| 204 | 0.518 | ABC94602 | UDP-glycosyltransferase-like protein | 8/–38 | 49.3 | |
| Electron transport | 224 | 0.244 | NP_001045608 | Ferredoxin-NADP(H) oxidoreductase | 3/–27 | 38.7 |
| 184 | 0.454 | NP_001045608 | Ferredoxin-NADP(H) oxidoreductase | 5/–30 | 38.7 | |
| 182 | 0.532 | NP_001045608 | Ferredoxin-NADP(H) oxidoreductase | 8/–46 | 38.7 | |
| Energy production | 43 | 0.378 | NP_001043900 | Mitochondrial F1-ATPase beta subunit | 50/–257 | 59.4 |
| 186 | 0.382 | AAA84588 | atpB gene product | 24/–147 | 53.9 | |
| 45 | 0.534 | P0C2Z7 | ATP synthase subunit beta | 133/–472 | 53.9 | |
| Amino-acid metabolism | 213 | 0.548 | NP_001049852 | Aspartate aminotransferase | 7/–61 | 49.8 |
| Metabolism | 198 | 0.549 | CAA81481 | S-adenosylmethionine synthetase, type 1 | 19/–108 | 43.2 |
| Protective proteins | 310 | 0.373 | BAA74702 | Germin-like protein 1 | 3/–17 | 21.8 |
| 344 | 0.530 | BAA74702 | Germin-like protein 1 | 16/–4 | 21.8 | |
| Unknown function | 243 | 0.514 | NP_001061403 | Hypothetical protein | 3/–21 | 27.1 |
| 205 | 0.526 | NP_001052894 | Hypothetical protein | 4/–27 | 47.6 | |
| 219 | 0.489 | NP_001043910 | Hypothetical protein | 89/–396 | 36.2 | |
| 242 | 0.369 | EAZ06282 | Hypothetical protein | 6/–37 | 27.1 |
As in Table 1, fold-change values were derived from heat/control spot volumes.
Spots of interest were excised and washed for 5 min with 100 mM NH4HCO3, followed by destaining with 50% acetonitrile (v/v) in 50 mM NH4HCO3 for 10 min twice. Reduction and alkylation required 10 mM DTT in 100 mM NH4HCO3, applied at 37 °C for 60 min followed by 55 mM iodacetamide in 100 mM NH4HCO3 for 45 min. Gels were digested with 20 μl of 12.5 ng μl−1 trypsin (Promega) in 50 mM NH4HCO3 on ice for 30 min then incubation at 37 °C overnight. Peptides were extracted by washing gel pieces with 30 μl of 50% acetonitrile and 2% formic acid (v/v) for 20 min on three occasions. Vacuum centrifugation was used to concentrate the peptide solution to 10 μl prior to loading of samples onto a ThermoFinnigan LCQ-Deca ion trap mass spectrometer for peptide identification through nano liquid chromatography on line with tandem mass spectrometry (nanoLC-MS/MS) as described by Medina et al. (2005). Tandem mass spectra were searched using the XTandem algorithm run under the GPM-XE interface (Craig and Beavis, 2003; Fenyo and Beavis, 2003). The default XTandem search parameters were used and sequences searched against a database of rice (O. sativa) protein sequences (March 2008 version), representing the complete rice genome, downloaded from NCBI (www.ncbi.nlm.nih.gov).
Semi-quantitative RT-PCR
Leaf blades of seedlings were instantly frozen in liquid nitrogen after 24 h of treatment and stored at –80 °C until RNA was extracted using a RNeasy Plant Mini Kit (Qiagen) following the manufacturer's instructions. RNA was converted to cDNA using a SuperScript VILO cDNA synthesis kit (Invitrogen) with 2.1 μg of RNA used as a template. PCR was performed using a GoTaq Green Master Mix (Promega), with a 25 μl reaction solution containing 2 μl of cDNA and gene-specific forward and reverse primers. Primers were designed using protein sequences identified by nanoLC-MS/MS. Protein sequences were BLAST searched through NCBI and the corresponding rice cDNA sequences used as the template for primer design. The primers are given in Supplementary Table S1 at JXB online. For PCR, an initial step of 95 °C for 2 min was followed by 22, 26 or 30 cycles of 95 °C for 30 s, 52 °C for 30 s, and a final step of 72 °C for 20 s. PCR products were visualized on a 2.5% agarose gel stained with Gel Red (Biotium) and imaged with GeneSnap version 6.00.26 software (Syngene, Frederick, MD). Band intensities were analysed using ImageJ software and normalized by comparison with actin. RNA was extracted from triplicate, biological (pot) replicates to calculate means.
Determination of thiamine concentration
Thiamine was extracted from leaf blades, converted to thiochrome and fluorescence measured by a method similar to that of Ohta (1993). Leaf blades were ground in liquid nitrogen and 0.6 g added to 10 ml of 0.1 M HCl, 40% methanol (Buffer A). Samples were vortexed and incubated at 60 °C for 30 min and centrifuged at 4000 g for 15 min, supernatant was filtered (0.45 μm) and an equal volume added to 0.1% potassium hexacyanoferrate (III) in 15% NaOH (Buffer B). This 50:50 mixture was diluted a further 8-fold again in equal volumes of Buffer A and Buffer B, before fluorescence measurements were taken to overcome a quenching effect that, at higher concentrations, resulted in an underestimation of thiamine levels (see Supplementary Fig. S3 at JXB online). To confirm the accuracy of the assay, a sample was spiked with a known quantity of thiamine, resulting in a 99.5% recovery (see Supplementary Fig. S4 at JXB online). Fluorescence was measured (Ex. 375 nm, Em. 455 nm, and 10 nm bandpass) on a PerkinElmer LS 55 luminescence spectrometer. Thiamine concentration was determined by comparing results with a standard curve between 0.02 ng μl−1 and 0.625 ng μl−1 (R2=0.9999).
