Abstract
Low-oxygen (hypoxia) stress associated with natural phenomena such as waterlogging, results in widespread transcriptome changes and a metabolic switch from aerobic respiration to anaerobic fermentation. High-throughput sequencing of small RNA libraries obtained from hypoxia-treated and control root tissue identified a total of 65 unique microRNA (miRNA) sequences from 46 families, and 14 trans-acting small interfering RNA (tasiRNA) from three families. Hypoxia resulted in changes to the abundance of 46 miRNAs from 19 families, and all three tasiRNA families. Chemical inhibition of mitochondrial respiration caused similar changes in expression in a majority of the hypoxia-responsive small RNAs analysed. Our data indicate that miRNAs and tasiRNAs play a role in gene regulation and possibly developmental responses to hypoxia, and that a major signal for these responses is likely to be dependent on mitochondrial function.
Keywords: Abiotic stress, Arabidopsis thaliana, deep sequencing, hypoxia, microRNA, mitochondria, trans-acting small interfering RNA
Introduction
Waterlogging is major cause of reduced crop yields worldwide (Boyer, 1982). Initially, the foremost detrimental effect of waterlogging is the reduced access to oxygen in the rhizosphere, which is essential for mitochondrial respiration (Agarwal and Grover, 2006). Genome-wide expression studies in model plant species such as Arabidopsis thaliana have found that low oxygen (hypoxia) causes widespread changes in 5–10% of the transcriptome (Klok et al., 2002; Branco-Price et al., 2005; Liu et al., 2005; Loreti et al., 2005; Lasanthi-Kudahettige et al., 2007; Kreuzwieser et al., 2009). Co-incident with these mRNA changes are dramatic changes to protein synthesis and degradation, and a metabolic switch from aerobic respiration to anaerobic fermentation (Bailey-Serres and Voesenek, 2008).
Despite the wealth of molecular and phenotypic data on plant responses to waterlogging, very little is known about how declining oxygen levels are sensed, and how the complex and extensive expression changes are controlled (Bailey-Serres and Chang, 2005). It is also not understood whether mitochondria play a passive role or are active in responding to decreased oxygen by retrograde regulation of nuclear genes (Rhoads and Subbaiah, 2007). To date only a few regulators of gene expression have been implicated in the hypoxia response in plants. Rice plants can increase their chance of surviving submergence in two antithetical pathways that are initiated by ethylene. Over-expression of SUBMERGENCE 1A (SUB1A) an AP2-EREBP type transcription factor aids survival under short-term flooding by conserving energy and carbohydrate consumption via the regulation of a suite of genes associated with carbohydrate metabolism, ethanolic fermentation, and cell expansion (Fukao et al., 2006; Xu et al., 2006), whereas, deepwater rice outgrows rising floodwaters aided by the expression of two AP2-ERFBP genes called SNORKEL1,2 (SK1, SK2) that trigger internode elongation (Hattori et al., 2009). In Arabidopsis, the transcription factor AtMYB2 is induced early following hypoxia stress and trans-activates the ALCOHOL DEHYDROGENASE (ADH) promoter via the GT-motif (Hoeren et al., 1998), aiding the metabolic shift to fermentation. Recently, VERNALIZATION INSENSITIVE 3 (VIN3) a chromatin remodelling protein was found to be required for the survival of Arabidopsis seedlings to hypoxic stress (Bond et al., 2009), implicating histone modification as an important process in hypoxia. The Arabidopsis NAC domain containing gene ANAC102 has been found to be necessary for efficient seed germination following hypoxia, but not for seedling survival (Christianson et al., 2009), indicating that the response to hypoxia differs depending on developmental stage.
Small ribonucleic acids (smRNAs) have been identified as important post-transcriptional regulators of gene expression. Endogenous smRNAs in plants can be divided into two broad classes, microRNAs (miRNAs) and small interfering RNAs (siRNAs), based on their biogenesis and function. The best characterized are the miRNAs which are ∼21 nucleotides (nt) in size and generated from non-coding transcripts capable of forming stable secondary structures. In plants, miRNAs often regulate groups of transcription factors, and are thought to act by targeting almost perfectly complementary mRNAs for endonucleolytic cleavage and translational inhibition (Vazquez, 2006; Brodersen et al., 2008). siRNAs can be divided into a number of classes, including cis-acting siRNA (casiRNAs), trans-acting siRNA (tasiRNA), long siRNA (lsiRNA), and natural antisense transcript siRNA (natsiRNA) (for a review see Ghildiyal and Zamore, 2009). tasiRNAs and natsiRNAs are thought to regulate target transcripts in a similar manner to miRNAs, whereas lsiRNAs are thought to destabilize target mRNAs causing rapid degradation. casiRNAs promote heterochromatin formation by directing DNA methylation and histone modification.
Although the majority of characterized smRNAs have been associated with plant development (Bonnet et al., 2006; Sunkar et al., 2007) a growing number of smRNAs have been demonstrated to be involved in abiotic and biotic stresses such as cold (Sunkar and Zhu, 2004; Lu et al., 2008; Zhou et al., 2008), salt (Sunkar and Zhu, 2004; Borsani et al., 2005; Lu et al., 2008; Ding et al., 2009), heat (Lu et al., 2008), dehydration (Sunkar and Zhu, 2004; Reyes and Chua, 2007), oxidative stress (Sunkar et al., 2006), mechanical stress (Lu et al., 2008), pathogen infection (Katiyar-Agarwal et al., 2006), and submergence in maize (Zhang et al., 2008). The regulation of smRNAs appears to play an important role in transcriptome adjustments to life-threatening stresses.
In this study, high throughput sequencing technology was used to investigate the role of smRNAs in the response of Arabidopsis to hypoxia. Our results identified a total of 65 unique miRNA sequences (from 46 families) and 14 tasiRNAs (from three families) present in hypoxia treated and control root tissue. Of these, 25 miRNA (19 families) and 10 tasiRNAs (three families) were found to have significantly changed abundance upon hypoxia. Inhibition of mitochondrial respiration caused similar changes to the levels of several smRNAs, indicating that these smRNAs may play a significant and specific role in plant responses to hypoxia.
Materials and methods
Arabidopsis Columbia-0 seeds were surface-sterilized in 0.5% (w/v) sodium hypochlorite. Seeds were sown on 100 mm Petri plates containing a modified Murashige-Skoog (MS) medium (Murashige and Skoog, 1962) containing 3% sucrose (w/v) and 0.8% (w/v) agar and were subjected to at least 24 h of cold treatment in the dark (4 °C) to break dormancy. Each plate contained approximately 40 seeds and were grown at 21±1 °C under cool-white fluorescent lights (120 μM m−2 s−1) with a 16 h photoperiod.
Hypoxia assay
Hypoxia assays were performed as outlined previously (Christianson et al., 2009). Briefly, 15 2.5-week-old plants were transferred to 100 mm Petri plates containing 15 ml of MS liquid media supplemented with 3% sucrose 1 d prior to treatment. Immediately prior to treatment, plants were transferred to liquid media which had been sparged with 0.1% O2 and then placed in 3.5 l anaerobic chambers (Oxoid, Adelaide, SA, Australia) and purged with 0.1% O2, 99.9% N2 at a flow rate of ∼5 l min−1 for 15 min. The plants were left in 0.1% O2, in the dark with gentle shaking for a designated period. Control plants were subjected to the same treatment except that they were not transferred to 0.1% O2 sparged media, and the chambers were not sparged with 0.1% O2. Immediately after the treatment, plants were removed from the chamber and a scalpel used to isolate root tissue, which was then placed in liquid nitrogen, and stored at −80 °C.
