Abstract
Cotton fibres are hair-like single-cells that elongate to several centimetres long after their initiation from the ovule epidermis at anthesis. The accumulation of malate, along with K+ and sugars, is thought to play an important role in fibre elongation through osmotic regulation and charge balance. However, there is a lack of evidence for or against such an hypothesis. Phosphoenolpyruvate carboxylase (PEPC) is a key enzyme responsible for the synthesis of malate. The potential role of PEPC in cotton fibre elongation is examined here. Developmentally, PEPC activity was higher at the rapid elongation phase than that at the slow elongation stage. Genotypically, PEPC activity correlated positively with the rate of fibre elongation and the final fibre length attained. Importantly, suppression of PEPC activity by LiCl that reduces its phosphorylation status decreased fibre length. To examine the molecular basis underlying PEPC activity, two cDNAs encoding PEPC, GhPEPC1 and 2, were cloned, which represents the major PEPC genes expressed in cotton fibre. RT-PCR analyses revealed that GhPEPC1 and 2 were highly expressed at the rapid elongation phase but weakly at the slow-to-terminal elongation period. In situ hybridization detected mRNA of GhPEPC1 and 2 in 1 d young fibres but not in the ovule epidermis prior to fibre initiation. Collectively, the data indicate that cotton fibre elongation requires high activity of PEPC, probably through the expression of the GhPEPC1 and 2 genes.
Keywords: Cell expansion, cotton, fibre elongation, phosphoenolpyruvate carboxylase
Introduction
Cotton is the most important textile crop worldwide as a result of its long cellulose-enriched mature fibres (Basra and Malik, 1984; Ruan, 2007). The quality and productivity of cotton fibre depend mainly on three processes: fibre initiation, elongation, and secondary cell wall cellulose synthesis, which determines, in turn, the number of fibres present on each ovule, the final fibre length attained, and the fibre strength at maturity (Ruan, 2005; Lee et al., 2007). Cotton fibres are single-celled hairs originating from the epidermis of ovular outer integuments at or just prior to anthesis (Basra and Malik, 1984). After initiation, each cotton fibre can elongate to several centimetres in length in about 20 d, rendering it among the fastest growing and the longest single-cell known in the plant kingdom (Ruan et al., 2004). Owing to their rate and magnitude of elongation and their accessibility for experimentation in vivo, cotton fibre represents an excellent model to study the mechanisms of rapid plant cell expansion at the single-cell level (Ruan and Chourey, 1998; Martin et al., 2001; Pfluger and Zambryski, 2001; Ruan et al., 2001).
Plant cell expansion is controlled by the concerted interaction of cell turgor and cell wall extensibility (Cosgrove, 2005). In cotton fibre, cell turgor is achieved through the influx of water driven by the accumulation of osmotically active solutes, mainly, hexoses, K+, and malate, which together accounts for about 80% of fibre sap osmolality (Dhindsa et al., 1975; Ruan, 2005). In this context, substantial progress has been made in understanding the control of sugar and K+ transport to, and utilization within, cotton fibre (Ruan et al., 1997, 2001, 2003). By contrast, however, little is known about the mechanisms regulating malate synthesis in these cells. Thus, this study aims to fill in this knowledge gap by exploring the role of phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) in fibre elongation, which is a key enzyme responsible for the synthesis of malate (Chollet et al., 1996; Johnson et al., 1996; Izui et al., 2004).
PEPC exists in bacteria, cyanobacteria, green algae, and in higher plants (Izui et al., 2004). It catalyses the irreversible β-carboxylation of phosphoenolpyruvate (PEP) in the presence of HCO3− and Mg2+ to yield oxaloacetate (OAA) and inorganic phosphate (Chollet et al., 1996). OAA is then converted to malate catalysed by malate dehydrogenase (Buchanan et al., 2000). Plant PEPC activities are subject to allosteric control by a variety of positive (e.g. glucose 6-P) and negative (e.g. L-malate, Asp) metabolite effectors (Chollet et al., 1996; Izui et al., 2004) and are up-regulated by the phosphorylation of conserved serine residues at the N terminus (Vidal and Chollet, 1997) by PEPC kinases (Giglioli-Guivarc'h et al., 1996; Coursol et al., 2000). A recent observation that LiCl treatment reduces PEPC phosphorylation status and activity by inhibiting PEPC kinase activity (Monreal et al., 2007) offers new opportunities to probe the roles of PEPC in plant development.
PEPCs appear to be involved in diverse physiological and cellular processes (Lepiniec et al., 1994; Gehrig et al., 2001). The enzyme is required for C4 and Crassulacean acid metabolism (CAM) photosynthesis by catalysing the initial carboxylation step in a C4 acid cycle that functions as a CO2 concentrating mechanism (Rao et al., 2002). In non-photosynthetic tissues, PEPCs function in the refixation of HCO3− released from dark respiration to produce C4-dicarboxylic acids, providing carbon skeletons for the synthesis of compounds that serve in processes such as C/N partitioning and nitrogen fixation in legumes (Lepiniec et al., 1994; Leegood, 2002; Rao et al., 2002). PEPC activity is also implicated in plant cell expansion through an osmotic effect from the resultant malate. A typical example is the involvement of PEPC in guard cell movement during stomatal opening (Tarczynski and Outlaw, 1990). However, direct evidence is lacking regarding the role of PEPC in plant cell expansion, the focus of this study.