Statistical analysis
When applying statistics a one-way analysis of variance was used with a 5% LSD test in cases of multiple comparisons. The statistical analysis was carried out using SPSS statistical analysis software (Ver. 16.0.1, SPSS Inc). Values are based on the means ±SE of 3–4 experimental replicates.
Results
Growth, photosynthesis and water relations in response to temperature
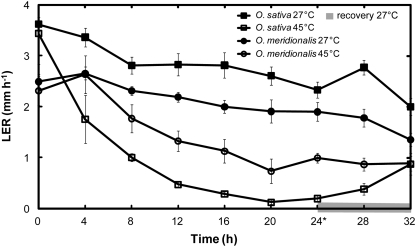
Third leaves of O. sativa elongated faster than O. meridionalis at 27 °C, however, when exposed to 45 °C, O. meridionalis elongated faster than O. sativa (Fig. 1). After 4 h of heat, LER of O. sativa was halved whereas leaf growth was not affected in O. meridionalis. Furthermore, 24 h at 45 °C caused a 91% decrease in the LER of O. sativa but only a 47% decline in O. meridionalis (Table 1). There was a noticeable increase in the growth rates of both species towards the end of the 24 h heat treatment, possibly connected to the second light photoperiod which began 22 h into the heat treatment.
Fig. 1.
Leaf elongation rates (LER) of third leaves beginning 1 d after emergence in O. sativa ssp. japonica (cv. Amaroo) (squares) and O. meridionalis (circles). Seedlings were grown in a growth chamber at 27 °C with a 12 h photoperiod and illumination of 300 μmol m−2 s−1. Measurements were taken by a Linear Variable Displacement Transducer (LVDT) placed within the chamber for 24 h at control, 27 °C (closed symbols) and heat, 45 °C (open symbols), followed by an 8 h recovery period (shaded grey) where heat-treated plants where returned to 27 °C. The first light period commenced 2 h before heat was applied and continued 10 h into the heat treatment; the second light period commenced 22 h into the heat period. An asterisk denotes 24 h of heat treatment when protein, gene and photosynthetic observations were made. Values are means ±SE, n=4.
Table 1.
Impact of temperature on leaf elongation, gas exchange and leaf thiamine concentrations
|
Oryza sativa |
Oryza meridionalis |
|||
| 27 °C | 45 °C | 27 °C | 45 °C | |
| LER (mm h−1) | 2.3±0.2a | 0.2±0.1b | 1.9±0.2a | 1.0±0.1c |
| NPR (μmol CO2 m−2 s−1) | 26.6±1.2a | 12.6±1.3c | 22.4±1.6b | 13±0.6c |
| Dark respiration rate (μmol CO2 m−2 s−1) | 0.7±0.09a | 1.2±0.07bc | 1.0±0.03ab | 1.4±0.1c |
| gs (mol H2O m−2 s−1) | 0.27±0.02a | 0.19±0.02b | 0.26±0.02a | 0.18±0.00b |
| Ci (μmol CO2 mol−1) | 193±4a | 232±11b | 210±2ac | 220±4bc |
| T (mmol H2O m−2 s−1) | 6.25±0.28a | 12.7±0.74b | 5.84±0.42a | 11.6±0.23b |
| Ψleaf (MPa) | –0.98±0.03a | –0.59±0.08b | –1.01±0.17a | –0.52±0.07b |
| RWC (%) | 97.1±0.7a | 96.4±1.9a | 97.9±0.04a | 97.6±0.01a |
| Tleaf (°C) | 27.0±0.02a | 45.0±0.4b | 26.9±0.03a | 45.0±0.39b |
| Thiamine concentration (μg g−1 FW) | 18.3±0.5a | 15.4±0.7b | ||
All measurements were taken 24 h into treatment. LER, leaf elongation rate; NPR, net photosynthetic rate; gs, stomatal conductance; Ci, intercellular CO2; T, transpiration rate; Ψleaf, leaf water potential; RWC, relative water content; Tleaf, leaf temperature. Values are means ±SE, n=3–4. Superscript letters indicate significant differences (LSD multiple comparison test, P <0.05).
Although 45 °C is a severe temperature for an herbaceous plant, neither species showed physical leaf symptoms of temperature stress such as wilting, necrosis or loss of pigmentation after heat exposure, which was consistent with sustained leaf elongation following heat application. In particular, the more heat-sensitive O. sativa had a LER equal to O. meridionalis after 8 h of recovery (Fig. 1).
There was a significant difference in the net photosynthetic rate between O. sativa and O. meridionalis at 27 °C but not at 45 °C (Table 1) and, therefore, the impact of heat on net photosynthesis was greater for O. sativa (53% fall) than for O. meridionalis (42% fall). There was a significant increase in the dark respiration rates of both species at 45 °C with a slightly greater increase recorded in O. sativa.
There was no difference between the species in transpiration rates, RWC or Ψleaf at 45 °C (Table 1). While transpiration rates increased to the same extent in both species when exposed to 45 °C, Ψleaf increased simultaneously, which could only occur if soil was able to maintain water supply and hydraulic function remained unimpaired. Thus, the 45 °C treatment imposed heat stress without any leaf water deficit.