Mitochondrial inhibitor treatment
The mitochondrial inhibitor assay was performed as outlined previously (Bond et al., 2009) except DMSO was used as a solvent for AA (Sigma-Aldrich, St Louis, Missouri, USA). Fifteen 2-week-old plants were transferred to 100 mm Petri plates containing 15 ml of MS liquid media supplemented with 3% sucrose 1 d prior to treatment. The media were replaced with MS liquid media containing 3% sucrose, 25 μM AA, and 5 mM of SHAM (Sigma-Aldrich, St Louis, Missouri, USA) and 0.4% DMSO. The plants were gently shaken for 5 h. The control treatment consisted of MS liquid media supplemented with 3% sucrose and 0.4% DMSO. Immediately after the treatment, root tissue was collected, placed in liquid nitrogen, and stored at −80 °C.
RNA extraction
Frozen root tissue was ground to a fine white powder using a mortar and pestle. Total RNA extraction was performed using TRI Reagent (Molecular Research Center Inc., Cincinnati, Ohio, USA) following manufacturer's instructions.
The smRNA fraction was isolated from total RNA by selectively precipitating HMW RNA in 0.5 vol. of 3 M NaCl/30% PEG (MW 8000) (Lu et al., 2007). The smRNA fraction obtained represented approximately 5% of the total RNA.
High throughput sequencing
Size-selected small RNAs (16–30 nt) was obtained from total RNA by size fractionation and sequenced by GeneWorks Pty Ltd. (Hindmarsh, SA, Australia) using high throughput pyrosequencing technology developed by Illumina, using an Illumina 1G Genome Analyzer, following the manufacturers instruction's. All small RNA sequences have been deposited in the Gene Expression Omnibus in GenBank under the identifier GSE16971 and GPL8810.
Bioinformatic analysis of small RNAs
Ilumina sequencing data were received in FASTQ format, with all reads shown as 36 mers prior to the removal of the 3′ adapter sequence. A custom script was created to find and trim the adaptor sequence from each sequence (Zhu et al., 2008). SOAP (Li et al., 2008) was used to align the reads to the Arabidopsis genome sequence (TAIR8), allowing for one mismatch and each mapped location was annotated with genomic region and/or gene names. A summary of the origin of the small RNA fragments can be found in Supplementary Table S1 at JXB online. Reads perfectly matching known Arabidopsis miRNA or miRNA* from the miRBase (Griffiths-Jones, 2004) and/or miRNA from the ASRP database (Gustafson et al., 2005) were identified.
Statistical analysis of miRNA frequency
To determine whether differences in miRNA frequency between control and hypoxia treated samples was significant; a χ-squared test was performed using total sequence numbers as previously described by Qiu et al. (2009).
Promoter analysis
Promoter regions from selected miRNAs were screened for the presence of common motifs using the web-based analysis tool PlantPAN (Chang et al., 2008). Since mature miRNAs of the same sequence can originate from different loci, only the promoter regions from unique mature miRNAs whose frequency was altered ≥1.5-fold in the hypoxia treatment were used for promoter analysis. Promoter sequences for miRNAs were extracted from TAIR8 (Rhee et al., 2003). In each case 1000 bp upstream (minus any coding region from upstream genes) from the designated start of the miRNA stem-loop was used, and miRNA promoter regions with ≤50 bp were discarded. A total of 13 miRNA promoters were used for the analysis (miR156g, miR157d, miR158a, miR158b, miR159a, miR161, miR167d, miR169a, miR173, miR391, miR775, miR824, miR829). The default settings on PlantPan were used at all times, for both co-occurrence of transcription factor binding sites in gene group.
Confirmation of mature miRNA expression by stem-loop quantitative PCR (SL-PCR)
The expression profiles of mature miRNAs and tasiRNAs were assayed by SL-PCR and performed as described previously (Varkonyi-Gasic et al., 2007). The stem–loop reverse transcription primers were designed following the methods previously described (Chen et al., 2005; Varkonyi-Gasic et al., 2007) and are listed in Supplementary Table S3 at JXB online. The stem-loop reverse transcriptase primer for each miRNA consists of a selfed stem-loop sequence (GTCGTATCCAGTGCAGGGTCCGAGGT ATTCGCACTGGATACGAC) with the specificity conferred by a six nucleotide extension at the 3′ end, that is complementary to the last six nucleotides at the 3′ end of mature miRNA. The RT reactions were performed on 100 ng LMW RNA using Superscript III (Invitrogen Australia Pty Ltd., Vic, Australia) and carried out according to the manufacturer's instructions. For each LWM RNA sample two cDNA reactions were performed. One containing all miRNA and tasiRNA stem-loop RT primers, and one for the housekeeping control U6 (see Supplementary Table S4 at JXB online). The reverse transcription product was amplified using a miRNA or tasiRNA specific forward primer and a universal reverse primer on a Rotor-Gene 6000 (QIAGEN Pty Ltd, Victoria, Australia). Reactions were performed in triplicate on three independent biological replicates, and amplicons from representative samples were sequenced to confirm the specificity of the amplification. The comparative quantification procedure was used to determine relative expression levels as previously described (Wilson et al., 2005).
Quantitative reverse transcriptase PCR (QRT-PCR) of miRNA target gene expression
The expression of experimentally determined and predicted target genes were assayed by QRT-PCR as previously described by Wilson et al. (2005). The reverse transcription product was amplified using gene-specific primers that generated amplicons that overlapped the known or predicted cleavage site (see Supplementary Table S5 at JXB online). Reactions were performed in triplicate on a Rotor-Gene 6000 (Qiagen Pty Ltd, Victoria, Australia). Data were normalized to AT5G08290 (Czechowski et al., 2005) and analysed using a comparative quantification procedure (Wilson et al., 2005).
Results
High throughput sequencing reveals a complex small RNA population in plant roots
To investigate the role of smRNAs in hypoxia stress in Arabidopsis, size fractionated RNA (16–30 nucleotides) was isolated from root tissue subjected to 5 h of either 0.1% oxygen or normoxic conditions, and sequenced using Illumina high throughput sequencing. Five hours of hypoxia treatment was chosen as hypoxia responses in the roots are significant at this time as indicated by the 88-fold induction of the hypoxia responsive gene, ADH (see Supplementary Fig. S1 at JXB online), but does not result in root tip death (Ellis et al., 1999). The total number of sequence reads was 2 550 664 for the normoxic sample and 2 998 711 for the hypoxia stressed sample (Table 1). After removing adapter only sequences and sequences of sizes <18 nt there were 383 969 and 390 392 unique sequences, respectively. This totalled 698 762 unique RNA sequences across the two samples. The percentage of unique RNA sequences with only a single read (singleton) provides an indication of sequence coverage and the complexity of the sample. The unstressed sequences comprised 80% singletons, whereas hypoxia-stressed sequences contained 81% singletons, indicating that the sequencing of the RNA population had not reached saturation.
Table 1.