Cotton fibres are capable of fixing HCO3− into malate, the major anion in these single-cells (Dhindsa et al., 1975). This, together with their extraordinary rate of cell elongation (Ruan, 2007), provides a unique opportunity to examine the potential role of PEPC in cell expansion. By using a combination of biochemical, pharmacological, and molecular approaches, data have been obtained to show that (i) PEPC activity correlated with fibre elongation both developmentally and genotypically; (ii) inhibition of PEPC activity pharmacologically reduced fibre elongation; and (iii) high activity of PEPC in fibres appeared to be achieved through expression of GhPEPC1 and 2 genes.
Materials and methods
Plant materials
Cotton (Gossypium hirsutum cv. Coker 315) plants were grown under greenhouse conditions. Flowers were tagged at anthesis. Cotton bolls were collected at specified time points.
For ovule culture, the bolls were collected at 2 DAA, surface-sterilized with 70% (v/v) ethanol for 30–60 s followed with treatment of 6% (v/v) sodium hypochlorite (NaOCl) for 20 min. Thereafter, the bolls were washed with sterile water to remove residual NaOCl. The seeds were cultured on BT medium (Beasley and Ting, 1973) in the dark at 30 °C without shaking. For RNA extraction and enzyme assay, samples were frozen in liquid N2 and stored at –70 °C until analysis. Fresh samples were fixed for in situ hybridization.
For treatment with LiCl and other salts, BT media containing salt were prepared and autoclaved. For treatment with a high concentration of salts, cotton seeds at 2 d after anthesis (DAA) were cultured for 3 d in BT medium containing 100 mM of LiCl, NaCl, or KCl. Alternatively, the 2 d seeds were cultured in BT medium containing 10 mM LiCl for 12 d. Sorbitol was chosen as an osmotic control in those experiments.
Semi-quantitative RT-PCR analyses
Total RNA was extracted according to Ruan et al. (1997). One microgram of RNA was treated by RQ1 DNAase (Promega), and the DNA-free RNA was used for cDNA synthesis using dT18 oligonucleotides with MMLV Reverse Transcriptase (Toyoba) following the manufacturer's instructions.
The following two primers were used to amplify a specific fragment of PEPC1: P1SpeSF, 5′-TAAAACTTGCGGATTG-3′ and P1SpeSR, 5′-ATACCAGACTGACAAC-3′. The primers of P2SpeSF (5′-AAATTACAGCGTGAAGTTGC-3′) and P2SpeSR (5′-CCTATAAGAAAGTTTTTGCCG-3′) were used to amplify PEPC2.
For internal control, a primer set of GhActinF (5′-CTACGGTAACATTGTGCTCA-3′) and GhActinR 5′-CACCATTAAGATGATGGGTC-3′) was used to amplify GhActin, which shows a relatively constant expression during fibre elongation (Sun et al., 2005). The cDNAs were amplified by using the following conditions: 25 cycles of 30 s at 94 °C, 30 s at 55 °C, and 30 s at 72 °C. A final extension step of 5 min at 72 °C was used to terminate the reaction.
In situ hybridization
In situ hybridization was carried out using our established protocol (Jin et al., 2009). To amplify gene-specific fragments of PEPC1 and PEPC2, the following two primer sets were used, respectively: P1PF: 5′-AATGAATTCTGAATACTTCCGCCTA-3′ and P1PR: 5′-AATAAGCTTCATACTCGCTTGTAGGG-3′; P2PF: 5′-GAATTCGGATCTTTGCCTGGACACAGA-3′ and P2PR: 5′-TCTAGAGATATGTGGCCGCAACT-3′. The PEPC1 and 2 fragments were cloned into pBluescript. The plasmids harbouring the PEPC1 cDNA were linearized with HindIII or EcoRI for the generation of sense and antisense probes, respectively, whereas plasmid containing PEPC2 was digested with EcoRI for making a sense probe and XbaI for the antisense probe. Sense and antisense RNA transcripts were synthesized by T3 and T7 RNA polymerase with digoxigenin-UTP (Roche Diagnostics) as the label. The linerlized transcripts were subject to alkaline degradation to obtain a probe of approximately 200 bp (Cox and Goldberg, 1988).
Assay of PEPC activity
Frozen samples were ground in liquid nitrogen with extraction buffer containing 200 mM HEPES–NaOH (pH 7.0), 10 mM MgCl2, 5 mM DTT, and 2% PVP-40 (Hatch and Oliver, 1978; Magnin et al., 1997). The crude extracts were then transferred to prechilled Eppendorf tubes and centrifuged at 12 000 g for 5 min. The supernatant was immediately used for the PEPCase assay according to Jiao and Chollet (1988). In short, the samples were incubated for 30 min at 25 °C in 1 ml of assay buffer (100 mM HEPES–NaOH pH 8.0, 10 mM MgCl2, 5 mM DTT, 2 mM NADH, 10 mM NaHCO3, 2 mM PEP-K, and 2 mM malate dehydrogenase). The reaction was initiated by adding PEP-K a final concentration of 2 mM and terminated by incubation at 95 °C for 10 min and the reaction absorbance was measured at 340 nm. Controls containing boiled extracts were used as blanks. The protein content was measured according to Bradford (1976).