Identification of proteins associated with heat stress
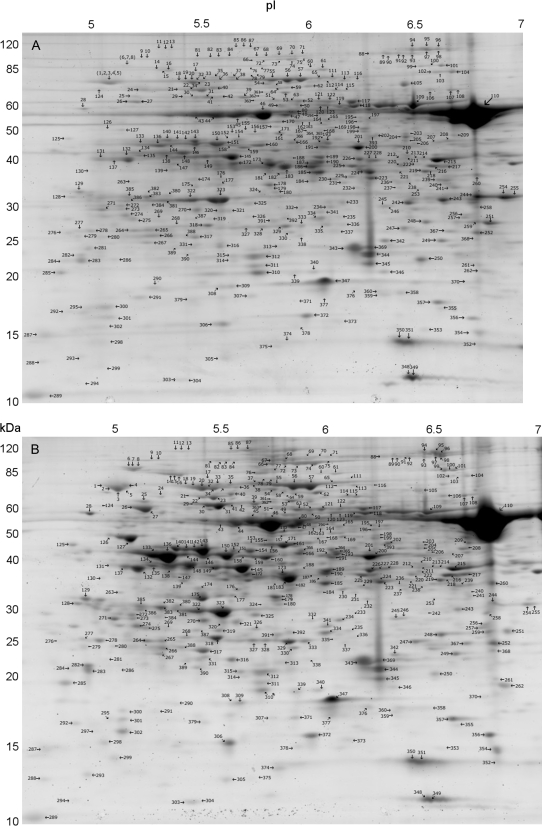
In total, 392 individual spots were matched and labelled across all replicates and treatments (Fig. 2; see Supplementary Fig. S1 at JXB online). Spots that did not have a unidirectional change across all replicates were excluded from further analysis. Protein identification was made of 50 spots by nanoLC-MS/MS. This includes all spots which showed an expression level change of greater than 1.8-fold between the two conditions. Table 2 contains identification and abundance measurements for 23 spots that showed greater than 1.8-fold increases under heat stress, along with two additional high-abundance spots which were consistently increased, but by less than 1.8-fold. Table 3 contains identification and abundance measurements for 25 spots showing a greater than 1.8-fold decrease in abundance due to heat stress.
Fig. 2.
Two-dimensional electrophoresis gels of 22-d-old O. meridionalis seedlings grown under either control, 27 °C (A) or 45 °C, 24 h heat treatment (B). Approximately 250 μg of leaf blade protein was initially run on pH 4–7 IPG strips followed by SDS-PAGE electrophoresis using 8–16 gradient Criterion gels. Labelled spots are those that were found in all replicates. The noticeable protein spot (spot 110) found at between 50 kDa and 60 kDa and a pI of just over 6.5 was identified as the Rubisco large subunit.
Due to the overlap of spots one to five, this cluster was considered as a single unit for the purpose of image analysis and mass spectrometry. Similarly, spots six to eight were treated as a single spot cluster rather than individual spots (see Supplementary Fig. S2 at JXB online). Many proteins were identified in multiple spots, which is not uncommon and suggestive of alternative RNA splicing and post-translational modifications such as glycosylation (Rodríguez-Piñeiro et al., 2007).
Photosynthetic metabolism proteins
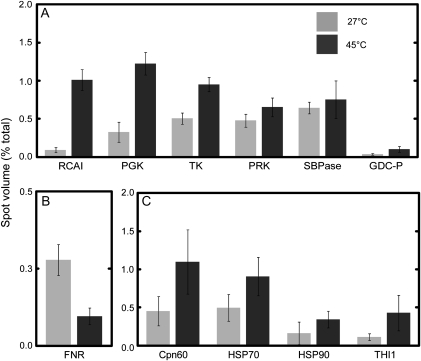
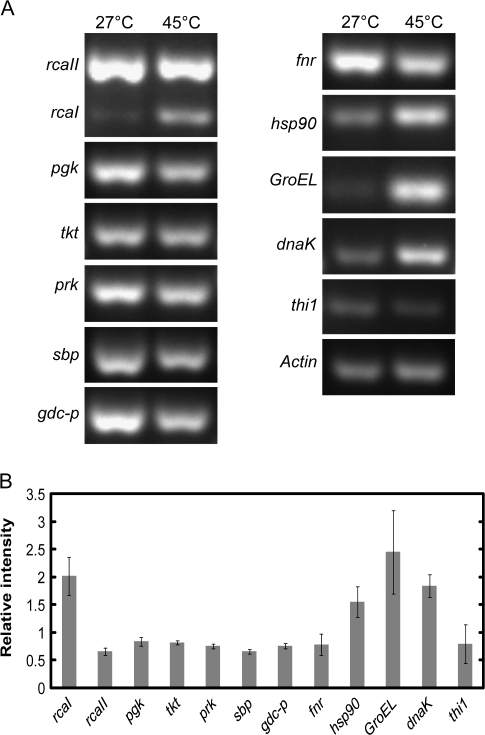
Many proteins of O. meridionalis associated with the dark reaction of photosynthesis increased in abundance during heat stress (Fig. 3A). The large isoform of Rubisco activase (RCAI) increased in relative abundance more than any other protein, while the small isoform of Rubisco activase (RCAII) was found in multiple spots that both increased and decreased (Tables 2, 3). In rice, there are two isoforms of Rubisco activase, a 45 kDa large isoform and a 41 kDa small isoform (To et al., 1999). The expression level of the mRNA encoding RCAI increased substantially after 24 h of heat while RCAII gene expression declined (Fig. 4).
Fig. 3.
Comparison of percentage spot volume on 2-DE gels between 27 °C (light shade) and 45 °C (dark shade) treated O. meridionalis seedlings. (A) Proteins associated with the dark reaction of photosynthesis including the Calvin Cycle enzymes, phosphoglycerate kinase, chloroplast precursor (PGK), transketolase, chloroplast precursor (TK), phosphoribulokinase (PRK), sedoheptulose-1,7-bisphosphatase, chloroplast precursor (SBPase), the photorespiration enzyme glycine dehydrogenase (GDC-P), and Rubisco activase, large isoform (RCAI). (B) The light reaction of photosynthesis represented by ferredoxin-NADP(H) oxidoreductase (FNR). (C) Protective proteins chaperone 60 (Cpn60), heat shock proteins 70 and 90 (HSP70, HSP90), and the thiamine biosynthesis protein THI1. Seedlings were grown in growth chambers at 27/22 °C with a 12 h photoperiod and illumination of 500 μmol m−2 s−1. Seedlings were either harvested under control conditions or exposed to 45 °C for a 24 h period prior to harvesting 2 h into a light period. Spots were analysed using ImageJ software with integrated density used as a determinant of spot volume. Values are mean ±SD, n=3.