Summary of small RNA sequencing data from control and hypoxia treated roots
| Untreated sample | Treated sample | |
| Total reads | 2 550 664 | 2 998 711 |
| Reads containing N | 32 278 | 7 152 |
| Reads without N | 2 518 386 | 2 991 559 |
| Adaptor only sequences | 268 521 | 334 287 |
| Low quality sequences removed | 4 186 | 8 215 |
| Sequences with small insert (<18 nt) | 444 444 | 997 892 |
| Sequences with large insert (>26 nt) | 406 465 | 164 682 |
| Unique sequences | 383 969 | 390 392 |
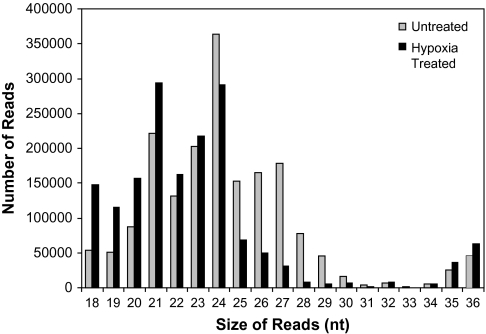
The RNA sequences were mainly within a range of 20–24 nt (61%) the size of most known smRNAs (Fig. 1). Hypoxia stress resulted in fewer 24 nt sequences and an increase in 21 nt sequences. Overall, the size distribution was similar to other previous studies on angiosperms (Fahlgren et al., 2007; Nobuta et al., 2008; Szittya et al., 2008; Zhu et al., 2008).
Fig. 1.
Size distribution of sequenced reads from control and hypoxia-treated roots. Small RNAs (16-30 nt) was isolated from untreated (grey bars) and hypoxia-treated (0.1% O2 for 5 h (black bars) root samples and sequenced using a Illumina 1G Genome Analyzer. Sequences were trimmed of their adapter sequence.
Sixty-five unique sequences match known miRNAs
The small RNA sequences from the hypoxia-stressed and control samples were analysed for the presence of previously characterized miRNAs. Only sequence reads that perfectly matched known Arabidopsis miRNA or miRNA* from miRBase (Griffiths-Jones, 2004) were considered. A total of 65 unique sequences from the combined RNA sequence reads matched known miRNAs (Table 2). These miRNAs belonged to 46 miRNA families and represent 45% of all known unique Arabidopsis miRNA sequences and 41% of all known miRNA families. Comparing normalized root miRNA frequencies (combined control and stressed RNAs) from this study to leaf and whole plant populations present in the Arabidopsis Small RNA Project (ASRP) database (Gustafson et al., 2005) showed that most miRNA families have tissue-specific expression (see Supplementary Table S2 at JXB online). For example, miRNA families 156, 157, 158, 165, and 829 are observed more frequently in roots than in leaf or whole plant tissue, whereas families 167, 169, 170, 171a, 172, 319, 396, 397, and 398 are low or absent in roots, but are numerous in leaf or whole plant samples.
Table 2.
Known miRNAs present in hypoxia treated and control roots
| miRNA family | miRNA | Sequence | Untreated reads | Hypoxia reads | Relative changea |
| 156 | 156a,b,c,d,e,f | UGACAGAAGAGAGUGAGCAC | 4 382 | 6 958 | 1.35* |
| 156g | CGACAGAAGAGAGUGAGCAC | 17 | 55 | 2.75* | |
| 156h | UGACAGAAGAAAGAGAGCAC | 2 | 5 | 2.13 | |
| 157 | 157a,b,c | UUGACAGAAGAUAGAGAGCAC | 4 119 | 8 409 | 1.74* |
| 157d | UGACAGAAGAUAGAGAGCAC | 58 | 282 | 4.14* | |
| 158 | 158a | UCCCAAAUGUAGACAAAGCA | 2 427 | 7 981 | 2.80* |
| 158b | CCCCAAAUGUAGACAAAGCA | 34 | 69 | 1.73* | |
| 159 | 159a | UUUGGAUUGAAGGGAGCUCUA | 73 | 162 | 1.89* |
| 159b | UUUGGAUUGAAGGGAGCUCUU | 11 | 14 | 1.08 | |
| 160 | 160a,b,c | UGCCUGGCUCCCUGUAUGCCA | 1 | 2 | 1.70 |
| 161 | 161.1 | UGAAAGUGACUACAUCGGGGU | 39 | 65 | 1.42* |
| 161.2 | UCAAUGCAUUGAAAGUGACUA | 20 | 119 | 5.06* | |
| 163 | 163 | UUGAAGAGGACUUGGAACUUCGAU | 2 | 4 | 1.70 |
| 164 | 164a,b | UGGAGAAGCAGGGCACGUGCA | 2 362 | 3 151 | 1.13* |
| 164c | UGGAGAAGCAGGGCACGUGCG | 452 | 550 | 1.04 | |
| 165 | 165a,b | UCGGACCAGGCUUCAUCCCCC | 14 977 | 15 750 | 0.89* |
| 166 | 166a,b,c,d,e,f,g | UCGGACCAGGCUUCAUUCCCC | 79 589 | 84 844 | 0.91* |
| 167 | 167a,b | UGAAGCUGCCAGCAUGAUCUA | 1 429 | 1 709 | 1.02 |
| 167d | UGAAGCUGCCAGCAUGAUCUGG | 1 | 4 | 3.40* | |
| 168 | 168a,b | UCGCUUGGUGCAGGUCGGGAA | 7 584 | 9 932 | 1.11* |
| 169 | 169a | CAGCCAAGGAUGACUUGCCGA | 10 | 3 | 0.26* |
| 169b,c | CAGCCAAGGAUGACUUGCCGG | 1 | 8 | 6.80* | |
| 169d,e,f,g | UGAGCCAAGGAUGACUUGCCG | 92 | 61 | 0.56* | |
| 169h,i,j,k,l,m,n | UAGCCAAGGAUGACUUGCCUG | 3 | 6 | 1.70 | |
| 171 | 171a | UGAUUGAGCCGCGCCAAUAUC | 0 | 2 | NA** |
| 172 | 172a, b | AGAAUCUUGAUGAUGCUGCAU | 15 | 93 | 5.27* |
| 172c,d | AGAAUCUUGAUGAUGCUGCAG | 2 | 2 | 0.85 | |
| 172e | GGAAUCUUGAUGAUGCUGCAU | 4 | 2 | 0.43 | |
| 173 | 173 | UUCGCUUGCAGAGAGAAAUCAC | 213 | 375 | 1.50* |
| 319 | 319a,b | UUGGACUGAAGGGAGCUCCCU | 1 | 0 | NA |
| 390 | 390a,b | AAGCUCAGGAGGGAUAGCGCC | 1 652 | 3 124 | 1.61* |
| 391 | 391 | UUCGCAGGAGAGAUAGCGCCA | 25 | 77 | 2.62* |
| 395 | 395a,d,e | CUGAAGUGUUUGGGGGAACUC | 1 | 2 | 1.70 |
| 395b,c,f | CUGAAGUGUUUGGGGGGACUC | 2 | 5 | 2.13 | |
| 399 | 399a | UGCCAAAGGAGAUUUGCCCUG | 1 | 1 | 0.85 |
| 399b,c | UGCCAAAGGAGAGUUGCCCUG | 4 | 3 | 0.64 | |
| 399d | UGCCAAAGGAGAUUUGCCCCG | 5 | 1 | 0.17 | |
| 399f | UGCCAAAGGAGAUUUGCCCGG | 11 | 12 | 0.93 | |
| 400 | 400 | UAUGAGAGUAUUAUAAGUCAC | 2 | 1 | 0.43 |
| 403 | 403 | UUAGAUUCACGCACAAACUCG | 2 | 4 | 1.70 |
| 408 | 408 | AUGCACUGCCUCUUCCCUGGC | 125 | 138 | 0.94 |
| 447 | 447a,b | UUGGGGACGAGAUGUUUUGUUG | 5 | 3 | 0.51 |
| 775 | 775 | UUCGAUGUCUAGCAGUGCCA | 9 | 27 | 2.55* |
| 780 | 780.1 | UCUAGCAGCUGUUGAGCAGGU | 0 | 1 | NA |
| 822 | 822 | UGCGGGAAGCAUUUGCACAUG | 115 | 121 | 0.89 |
| 823 | 823 | UGGGUGGUGAUCAUAUAAGAU | 6 | 10 | 1.42 |