Fig. 4.
Semi-quantitative RT-PCR temperature comparisons for selected O. meridionalis genes. RNA was initially extracted from the leaf tissue of seedlings exposed to either control (27 °C) or 24 h of high temperature (45 °C). (A) PCR products were run on a 2.5% agarose gel and stained with Gel Red (Biotium). Abbreviated gene names are; Rubisco activase large (RcaI) and small (RcaII) isoform genes, phosphoglycerate kinase (pgk), transketolase (tkt), phosphoribulokinase (prk), sedoheptulose-1,7-bisphosphatase (sbp), glycine dehydrogenase (gdc-p), ferredoxin-NADP(H) oxidoreductase (fnr), Heat shock protein 90 (hsp90), chaperone 60 (GroEL), heat shock protein 70 (dnaK), and thiamine biosynthesis protein (thi1). All genes underwent 26 cycles of PCR except RcaI, RcaII, and GroEL that underwent 22 cycles and dnaK which required 30 cycles. (B) Spot intensities of bands were compared and relative gene expression after 24 h at 45 °C (heat/control) was calculated with actin used as an internal standard. Values are mean ±SD, n=3.
Enzymes involved in the Calvin Cycle; chloroplastic phosphoglycerate kinase, transketolase, phosphoribulokinase (PRK), and chloroplastic sedoheptulose-1,7-bisphosphatase consistently increased in abundance with heat. However, expression of the genes encoding all of these proteins declined.
Glycine dehydrogenase (GDC-P) increased substantially with heat. Glycine metabolism is an essential part of photorespiration (Douce et al., 2001). Previously, reduced expression of GDC-P in chilled rice has been viewed as a loss of photorespiratory function (Yan et al., 2006). An increase in GDP-C, therefore, suggests an up-regulation of photorespiration by O. meridionalis during heat exposure.
The photosynthetic light reaction protein ferredoxin-NADP(H) oxidoreductase (FNR) displayed a substantial decline in abundance over the heat period (Fig. 3B). Three of the spots that decreased in volume with heat were identified as FNR, including spot 224 which decreased to the greatest extent (Table 3). The mRNA levels for this gene also declined.
Heat-induced protective proteins
The protective proteins chaperone 60 (Cpn60), heat shock protein 70 (HSP70), and heat shock protein 90 (HSP90) had increased levels of protein and gene expression following heat treatment (Figs 3C, 4). Proteins homologous to Cpn60 increased in multiple spots. A protein homologous to a germin-like protein was found in lower amounts with heat. Although germin-like proteins are protective proteins they seem to be associated with pathogen response in plants rather than abiotic stress such as heat (Byron, 2002; Miche et al., 2006; Zimmermann et al., 2006; Elvira et al., 2008). Soluble germin-like protein in barley has previously been found in reduced amounts when plants were subjected to heat (Vallelian-Bindschedler et al., 1998).
A thiamine biosynthesis protein homologous to THI1 found in Arabidopsis (Arabidopsis thaliana), increased in abundance with heat (Table 2; Fig. 3C) while the mRNA level dropped slightly (Fig. 4B). Results showed a small but significant decline in the amount of thiamine present in O. meridionalis after exposure to heat (Table 1). This is in contrast to the increase in expression of the THI1 homologue protein involved in thiamine synthesis.
Heat-suppressed proteins
The major component of Rubisco, easily identified as it accounted for an average of 22% of all protein in each gel, did not show substantial or consistent change across replicates. However, certain isoforms of the Rubisco large subunit (spots 250, 254, 255) were substantially reduced with heat (Table 3). The ATPase beta subunit was identified in multiple spots that both increased and decreased with heat. Of the other proteins identified as having a greater than 1.8-fold reduction under high temperatures, many were proteins of unknown function, while others were proteins of broad metabolic function (spots 213, 243, 205, 219, 242, 220, 204, 198).
Discussion
Many of the heat-induced proteins found in the wild rice species O. meridionalis have previously been shown to increase in O. sativa seedlings exposed to 42 °C, as determined by two-dimensional gel electrophoresis by Lee et al. (2007). For example, transketolase, HSP70, Cpn60, GDC-P, and the putative thiamine biosynthesis protein reported here all became more abundant over a similar 24 h period in O. sativa. Similar to the findings for O. meridionalis, there was a reduced abundance of FNR in O. sativa. By contrast, PRK declined in O. sativa during the 24 h heat stress, whereas it increased substantially with heat in O. meridionalis.
More than any other functional group, photosynthesis-related proteins were differentially expressed in O. meridionalis during heat stress, as is the case in cold-stressed rice seedlings (Yan et al., 2006) suggesting a specific connection between photosynthetic enzymes and temperature stress in rice.
A complex expression profile of Rubisco activase was observed in O. meridionalis under heat stress. The increased overall abundance of Rubisco activase with heat was caused by specific subunits of the multimeric protein. Rubisco activase is a member of the AAA+ protein family, as are most chaperones. Through ATP hydrolysis, Rubisco activase regenerates Rubisco that has been deactivated by bound non-substrate sugar phosphates, or RuBP bound to uncarbamylated active sites (Spreitzer and Salvucci, 2002; Portis, 2003). Rubisco activase seems to be heat-labile in many plant species, limiting photosynthetic capacity during heat stress (Law and Crafts-Brandner, 1999; Salvucci and Crafts-Brandner, 2004; Kurek et al., 2007).
Specifically, RCAI was almost undetectable at 27 °C but the protein increased in abundance at 45 °C through transcriptional up-regulation: preferential expression of the RCAI isoform therefore occurs at high temperatures in O. meridionalis. The high levels of RCAI observed are consistent with the heat tolerance of Rubisco activase isoforms in spinach (Spinacea oleracea L.), where the optimum temperature for ATP hydrolysis was 45 °C for the large isoform compared with 32 °C for the small isoform (Crafts-Brandner et al., 1997). Although the thermotolerance of RCAI has not previously been established in rice, under non-stressed conditions, transgenic rice over-expressing RCAI show greater photosynthetic capacity, through improvements in both dark and light reactions (Wu et al., 2007).
The expression profile of RCAII was complex. RCAII was found in multiple protein spots that both increased and decreased in abundance while rcaII expression levels decreased after exposure to heat. In pea (Pisum sativum L.) and spinach, which also express large and small Rubisco activase polypeptides, the small form is believed to be the more labile at higher temperatures (Crafts-Brandner et al., 1997; Salvucci et al., 2001). This is attributed to observations that the small form denatured and formed insoluble aggregates at high temperature. If this were the case in rice, identification of multiple spots of RCAII could be expected after application of heat. However, in O. meridionalis, there were no new RCAII spots detected in heat-treated tissue. The protein is therefore not being degraded but, instead, changing conformation or shifting to new isoforms in a consistent manner. Expression of distinctive activase forms at high temperatures, which have not been attributed to loss of function and aggregation, have been previously noted in cotton (Gossypium hirsutum L.), spinach, and wheat (Law et al., 2001; Law and Crafts-Brandner, 2001; Rokka et al., 2001). Similar post-translational protein modifications in tomato (Solanum lycopersicum) and oilseed rape (Brassica napus L. Reston) have been observed, with a series of spots corresponding to isoforms of a given protein (Agrawal and Thelen, 2006; Hattrup et al., 2007).
The overall increase in Rubisco activase was not matched by an increase in Rubisco which did not consistently change. This implies an increase in the activase/Rubisco ratio. Previous analysis of cotton and tobacco leaves found that an increase in the activase/Rubisco ratio leads to comparatively higher Rubisco activation states at temperatures up to 42 °C (Crafts-Brandner and Salvucci, 2000). Furthermore, by maintaining higher activation states, photosynthetic rates were less inhibited by heat stress.
Unlike O. sativa (Lee et al., 2007), in the heat-tolerant O. meridionalis there is an increase in multiple components of the Calvin Cycle including a consistent increase across all replicates of PRK, the enzyme responsible for the final step in RuBP regeneration. Alone these results indicate up-regulation of the Calvin Cycle when O. meridionalis was heat-treated. However, this would require sustained energy output from electron transport to provide the substrates required for the reduction phases of the Calvin Cycle. Crafts-Brandner and Law (2000), Cen and Sage (2005), and Kubien and Sage (2008) showed that the electron transport pathway of photosynthesis is highly susceptible at severe temperatures above 40 °C. In O. meridionalis, there is a decrease in both the gene expression and protein levels of FNR which catalyses the last enzymatic step of the non-cyclic photosynthetic light reaction responsible for the reduction of NADP+ in the PSI complex (Hurley et al., 2002). It is therefore likely that, although O. meridionalis had increased abundances of Calvin Cycle enzymes, inhibition of electron transport at such a severe temperature would inhibit both the light and dark reactions of photosynthesis. This may explain the significant reduction in the photosynthetic rate of O. meridionalis (and O. sativa) after heat exposure. Analysis by Hajirezaei et al. (2002) found growth rate and CO2 assimilation were reduced in tobacco plants with reduced levels of FNR. However, it was noted in their study that levels of Rubisco activase and transketolase were not altered by the impact of lower FNR levels. This is consistent with the reduced FNR but increased Rubisco activase and transketolase abundances in O. meridionalis.
Gene expression and protein abundance of the protective chaperone HSP70, HSP90,and Cpn60 increase in O. meridionalis upon heat stress. Cpn60 is a form of chaperone found in mitochondria and chloroplasts of plants and believed to support protein folding (Wang et al., 2004). HSP70 and HSP90 have been associated with an array of protective functions including protein refolding, transportation, and protein signalling pathways (Wang et al., 2004). Mutational studies of Cpn60 demonstrate that both the alpha and beta subunits are necessary for effective chloroplast function and are thus important in heat tolerance (Apuya et al., 2001; Ishikawa et al., 2003). Similarly the HSP70 family is directly correlated with thermotolerance in plants (Lee and Schöffl, 1996; Sung and Guy, 2003).
Of particular interest is the dual increase in RCAI and Cpn60 in O. meridionalis upon heat stress. An interaction between Rubisco activase and the Cpn60-β subunit is likely, as the chaperone has been shown to bind to Rubisco activase during heat stress in what is thought to be a protective role (Salvucci, 2008).
A THI1 homologue was found in greater abundance with heat in O. meridionalis. Similarly, in the thermotolerant species Populus euphratica, THI1 increased 3-fold during the first 6 h of exposure to 42 °C (Ferreira et al., 2006). In Arabidopsis, thi1 gene expression increased in roots subjected to hypoxia and in roots and rosettes subjected to high salt concentrations (Ribeiro et al., 2005). The most likely mode of action for THI1 would be an increased abundance of its known product, thiamine. In support of this, the application of thiamine to rice resulted in protection against a wide range of pathogens, with mutational studies attributing this to interaction between the thiamine and the salicylic-acid pathway (Ahn et al., 2005). Recently, abiotic stress through the application of polyethylene glycol, NaCl, and H2O2 in maize (Zea mays L.) lead to an increase in leaf thiamine concentration, again supporting a direct role of thiamine in the stress response (Rapala-Kozik et al., 2008).