| 824 | 824 | UAGACCAUUUGUGAGAAGGGA | 20 | 39 | 1.66* |
| 827 | 827 | UUAGAUGACCAUCAACAAACU | 10 | 20 | 1.70 |
| 829 | 829.1 | AGCUCUGAUACCAAAUGAUGGAAU | 1 564 | 1 228 | 0.67* |
| 829.2 | CAAAUUAAAGCUUCAAGGUAG | 1 | 0 | NA | |
| 830 | 830 | UAACUAUUUUGAGAAGAAGUG | 1 | 0 | NA |
| 837 | 837-3p | AAACGAACAAAAAACUGAUGG | 14 | 27 | 1.64* |
| 837-5p | AUCAGUUUCUUGUUCGUUUCA | 1 | 1 | 0.85 | |
| 838 | 838 | UUUUCUUCUACUUCUUGCACA | 0 | 1 | NA |
| 841 | 841 | UACGAGCCACUUGAAACUGAA | 1 | 0 | NA |
| 842 | 842 | UCAUGGUCAGAUCCGUCAUCC | 23 | 24 | 0.89 |
| 843 | 843 | UUUAGGUCGAGCUUCAUUGGA | 0 | 2 | NA |
| 846 | 846 | UUGAAUUGAAGUGCUUGAAUU | 3 | 3 | 0.85 |
| 848 | 848 | UGACAUGGGACUGCCUAAGCUA | 10 | 14 | 1.19 |
| 852 | 852 | AAGAUAAGCGCCUUAGUUCUG | 1 | 2 | 1.70 |
| 860 | 860 | UCAAUAGAUUGGACUAUGUAU | 0 | 2 | NA |
| 861 | 861-5p | CCUUGGAGAAAUAUGCGUCAA | 1 | 0 | NA |
| 862 | 862-5p | UCCAAUAGGUCGAGCAUGUGC | 0 | 3 | NA |
| 869 | 869.2 | UCUGGUGUUGAGAUAGUUGAC | 50 | 57 | 0.97 |
| 1888 | 1888 | UAAGUUAAGAUUUGUGAAGAA | 0 | 2 | NA |
Relative change is the ratio of hypoxia-treated sample to the control and takes into account the differences in total read between samples. *P value ≤0.01. **Relative change was not calculated as they contained 0 reads in one sample.
High throughout sequencing can identify not only miRNAs but can also accurately measure miRNA expression (’t Hoen et al., 2008). The abundance of miRNAs was found to vary greatly within each sample and within miRNA families (Table 2). The most abundant miRNA identified was the miR166 family (n=164 433), although most miRNAs were little expressed, as almost half of the miRNAs identified had fewer than 10 reads in total. Statistical analysis of the frequency of miRNAs (Qiu et al., 2009) between the two treatments revealed that hypoxia treatment significantly changed the abundance of 25 miRNAs from 19 miRNA families (P value ≤0.01) with the majority of those changing by ≥1.5-fold. In most cases, this was an increase in miRNA frequency, with only five miRNAs (from four miRNA families) in the hypoxia sample experiencing a significant decrease in expression (Table 2). Apart from miR169, multiple members of a miRNA family were regulated similarly (e.g. miR156a–f and g, and miR158a, b). These results indicate that hypoxia alters the expression of a specific subset of miRNAs.
Common cis-acting elements within promoter regions of miRNAs affected by hypoxia
The promoter regions of miRNAs have the same type of promoters as protein coding genes and are transcribed by RNA polymerase II (Megraw et al., 2006; Zhou et al., 2007). However, analysis of the promoters of miRNAs is difficult, as mature miRNAs with identical sequence can originate from different loci. In order to determine whether miRNAs with altered expression under hypoxia possessed common regulatory motifs, promoter regions from unique mature miRNAs sequences whose frequency was altered ≥1.5-fold in the hypoxia treatment were used for motif analysis (12 miRNAs). Analysis using Plant Promoter Analysis Navigator (PlantPAN) (Chang et al., 2008) indicated that two known variants of the homeobox domain leucine zipper class 1 (HDZip I) promoter motif, ATHB-5 (CAATTATTG, P value=4E−6) and ATHB1 (NYNCAATTATTGSA, P value=4E−9) were significantly overrepresented.
tasiRNAs change under hypoxia stress
Another major class of smRNA capable of altering the expression of genes is tasiRNA; although tasiRNA expression has not been found to alter with abiotic stress. Sequences were compared against known Arabidopsis tasiRNAs and 14 unique sequences (61% of total known sequences) were recognized as fully matching known tasiRNAs (Table 3). The tasiRNAs identified belonged to three of the four known tasiRNA families, TAS1, TAS2, and TAS3. All of the tasiRNAs (n=10) that were found to change significantly under hypoxia stress showed increased abundance under hypoxia.
Table 3.
Known tasiRNA present in control and hypoxia treated roots
| tasiRNA | TAS locus | Sequence | Untreated reads | Hypoxia reads | Relative changea |
| 1786 | TAS1a | UGAUAUUUGUAGUAAUGGCG | 1 | 6 | 5.10* |
| 1852 | TAS1a | AUGAUAUUUGUAGUAAUGGCG | 430 | 863 | 1.71* |
| 255 | TAS1a,b,c | UUCUAAGUCCAACAUAGCGUA | 20 | 61 | 2.59* |
| 289 | TAS1a,b,c | UUCUAAGUCCAACAUAGCAUA | 3 | 16 | 4.54* |
| 619 | TAS1c | AUAUUCCAGGAUAUGCAAAAG | 0 | 1 | NA** |
| 850 | TAS1c | UUCUAAGUUCAACAUAUCGAC | 0 | 2 | NA |
| 1413 | TAS1c | UUCUAAGUUCAACAUAUCGACG | 0 | 1 | NA |
| 143 | TAS2 | AUAAUCAAGUGAAUAGUUUAA | 3 | 9 | 2.55* |
| 614 | TAS2 | GAUGGUAGUUCAAGUAUUCCA | 7 | 21 | 2.55* |
| 1511 | TAS2 | UCCAAGCGAAUGAUGAUACUU | 25 | 90 | 3.06* |
| 1767 | TAS2 | UUUGAACUUGUGUAUUUUGAA | 10 | 26 | 2.21* |
| 1940 | TAS2 | GAAUACUUGAACUACCAUCUA | 6 | 4 | 0.57 |
| 2140 | TAS2 | AUAUCCCAUUUCUACCAUCUG | 2 | 14 | 5.95* |
| 2142 | TAS3a | UUCUUGACCUUGUAAGACCCC | 337 | 469 | 1.18* |
Relative change is the ratio of hypoxia-treated sample to the untreated control and takes into account differences in total read between samples. *P value≤0.01; ** Relative change was not calculated as they contained 0 reads in one sample.