By contrast, the concentration of thiamine in leaves of O. meridionalis fell significantly during heat stress, in spite of the enzyme THI1, which is responsible for its synthesis, increasing substantially. Findings in Arabidopsis and yeast suggest that THI1 might fulfil a function distinct from thiamine biosynthesis during heat stress. Specifically, Arabidopsis THI1 protein and THI4 found in yeast appear to be involved in the protection and repair of damaged mitochondrial DNA (Machado et al., 1997; Chabregas et al., 2001). In yeast, a THI4 mutant is more susceptible to oxidative stress under high temperatures even though the cultures were supplemented with thiamine (Medina-Silva et al., 2006). Alternatively, if thiamine were degraded faster at high temperatures, an increase in THI1 may simply be indicative of an increase in thiamine turnover.
Conclusions
The higher growth rate of O. meridionalis at 45 °C compared with O. sativa ssp. japonica, as well as a lesser impact of heat on photosynthesis, indicated tolerance of O. meridionalis to the extreme heat typical of its natural range. Rubisco activase and the regulation of the large and small isoforms found in rice are a striking aspect of the heat stress response of O. meridionalis. The Rubisco activase large isoform, in particular, is selectively up-regulated in response to heat. Multiple enzymes of the Calvin Cycle increased in abundance with heat. A fall in FNR, an important component of the light reaction, implies a susceptibility of electron transport at 45 °C for O. meridionalis. The consistent increase in expression of a THI1 homologue at high temperatures was notable because both THI1, an enzyme involved in thiamine biosynthesis, and thiamine have been linked to the heat stress response in plants. Interestingly, thiamine levels fell in heat-stressed O. meridionalis even though the abundance of THI1 increased.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Primers used in semi-quantitative RT-PCR.
Supplementary Fig. S1. 2-DE triplicate gels.
Supplementary Fig. S2. 2-DE spot clusters.
Supplementary Fig. S3. Serial dilution of leaf thiamine extract.
Supplementary Fig. S4. Accuracy of thiamine quantification assay.
Supplementary Material
Acknowledgments
The authors would like to thank Artur Sawicki, Mohammad Masood, Karlie Neilson, Tony Jerkovic, Ron Bradner, Robert Willows, Juliet Suich, Phyllis Farmer, and Thomas Roberts. PH acknowledges support from the NSW Office of Science and Medical Research in the form of a Biofirst Fellowship.
Glossary
Abbreviations
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- RCAI
Rubisco activase large isoform
- RCAII
Rubisco activase small isoform
- FNR
ferredoxin-NADP(H) oxidoreductase
- THI1
thiamine biosynthesis protein
- HSP
heat shock protein
- dnaK
heat shock protein 70 gene
- GroEL
chaperone 60 gene
- PGK
phosphoglycerate kinase gene
- PRK
phosphoribulokinase gene
- GDC-P
glycine dehydrogenase gene
- TK
transketolase
- tkt
transketolase gene
- SBPase
sedoheptulose-1,7-bisphosphatase
- sbp
sedoheptulose-1,7-bisphosphatase gene
- RuBP
ribulose-1,5-bisphosphate
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Agrawal GK, Thelen JJ. Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Molecular Cell Proteomics. 2006;5:2044–2059. doi: 10.1074/mcp.M600084-MCP200. [DOI] [PubMed] [Google Scholar]
- Ahn I-P, Kim S, Lee Y-H. Vitamin B1 functions as an activator of plant disease resistance. Plant Physiology. 2005;138:1505–1515. doi: 10.1104/pp.104.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AJ, Xu JL, Ismail AM, Fu BY, Vijaykumar CHM, Gao YM, Domingo J, Maghirang R, Yu SB, Gregorio G. Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Research. 2006;97:66–76. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB. The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60α gene. Plant Physiology. 2001;126:717–730. doi: 10.1104/pp.126.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar DS, Khush GS. Alien introgression in rice. Plant Molecular Biology. 1997;35:35–47. [PubMed] [Google Scholar]
- Byron GL. Oxalate, germins, and higher-plant pathogens. IUBMB Life. 2002;53:67–75. doi: 10.1080/15216540211474. [DOI] [PubMed] [Google Scholar]
- Cen Y-P, Sage RF. The regulation of Rubisco activity in response to variation in temperature and atmospheric CO2 partial pressure in sweet potato. Plant Physiology. 2005;139:979–990. doi: 10.1104/pp.105.066233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabregas SM, Luche DD, Farias LP, Ribeiro AF, van Sluys M-A, Menck CFM, Silva-Filho MC. Dual targeting properties of the N-terminal signal sequence of Arabidopsis thaliana THI1 protein to mitochondria and chloroplasts. Plant Molecular Biology. 2001;46:639–650. doi: 10.1023/a:1011628510711. [DOI] [PubMed] [Google Scholar]
- Cheng S-H, Zhuang J-Y, Fan Y-Y, Du J-H, Cao L-Y. Progress in research and development on hybrid rice: a super-domesticate in China. Annals of Botany. 2007;100:959–966. doi: 10.1093/aob/mcm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Law RD. Effect of heat stress on the inhibition and recovery of the ribulose-1,5-bisphosphate carboxylase/oxygenase activation state. Planta. 2000;212:67–74. doi: 10.1007/s004250000364. [DOI] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Salvucci ME. Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. Proceedings of the National Academy of Sciences, USA. 2000;97:13430–13435. doi: 10.1073/pnas.230451497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, van de Loo FJ, Salvucci ME. The two forms of ribulose-1,5-bisphosphate carboxylase/oxygenase activase differ in sensitivity to elevated temperature. Plant Physiology. 1997;114:439–444. doi: 10.1104/pp.114.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Communications in Mass Spectrometry. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F. The glycine decarboxylase system: a fascinating complex. Trends in Plant Science. 2001;6:167–176. doi: 10.1016/s1360-1385(01)01892-1. [DOI] [PubMed] [Google Scholar]
- Elvira MI, Galdeano MM, Gilardi P, Garcia-Luque I, Serra MT. Proteomic analysis of pathogenesis-related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum chinense L3 plants. Journal of Experimental Botany. 2008;59:1253–1265. doi: 10.1093/jxb/ern032. [DOI] [PubMed] [Google Scholar]
- Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Analytical Chemistry. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- Ferreira S, Hjerno K, Larsen M, Wingsle G, Larsen P, Fey S, Roepstorff P, Salome Pais M. Proteome profiling of Populus euphratica Oliv. upon heat stress. Annals of Botany. 2006;98:361–377. doi: 10.1093/aob/mcl106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Sang T, Lu B-R, Hong D-Y. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proceedings of the National Academy of Sciences, USA. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biology. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirezaei M-R, Peisker M, Henning M, Tschiersch J, Palatnik F, Valle EM, Carrillo N, Sonnewald U. Small changes in the activity of chloroplastic NADP+-dependent ferredoxin oxidoreductase lead to impaired plant growth and restrict photosynthetic activity of transgenic tobacco plants. The Plant Journal. 2002;29:281–293. doi: 10.1046/j.0960-7412.2001.01209.x. [DOI] [PubMed] [Google Scholar]
- Hattrup E, Neilson KA, Breci L, Haynes PA. Proteomic analysis of shade-avoidance response in tomato leaves. Journal of Agricultural and Food Chemistry. 2007;55:8310–8318. doi: 10.1021/jf0713049. [DOI] [PubMed] [Google Scholar]
- Hurley JK, Morales R, Martínez-Júlvez M, Brodie TB, Medina M, Gómez-Moreno C, Tollin G. Structure–function relationships in Anabaena ferredoxin/ferredoxin:NADP+ reductase electron transfer: insights from site-directed mutagenesis, transient absorption spectroscopy and X-ray crystallography. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2002;1554:5–21. doi: 10.1016/s0005-2728(02)00188-3. [DOI] [PubMed] [Google Scholar]
- IPCC. Climate change 2007: the physical science basis. Contribution of Working group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. 2007. In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, Miller HL, eds. United Kingdom and New York, NY, USA: Cambridge University Press, Cambridge. [Google Scholar]
- Ishikawa A, Tanaka H, Nakai M, Asahi T. Deletion of a chaperonin 60β gene leads to cell death in the Arabidopsis lesion initiation 1 mutant. Plant Cell Physiology. 2003;44:255–261. doi: 10.1093/pcp/pcg031. [DOI] [PubMed] [Google Scholar]
- Juliano AB, Naredo ME, Lu B-R, Jackson MT. Genetic differentiation in Oryza meridionalis Ng based on molecular and crossability analyses. Genetic Resources and Crop Evolution. 2005;V52:435–445. [Google Scholar]
- Komatsu S, Konishi H, Shen S, Yang G. Rice proteomics: a step toward functional analysis of the rice genome. Molecular and Cellular Proteomics. 2003;2:2–10. doi: 10.1074/mcp.r200008-mcp200. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Tanaka N. Rice proteome analysis: a step toward functional analysis of the rice genome. Proteomics. 2004;4:938–949. doi: 10.1002/pmic.200401040. [DOI] [PubMed] [Google Scholar]
- Kubien DS, Sage RF. The temperature response of photosynthesis in tobacco with reduced amounts of Rubisco. Plant, Cell and Environment. 2008;31:407–418. doi: 10.1111/j.1365-3040.2008.01778.x. [DOI] [PubMed] [Google Scholar]
- Kurek I, Chang TK, Bertain SM, Madrigal A, Liu L, Lassner MW, Zhu G. Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. The Plant Cell. 2007;19:3230–3241. doi: 10.1105/tpc.107.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafitte HR, Li ZK, Vijayakumar CHM, et al. Improvement of rice drought tolerance through backcross breeding: evaluation of donors and selection in drought nurseries. Field Crops Research. 2006;97:77–86. [Google Scholar]
- Law D, Crafts-Brandner S, Salvucci M. Heat stress induces the synthesis of a new form of ribulose-1,5-bisphosphate carboxylase/oxygenase activase in cotton leaves. Planta. 2001;214:117–125. doi: 10.1007/s004250100592. [DOI] [PubMed] [Google Scholar]
- Law RD, Crafts-Brandner SJ. Inhibition and acclimation of photosynthesis to heat stress Is closely correlated with activation of ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Physiology. 1999;120:173–182. doi: 10.1104/pp.120.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RD, Crafts-Brandner SJ. High temperature stress increases the expression of wheat leaf ribulose-1,5-bisphosphate carboxylase/oxygenase activase protein. Archives of Biochemistry and Biophysics. 2001;386:261–267. doi: 10.1006/abbi.2000.2225. [DOI] [PubMed] [Google Scholar]
- Lee D-G, Ahsan N, Lee S-H, Kang KY, Bahk JD, Lee I-J, Lee B-H. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics. 2007;7:3369–3383. doi: 10.1002/pmic.200700266. [DOI] [PubMed] [Google Scholar]
- Lee J, Schöffl F. An Hsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana. Molecular and General Genetics. 1996;252:11–19. doi: 10.1007/s004389670002. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in arabidopsis. The Plant Cell. 2004;16:1378–1391. doi: 10.1105/tpc.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B-R, Silitonga TS. Wild rice Oryza meridionalis was first found in Indonesia (notes from field) International Rice Research Notes. 1999;24:28–30. [Google Scholar]
- Machado CR, Praekelt UM, de Oliveira RC, Barbosa ACC, Byrne KL, Meacock PA, Menck CFM. Dual role for the yeast THI4 gene in thiamine biosynthesis and DNA damage tolerance. Journal of Molecular Biology. 1997;273:114–121. doi: 10.1006/jmbi.1997.1302. [DOI] [PubMed] [Google Scholar]
- Medina-Silva R, Barros MP, Galhardo RS, Netto LES, Colepicolo P, Menck CFM. Heat stress promotes mitochondrial instability and oxidative responses in yeast deficient in thiazole biosynthesis. Research in Microbiology. 2006;157:275–281. doi: 10.1016/j.resmic.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Medina ML, Haynes PA, Breci L, Francisco WA. Analysis of secreted proteins from Aspergillus flavus. Proteomics. 2005;5:3153–3161. doi: 10.1002/pmic.200401136. [DOI] [PubMed] [Google Scholar]