SL-PCR verifies deep sequencing results
As only one sample of stressed and unstressed RNA was sequenced, it was necessary to examine the expression of a selection of smRNAs to verify that the changes observed were biologically consistent. The SL-PCR protocol of Chen et al. (2005), that uses stem-loop primers in the reverse transcriptase reaction, was used to measure mature smRNA expression on at least three independent replicate samples from untreated and hypoxia-stressed roots. QRT-PCR was conducted for seven miRNAs, and one tasiRNA that showed significant increases in frequency under hypoxia stress based on the deep sequencing data. The QRT-PCR results were similar in magnitude to the deep sequencing results and confirm that smRNAs identified from high throughput sequencing change in response to hypoxia treatment (Table 4).
Table 4.
Expression levels of selected miRNAs and a tasiRNA under hypoxia stress and mitochondrial inhibitor treatment
| smRNA | Hypoxia |
Mitochondrial inhibitor | |
| Sequencing | QRT-PCRa | QRT-PCRa | |
| miR156g | 2.75 | 3.9b | 2.6 |
| miR157d | 4.14 | 3.2 | 2.5 |
| miR158a | 2.8 | 3.2 | 2.0 |
| miR159a | 1.89 | 3.8 | 2.0 |
| miR172a, b | 5.27 | 2.8 | 1.1 |
| miR391 | 2.62 | 4.0 | 2.0 |
| miR775 | 2.55 | 1.9 | 2.1 |
| tasiR289 (Tas1a,b,c) | 4.54 | 13 | 1.9 |
Average of at least three biological replications performed in triplicate.
Relative change is the ratio of hypoxia-treated sample to the control.
Inhibition of mitochondrial respiration induces the expression of hypoxia responsive miRNAs and tasiRNA
Hypoxia is associated with a reduction in mitochondrial respiration (for a recent review see Igamberdiev and Hill, 2009). Inhibitors of mitochondrial respiration pathways were used to determine whether the alterations in mitochondrial respiration that resulted, also cause induction to hypoxia-induced smRNAs. Antimycin A (AA) inhibits complex III of the cytochrome pathway (Ikuma and Bonner, 1967), while salicylhydroxamic acid (SHAM) inhibits the alternative oxidase (AOX) pathway (Schonbaum et al., 1971). In combination, these inhibitors induced ADH expression in roots by 10-fold, similar to changes previously observed by Bond et al. (2009). Of the set of seven miRNAs and one tasiRNA verified by SL-PCR to be induced by hypoxia, only one, miR172, was not induced by the mitochondrial inhibitors, suggesting that mitochondrial respiration may be a major factor in smRNA regulation under hypoxia (Table 4).
miRNA and tasiRNA target expression analysis
Genes targeted by miRNA and tasiRNAs are thought to be regulated in plants mainly via endonucleolytic cleavage of mRNAs due to their near-perfect complementarity to their targets, although recent studies indicate the existence of widespread translational inhibition (Brodersen et al., 2008). A large proportion of miRNAs target transcripts that encode transcription factors required for growth, development, and stress responses. The targets (either experimentally determined or predicted) of 42 of the verified hypoxia induced smRNAs were assayed by QRT-PCR (Table 5). The high molecular weight (HMW) fraction of RNA, taken from the same plants used for miRNA and tasiRNA QRT-PCR verification, was used. Two biological replicates of hypoxia-stressed and unstressed samples were tested.
Table 5.
QRT-PCR relative expression of experimentally determined or predicted gene targets of selected smRNAs
| smRNA | Target genes | Relative change by QRT-PCRa | Target gene details |
| miR156g | AT1G27360 | 1.27 | Squamosa promoter-binding-like 11 |
| & | AT1G27370 | 0.47 | Squamosa promoter-binding-like 10 |
| miR157d | AT1G53160 | 0.41 | Squamosa promoter-binding-like 4 |
| AT1G69170 | 1.29 | Squamosa promoter-binding-like 6 | |
| AT2G42200 | 0.62 | Squamosa promoter-binding-like 9 | |
| AT3G15270 | 1.64 | Squamosa promoter-binding-like 5 | |
| AT3G57920 | 0.31 | Squamosa promoter-binding-like 15 | |
| AT5G43270 | 0.55 | Squamosa promoter-binding-like 2 | |
| AT5G50570 | 1.16 | Squamosa promoter-binding-like 13A | |
| AT5G50670 | AS ABOVE* | Squamosa promoter-binding like 13B | |
| miR157d | AT2G33810 | 1.85 | Squamosa promoter-binding-like 3 |
| miR158a | AT1G64100 | 3.01 | Pentatricopeptide repeat-containing |
| AT3G03580 | 1.2 | Pentatricopeptide repeat-containing | |
| miR159a | AT2G26950 | 1.99 | Myb domain 104 |
| AT2G26960 | 4.57 | Myb domain 81 | |
| AT2G32460 | 2.56 | Myb domain 101 | |
| AT2G34010 | 0.24 | Unknown | |
| AT3G11440 | 1.72 | Myb domain 65 | |
| AT3G60460 | 4.68 | Duo Pollen 1 (R2R3 Myb domain) | |
| AT4G37770 | 0.85 | 1-Amino-cyclopropane-1-carboxylate synthase 8 | |
| AT5G06100 | 1.31 | Myb domain 33 | |
| AT5G18100 | 1.99 | Copper/Zinc superoxide dismutase 3 | |
| AT5G55020 | ND** | Myb domain 120 | |
| miR172a,b | AT2G28550 | 0.46 | Target of Eat1 1 |
| AT2G39250 | 1.85 | Schnarchzapfen | |
| AT3G54990 | 1.01 | Schlafmutze | |
| AT4G36920 | 1.07 | Apetala 2 | |
| AT5G60120 | 0.32 | Target of Eat1 2 | |
| AT5G67180 | 0.57 | Target of Eat1 3 | |
| miR391a | At1g50990 | 6.49 | Protein kinase |
| miR775 | AT1G53290 | 0.69 | Galactosyltransferase family |
| tasiR289 | At1g15940 | ND** | Unknown |
| (TAS1) | At1g51670 | 0.23 | Unknown, contains domain cysteine proteinases |
| At1g68840 | 2.57 | Regulator ATPase of the vacuolar membrane (RAP2.8) | |
| At2g42090 | 6.15 | Actin 9 | |
| At3g27230 | 0.31 | Unknown | |
| At3g61320 | 0.67 | Chloroplast precursor | |
| At4g05360 | 4.65 | Zinc knuckle (CCHC-type) family | |
| At4g29760 | ND** | Unknown, contains domain cysteine proteinases | |
| At4g29770 | 3.91 | Unknown | |
| At5g18040 | 2.44 | Unknown, contains domain cysteine proteinases | |
| At5g18065 | 5.13 | Unknown, contains domain cysteine proteinases | |
| At5g42670 | 1.09 | Agenet domain-containing | |
| At5g52070 | 1.25 | Agenet domain-containing |
Targets in bold have been verified experimentally.