- Miche L, Battistoni F, Gemmer S, Belghazi M, Reinhold-Hurek B. Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Molecular Plant–Microbe Interactions. 2006;19:502–511. doi: 10.1094/MPMI-19-0502. [DOI] [PubMed] [Google Scholar]
- Ng NQ, Hawkes JG, Williams JT, Chang TT. The recognition of a new species of rice (Oryza) from Australia. Botanical Journal of the Linnean Society. 1981;82:327–330. [Google Scholar]
- Nishikawa T, Vaughan DA, Kadowaki K-i. Phylogenetic analysis of Oryza species, based on simple sequence repeats and their flanking nucleotide sequences from the mitochondrial and chloroplast genomes. Theoretical and Applied Genetics. 2005;110:696–705. doi: 10.1007/s00122-004-1895-2. [DOI] [PubMed] [Google Scholar]
- Ohta H, Maeda M, Nogata Y, Yoza K, Takeda Y, Osajima Y. A simple determination of thiamine in rice (Oryza sativa L.) by high-performance liquid chromatography with post-column derivatization. Journal of Liquid Chromatography. 1993;16:2617–2629. [Google Scholar]
- Portis A. Rubisco activase: Rubisco's catalytic chaperone. Photosynthesis Research. 2003;75:11–27. doi: 10.1023/A:1022458108678. [DOI] [PubMed] [Google Scholar]
- Rapala-Kozik M, Kowalska E, Ostrowska K. Modulation of thiamine metabolism in Zea mays seedlings under conditions of abiotic stress. Journal of Experimental Botany. 2008;59:4133–4143. doi: 10.1093/jxb/ern253. [DOI] [PubMed] [Google Scholar]
- Ribeiro DT, Farias LP, de Almeida JD, Kashiwabara PM, Ribeiro AFC, Silva-Filho MC, Menck CFM, Van Sluys M-A. Functional characterization of the thi1 promoter region from Arabidopsis thaliana. Journal of Experimental Botany. 2005;56:1797–1804. doi: 10.1093/jxb/eri168. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Piñeiro AM, Álvarez-Chaver P, Martínez-Zorzano VS, Rodríguez-Berrocal FJ, De la Cadena MP. Relevance of protein isoforms in proteomic studies. Current Proteomics. 2007;4:235–252. [Google Scholar]
- Rokka A, Zhang L, Aro E-M. Rubisco activase: an enzyme with a temperature-dependent dual function? The Plant Journal. 2001;25:463–471. doi: 10.1046/j.1365-313x.2001.00981.x. [DOI] [PubMed] [Google Scholar]
- Salekdeh H, Leonard JJ, Ghareyazie W, Bennett J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics. 2002;2:1131–1145. doi: 10.1002/1615-9861(200209)2:9<1131::AID-PROT1131>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Salvucci ME. Association of Rubisco activase with chaperonin-60β: a possible mechanism for protecting photosynthesis during heat stress. Journal of Experimental Botany. 2008;59:1923–1933. doi: 10.1093/jxb/erm343. [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ. Relationship between the heat tolerance of photosynthesis and the thermal stability of Rubisco activase in plants from contrasting thermal environments. Plant Physiology. 2004;134:1460–1470. doi: 10.1104/pp.103.038323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, Osteryoung KW, Crafts-Brandner SJ, Vierling E. Exceptional sensitivity of Rubisco activase to thermal denaturation in vitro and in vivo. Plant Physiology. 2001;127:1053–1064. [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, Salvucci ME. RUBISCO: structure, regulatory interactions, and possibilities for a better enzyme. Annual Review of Plant Biology. 2002;53:449–475. doi: 10.1146/annurev.arplant.53.100301.135233. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Sopory SK, Kavi Kishor PB. Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene. 2007;388:1–13. doi: 10.1016/j.gene.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Sung DY, Guy CL. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiology. 2003;132:979–987. doi: 10.1104/pp.102.019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K-Y, Suen D-F, Chen S-CG. Molecular characterization of ribulose-1,5-bisphosphate carboxylase/oxygenase activase in rice leaves. Planta. 1999;209:66–76. doi: 10.1007/s004250050607. [DOI] [PubMed] [Google Scholar]
- Vallelian-Bindschedler L, Mösinger E, Métraux J-P, Schweizer P. Structure, expression and localization of a germin-like protein in barley (Hordeum vulgare L.) that is insolubilized in stressed leaves. Plant Molecular Biology. 1998;37:297–308. doi: 10.1023/a:1005982715972. [DOI] [PubMed] [Google Scholar]
- Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, Cresti M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis. 2003a;24:2369–2375. doi: 10.1002/elps.200305500. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003b;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wu H, Li L, Jing Y, Kuang T. Over-and anti-sense expressions of the large isoform of ribulose-1,5-bisphosphate carboxylase/oxygenase activase gene in Oryza sativa affect the photosynthetic capacity. Photosynthetica. 2007;45:194–201. [Google Scholar]
- Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, McCouch SR. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics. 1998;150:899–909. doi: 10.1093/genetics/150.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S-P, Zhang Q-Y, Tang Z-C, Su W-A, Sun W-N. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Molecular and Cellular Proteomics. 2006;5:484–496. doi: 10.1074/mcp.M500251-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zheng X, Luo J, Gaut BS, Ge S. Multilocus analysis of nucleotide variation of Oryza sativa and Its wild relatives: severe bottleneck during domestication of rice. Molecular Biology and Evolution. 2007;24:875–888. doi: 10.1093/molbev/msm005. [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Baumlein H, Mock H-P, Himmelbach A, Schweizer P. The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiology. 2006;142:181–192. doi: 10.1104/pp.106.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.