Relative change is the ratio of hypoxia-treated sample to the control. The value is an average of at least two biological replications performed in triplicate. *Primers amplified both these genes. **Not detected (ND).
Many of the potential target genes assayed were transcription factors of the Squamosa Promoter Binding-like (SPL), MYB, and AP2-EREBP classes. Twelve of the targets had more than a 33% decrease in expression, indicating that they may be actively cleaved by smRNAs induced by hypoxia (Table 5). Overall, there was no obvious pattern in the expression of target genes under hypoxia stress, as the magnitude of the change varied within predicted targets for specific smRNAs. For example, miR172a, b had six potential targets (five experimentally determined and one predicted); three showed decreased expression, one had increased expression, and two experienced almost no change (Table 5). However, the significant number of target genes down-regulated suggests that miRNA and tasiRNAs do play a role in regulating gene expression under hypoxia stress. The genes potentially regulated by hypoxia stress-responsive smRNAs, encode five SPL proteins, three AP2-EREBP proteins, and four proteins of unknown function.
Discussion
High throughput sequencing of smRNAs from roots subjected to hypoxia revealed a large and complex population of smRNAs. Apart from known miRNA and tasiRNA our smRNA datasets were also analysed for natsiRNAs that are implicated in gene regulation during stress (Borsani et al., 2005). Many putative natsiRNA were identified (see Supplementary Table S6 at JXB online), however, they usually contained very few reads and were not obviously hypoxia regulated.
Analysis of publically available information on miRNA frequency in leaf tissue and whole seedlings shows that the majority of the miRNAs identified in this study have tissue preferential expression (see Supplementary Table S2 at JXB online). Therefore, it is likely that miRNAs play an important role in root-specific processes. Hypoxia treatment on roots resulted in changes to 25 unique miRNAs (19 families) and 10 unique tasiRNAs (three families; Table 2), the majority of these increased under hypoxia, indicating that miRNAs are mainly involved in down-regulating gene expression under hypoxia. Of the hypoxia-regulated miRNAs, only three families (miR166, miR169, and miR172) have previously been reported to be responsive to another abiotic stress in Arabidopsis (cold stress) (Zhou et al., 2008). Of the five miRNAs originally reported by Sunkar and Zhu (2004), as either cold, dehydration, NaCl, or ABA responsive (miR393, miR397b, miR402, miR319c, and miR389a.1), none were sequenced in either root small RNA sample. Heterologous microarray analysis on low-oxygen-stressed maize root cells identified 15 submergence-responsive miRNA families (nine maize families and six families from three other plant species), four of which were common with this study (miR159, miR166, miR167, and miR168). Of these, two families have different predicted target genes from the Arabidopsis miRNAs, indicating that smRNAs regulated by hypoxia may be species specific. Until now, tasiRNAs have not been found to be responsive to abiotic stresses, although few studies have tracked their abundance. The uniqueness of these results may reflect the lack of other root-specific small RNA analyses, or indicate that many of these miRNAs may target hypoxia-specific processes. The latter is supported by the replication of some of the hypoxia responses by chemical inhibition of mitochondrial respiration (Table 4).
Although there was little overlap with other abiotic stresses in Arabidopsis, there is a surprising degree of overlap between hypoxia-responsive miRNAs and virus-induced miRNAs. Arabidopsis infected with tobacco mosaic virus-Cg (TMV-Cg) was found to induce 20 miRNAs families of which 15 are also hypoxia-responsive (Tagami et al., 2007); seven of these hypoxia-responsive miRNA families were also induced in tobacco infected with TMV (Bazzini et al., 2007). However, infection with TMV-Cg did not significantly affect the percentage of tasiRNAs (Tagami et al., 2007). These results indicate that either specific cross-talk exists between miRNA pathways for hypoxia and virus infection, or that the two stresses affect similar cellular processes such as energy production.
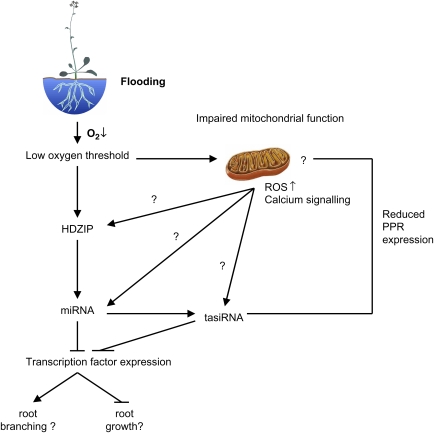
Analysis of hypoxia-responsive miRNAs with promoter sequences that could be unambiguously determined (12 miRNAs in total), revealed an over-representation of two HDZip I motifs (ATHB1 and ATHB-5). It is known that in order for HDZip I proteins to bind DNA, homo- or heterodimer formation with proteins of the same class is required, and that they can activate or repress transcription (Singh, 1998). Many HDZip I genes are regulated by environmental factors such as drought, extreme temperatures, osmotic stresses, and illumination conditions and are active in leaf light perception signalling, cotyledon and leaf development, and ABA-related and abiotic stress responses (Henriksson et al., 2005). Of the 16 HDZip I genes in Arabidopsis that have extensive publicly available gene expression information, 10 were found to be down-regulated and two up-regulated, early in hypoxia stress treatments (Zimmermann et al., 2004). Therefore, it is possible that members of this group may directly regulate the expression of a subset of these hypoxia-responsive miRNAs, and indicate that changes in miRNA frequencies occur after early hypoxia-responsive transcription factors (Fig. 2).
Fig. 2.
A hypothetical model of the regulation of hypoxia-responsive miRNAs and tasiRNAs and their potential roles on Arabidopsis root responses. Flooding reduces oxygen levels until it reaches a critical threshold that is sensed by an unknown mechanism either in the nucleus or mitochondria. Once sensed, a HD-ZIP is induced that regulates a suite of miRNAs. These miRNA are potentially involved in halting root growth and developmental responses in the root that occur after hypoxia such as root branching, as well as regulating tasiRNA expression that regulates PPRs involved in mitochondrial function. Impairing mitochondrial function can also regulate a subset of hypoxia-responsive miRNA and tasiRNAs possibly as a result of increased ROS levels and calcium signalling leading to retrograde regulation. Promotive effects are shown by ‘→’; repressive effects by ‘–’.
QRT-PCR verification of the putative targets for eight hypoxia-induced smRNAs confirmed a number of genes had the expected change in steady-state levels of mRNA if targeted for endonucleolytic cleavage (12 out of 42 targets). However, it has recently been shown that, in addition to RNA cleavage, miRNAs in plants have widespread translational repressor capability (Brodersen et al., 2008), so many of these targets may in fact be regulated through this translational pathway but not be detected using QRT-PCR and will require further validation. Hypoxia is known differentially to affect translational rates of specific mRNA, but few (3 out of 44) of the potential targets of hypoxia-induced smRNAs are known to possess altered translation rates based on polysome analysis (Branco-Price et al., 2008). Analysis of the spatial expression of miRNAs has shown that they often possess specific tissue patterns (Valoczi et al., 2006). For example, the hypoxia-responsive miR159a is strongly expressed in root tips and lateral roots. Putative miR159a target genes expressed throughout the root may not show a change in steady-state mRNA levels when whole roots are assayed.
The early events in a plant's response to hypoxia stress are characterized by a cascade of signalling events involving the differential expression of a wide range of transcription factor including the AP2-EREBP, MADS, MYB, Homeobox, WRKY, bZIP, C2H2 zinc-finger, bHLH, and NAC families (Klok et al., 2002; Liu et al., 2005; Christianson et al., 2009). As is characteristic of miRNAs, the majority of the putative targets for the hypoxia-responsive miRNA families are transcription factors mainly from the MYB, NAC, Homeobox, SPL, ARF, AP2, MADS, and CCAAT-HAP2 families. miRNAs may therefore play a role in regulating secondary signalling events, especially the SPL and AP2 genes that show inverse regulation to miR156/157 and miR172 expression (Tables 2, 4, 5). Unfortunately, the bulk of these transcription factors are poorly characterized with regard to their contribution to root development. However, SPL and AP2 genes have important roles in plant growth and floral development. Loss-of-function mutants of SPL9 and SPL15 have demonstrated that they are active in controlling juvenile-to-adult phase transition, vegetative plastochron and branching (Schwarz et al., 2008). Analysis of the AP2 proteins TOE1, and TOE2, indicates that they are involved in the transitions between developmental states of the shoot apical meristem as the toe1-1:toe2-1 double mutant flowers much earlier than the wild type, whereas overexpression of TOE1 leads to delayed flowering (Aukerman and Sakai, 2003). Initially, hypoxia stress results in the cessation of growth (Van Dongen et al., 2009) with longer term exposure causing cell death, especially at the metabolically active root tip, and aerenchyma formation in some species. Upon restoration of normal oxygen levels in Arabidopsis, adventitious roots form (Ellis et al., 1999). Overexpression of miR166 in Medicago truncatula results in decreased lateral root formation and changes in root vascular bundles, and this highly expressed family was one of the few to decrease under hypoxia stress (Boualem et al., 2008). miRNAs such as miR156/157, miR166, and miR172 may be involved in controlling or stopping root growth, and redirecting root development when normoxia resumes. These miRNAs may therefore link sub-lethal exposure to hypoxia with the developmental changes that occur during and after the stress (Fig. 2).
tasiRNAs are a specialized class of siRNAs that are generated by miRNA processing of a TAS gene transcript, resulting in the production of 21 nt RNAs that are phased with respect to the miRNA cleavage site. Four families of TAS genes have been identified in Arabidopsis, with TAS1 and TAS2 transcripts recognized by miR173, TAS3 recognized by miR390, and TAS4 targeted by miR828 (Allen et al., 2005; Rajagopalan et al., 2006). TAS1, TAS2, and TAS3 tasiRNAs all showed increased expression in the hypoxia-treated samples, whereas no TAS4 RNAs were identified (Table 3). These changes in tasiRNA levels are reflected in the changes to tasiRNA targeting miRNAs; both miR173 and miR390 showed increased expression (1.5-fold and 1.6-fold respectively), whereas miR828 was not sequenced (Table 2). The majority of the genes targeted by TAS1 and TAS2 (and as well as miR158) are of the pentatricopeptide repeat (PPR) family. PPRs are a large family containing 442 genes in Arabidopsis that are putative RNA-binding proteins, and can divided into two subfamilies; P and PLS (Lurin et al., 2004). Most of the PPRs targeted by hypoxia-responsive smRNAs are from the P subfamily that is predicted to localize to the mitochondria (Lurin et al., 2004). Relatively few of these proteins have described functions but they have been implicated in different stages of plastid gene expression, including transcription, splicing, processing, editing, translation, and stability, as well as mitochondrial induced fertility restoration (Andres et al., 2007). It is likely that some of the observed changes in tasiRNAs are the result of decreased mitochondrial function during hypoxia as TAS1 is induced by chemical inhibition of the cytochrome and alternative oxidase respiration pathways in the mitochondria (Fig. 2). The down-regulation of these PPR genes may either aid mitochondrial protection during hypoxia, or simply reflect a decreased requirement for these gene transcripts.
TAS3 targets members of the Auxin Response Factor (ARF) family that transmits auxin signals during growth and development. Specifically, TAS3 is thought to target ARF3 and 4 that are implicated in adult-juvenile transition functions, similar to the putative SPL and AP2 targets (Hunter et al., 2006). Recent evidence indicates that TAS3 could have a specific role in regulating root architecture (Maizel et al., 2008).
Oxygen-deprivation triggers an immediate and reversible rise in cytosolic Ca2+ that is co-localized around the mitochondria (Subbaiah and Sachs, 2003). It is known that Ca2+ signalling occurs before gene expression changes, and is required for the induction of important hypoxia-responsive genes such as ADH (Subbaiah et al., 1994a; b) and the transduction of ethylene signalling in the formation of aerenchyma (He et al., 1996). Anoxia causes increases in hydrogen peroxide formation in Arabidopsis seedlings (Baxter-Burrell et al., 2002) and is associated with RHO-LIKE SMALL G PROTEIN OF PLANTS (ROP) that regulate intracellular Ca2+ gradients. Chemical inhibition of the cytochrome pathway also results in increased mitochondrial reactive oxygen species (Maxwell et al., 1999). One hypothesis for oxygen sensing that would be shared by both plant and animals, is that changes in reactive oxygen species levels within and around the mitochondria stimulate Ca2+ signalling, and that this influences nuclear gene expression by retrograde communication (Semenza, 1999). Our data does not resolve the nature of sensing, but indicates that smRNAs play a role in gene regulation and possible developmental responses to hypoxia, and that a major signal for these responses is likely to be dependent on mitochondrial function and not on oxygen per se.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Relative expression of ADH during hypoxia stress.
Supplementary Table S1. Summary of the origin of small RNA sequencing data.
Supplementary Table S2. Comparison of read numbers of miRNAs from different tissues.
Supplementary Table S3. Stem-loop RT and QRT-PCR primer sequences.
Supplementary Table S4. Primers for U6 loading control for QRT-PCR analysis of mature smRNAs.
Supplementary Table S5. Target genes and primers used for QRT-PCR analysis.
Supplementary Table S6. Potential natsiRNA present in control and hypoxia stressed roots
Supplementary Material
Acknowledgments
Dov Moldovan was supported by a CSIRO Office of the Chief Executive postgraduate scholarship. BJP and DM acknowledge the support of the Australian Research Council Centre of Excellence in Plant Energy Biology (CE0561495). We would like to thank Tony Millar and Ian Small for fruitful discussions. We are also grateful to Chris Helliwell, Narayana Upadhyaya, and anonymous reviewers for their critical reading of the manuscript.
Glossary
Abbreviations
- smRNA
small RNA
- miRNA
microRNA
- tasiRNA
trans-acting small interfering RNA
- QRT-PCR
quantitative reverse transcriptase PCR
- SL-PCR
stem-loop quantitative PCR
References
- Agarwal S, Grover A. Molecular biology, biotechnology and genomics of flooding-associated low O2 stress response in plants. Critical Reviews in Plant Sciences. 2006;25:1–21. [Google Scholar]
- Allen E, Xie ZX, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Andres C, Lurin C, Small ID. The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiologia Plantarum. 2007;129:14–22. [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. The Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R. Sensing and signalling in response to oxygen deprivation in plants and other organisms. Annals of Botany. 2005;96:507–518. doi: 10.1093/aob/mci206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. Flooding stress: acclimations and genetic diversity. Annual Review of Plant Biology. 2008;59:313–339. doi: 10.1146/annurev.arplant.59.032607.092752. [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang ZB, Springer PS, Bailey-Serres J. RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science. 2002;296:2026–2028. doi: 10.1126/science.1071505. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Hopp HE, Beachy RN, Asurmendi S. Infection and co-accumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proceedings of the National Academy of Sciences, USA. 2007;104:12157–12162. doi: 10.1073/pnas.0705114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DM, Wilson IW, Dennis ES, Pogson BJ, Finnegan EJ. VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. The Plant Journal. 2009;59:576–587. doi: 10.1111/j.1365-313X.2009.03891.x. [DOI] [PubMed] [Google Scholar]
- Bonnet E, Van de Peer Y, Rouz P. The small RNA world of plants. New Phytologist. 2006;171:451–468. doi: 10.1111/j.1469-8137.2006.01806.x. [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F. MicroRNA166 controls root and nodule development in Medicago truncatula. The Plant Journal. 2008;54:876–887. doi: 10.1111/j.1365-313X.2008.03448.x. [DOI] [PubMed] [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJH, Larive CK, Bailey-Serres J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. The Plant Journal. 2008;56:743–755. doi: 10.1111/j.1365-313X.2008.03642.x. [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. Genome-wide analysis of transcript abundance and translation in arabidopsis seedlings subjected to oxygen deprivation. Annals of Botany. 2005;96:647–660. doi: 10.1093/aob/mci217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Chang WC, Lee TY, Huang HD, Huang HY, Pan RL. PlantPAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics. 2008;9:561. doi: 10.1186/1471-2164-9-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiology. 2009;149:1724–1738. doi: 10.1104/pp.108.131912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Zhang L, Wang H, Liu Z, Zhang Z, Zheng Y. Differential expression of miRNAs in response to salt stress in maize roots. Annals of Botany. 2009;103:29–38. doi: 10.1093/aob/mcn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiology. 1999;119:57–64. doi: 10.1104/pp.119.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA Genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Xu KN, Ronald PC, Bailey-Serres J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. The Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Reviews Genetics. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Research. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: the Arabidopsis Small RNA Project database. Nucleic Acids Research. 2005;33:D637–640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- He CJ, Morgan PW, Drew MC. Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiology. 1996;112:463–472. doi: 10.1104/pp.112.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E, Olsson ASB, Johannesson H, Johansson H, Hanson J, Engstrom P, Soderman E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiology. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics. 1998;149:479–490. doi: 10.1093/genetics/149.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig RS. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev AU, Hill RD. Plant mitochondrial function during anaerobiosis. Annals of Botany. 2009;103:259–268. doi: 10.1093/aob/mcn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuma H, Bonner WD. Properties of higher plant mitochondria. 3. Effects of respiratory inhibitors. Plant Physiology. 1967;42:1535–1544. doi: 10.1104/pp.42.11.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences, USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. The Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, Whelan J. Differential response of grey poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiology. 2009;149:461–473. doi: 10.1104/pp.108.125989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P. Transcript profiling of the anoxic rice coleoptile. Plant Physiology. 2007;144:218–231. doi: 10.1104/pp.106.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RQ, Li YR, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Liu FL, VanToai T, Moy LP, Bock G, Linford LD, Quackenbush J. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiology. 2005;137:1115–1129. doi: 10.1104/pp.104.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiology. 2005;137:1130–1138. doi: 10.1104/pp.104.057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Lu SF, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. The Plant Journal. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Marin E, Herz A, Crespi M. Activation of the TAS3-derived tasiRNA pathway in the root system of Arabidopsis. Comparative Biochemistry and Physiology Part A, Molecular and Integrative Physiology. 2008;150:S197–S197. [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proceedings of the National Academy of Sciences, USA. 1999;96:8271–8276. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG. MicroRNA promoter element discovery in Arabidopsis. RNA. 2006;12:1612–1619. doi: 10.1261/rna.130506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nobuta K, Lu C, Shrivastava R, et al. Distinct size distribution of endogenous siRNAs in maize: evidence from deep sequencing in the mop1-1 mutant. Proceedings of the National Academy of Sciences, USA. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Pan X, Wilson IW, Li F, Liu M, Teng W, Zhang B. High throughput sequencing technology reveals that the taxoid elicitor methyl jasmonate regulates microRNA expression in Chinese yew (Taxus chinensis) Gene. 2009;436:37–44. doi: 10.1016/j.gene.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes and Development. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Research. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion. 2007;7:177–194. doi: 10.1016/j.mito.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Schonbaum GR, Bonner WD, Storey BT, Bahr JT. Specific inhibition of cyanide-insensitive respiratory pathway in plant mitochondria by hydroxamic acids. Plant Physiology. 1971;47:124–128. doi: 10.1104/pp.47.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Molecular Biology. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Perspectives on oxygen sensing. Cell. 1999;98:281–284. doi: 10.1016/s0092-8674(00)81957-1. [DOI] [PubMed] [Google Scholar]
- Singh KB. Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiology. 1998;118:1111–1120. doi: 10.1104/pp.118.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Bush DS, Sachs MM. Elevation of cytosolic calcium precedes anoxic gene-expression in Maize suspension-cultured cells. The Plant Cell. 1994a;6:1747–1762. doi: 10.1105/tpc.6.12.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Sachs MM. Molecular and cellular adaptations of maize to flooding stress. Annals of Botany. 2003;91:119–127. doi: 10.1093/aob/mcf210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Zhang JK, Sachs MM. Involvement of intracellular calcium in anaerobic gene-expression and survival of maize seedlings. Plant Physiology. 1994b;105:369–376. doi: 10.1104/pp.105.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in Plant Science. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. The Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MPS, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. 2008;9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami Y, Inaba N, Kurihara Y, Kutsuna N, Watanabe Y. Small RNA expression profile of Arabidopsis thaliana infected with tobacco mosaic virus. Plant and Cell Physiology. 2007;48:S36–S36. doi: 10.1093/dnares/dsm022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ’t Hoen PAC, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RHAM, de Menezes RX, Boer JM, van Ommen G-JB, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Research. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi A, Varallyay E, Kauppinen S, Burgyan J, Havelda Z. Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. The Plant Journal. 2006;47:140–151. doi: 10.1111/j.1365-313X.2006.02766.x. [DOI] [PubMed] [Google Scholar]
- Van Dongen JT, Frohlich A, Ramirez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Annals of Botany. 2009;103:269–280. doi: 10.1093/aob/mcn126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu RM, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F. Arabidopsis endogenous small RNAs: highways and byways. Trends in Plant Science. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wilson IW, Kennedy GC, Peacock JW, Dennis ES. Microarray analysis reveals vegetative molecular phenotypes of arabidopsis flowering-time mutants. Plant and Cell Physiology. 2005;46:1190–1201. doi: 10.1093/pcp/pci128. [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wei L, Zou X, Tao Y, Liu Z, Zheng Y. Submergence-responsive microRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Annals of Botany. 2008;102:509–519. doi: 10.1093/aob/mcn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochimica et Biophysica Acta. 2008;1779:780–788. doi: 10.1016/j.bbagrm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Zhou XF, Ruan JH, Wang GD, Zhang WX. Characterization and identification of microRNA core promoters in four model species. PLos Computational Biology. 2007;3:412–423. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan LJ, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Research. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.