Malate extraction and measurements
Samples in ∼50 mg each were extracted for 1 h at 80 °C in 1.5 ml of 80% ethanol and 20% water containing 100 mM HEPES–KOH (pH 7.1) and 20 mM MgCl2. After cooling to room temperature, the extracts were centrifuged at 12 000 g for 5 min. The supernatant was recovered, mixed with 150 ml of charcoal suspension (100 mg ml−1) and centrifuged at 12 000 g for 5 min. The supernatant was stored at –20 °C until use (Famiani et al., 2000).
Malate content was measured using the enzyme-coupled method (Lowry and Passonneau, 1972; Famiani et al., 2000). The assay mixture contained, in 1 ml: 50 mM 2-amino-2-methylpropanol and 40 mM glutamate (pH 9.9), and 1 mM NAD+. The reaction was initiated by adding 10 U of glutamate oxalacetate transaminase and 0.7 U of malate dehydrogenase to the assay mixture. The absorbance at 340 nm was read after 1 h. A standard curve spanning 0–0.15 μM malate prepared in the assay medium was used to calculate the amounts of malate in samples.
Measurement of fibre length
Fibre length was measured according to Schubert et al. (1973). For consistency, the measurement was done from the chalazal end of the seed in all cases.
Results
PEPC activity correlates with cotton fibre elongation developmentally and genotypically
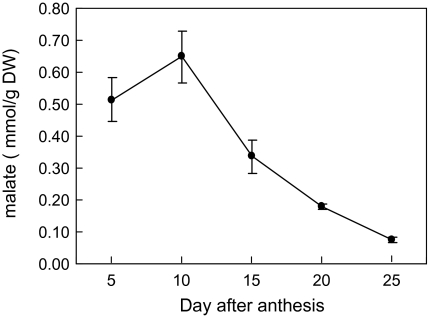
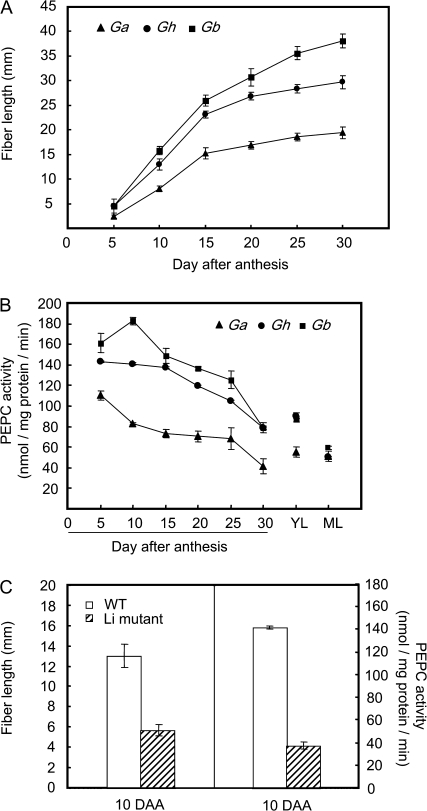
Measurement of malate in cotton fibre revealed its high content at the rapid phase of elongation at 5 d and 10 d after anthesis (DAA) and an evidently reduced content at 15 DAA onwards (Fig. 1), when fibre elongation significantly slowed (Ruan, 2007). These findings are consistent with previous observations from in vitro-cultured cotton fibre (Dhindsa et al., 1975). To determine the underlying biochemical basis of malate accumulation in rapidly elongating fibres, PEPC activity was assayed in fibres over the entire elongation period across three genotypes, Gb, Gh, and Ga that differ in the rate and extent of fibre elongation (Ruan et al., 2004). Figure 2A shows that fibre elongation displayed a similar temporal trend among the three genotypes, i.e. a rapid elongation phase from 5–15 DAA followed by a much slower elongation period from 15 DAA onwards. However, within a given time period, Gb elongated much faster than Gh, whereas Ga exhibited the slowest elongation rate (Fig. 2A). Corresponding to this genotypic difference, PEPC activity was also high, intermediate, and low in Gb, Gh, and Ga, respectively (Fig. 2B). Moreover, the fibre PEPC activity was significantly higher in the fast elongation phase (5–10 DAA) than that in the slow phase of 15 DAA onwards across the genotypes (Fig. 2B). By 25–30 DAA when elongation terminates (Fig. 2A; Basra and Malik, 1984), the PEPC activity dropped sharply to that observed in leaves (Fig. 2B).
Fig. 1.
Changes of malate content during cotton fibre elongation in G. hirsutum L. (Gh). Each value is the mean ±SE of three replicates.
Fig. 2.
Correlation between PEPC activity and fibre elongation among Genotypes of G. arboreum (Ga), G. hirsutum (Gh), and G. barbadense (Gb). (A) Kinetics of fibre elongation; (B) PEPC activity, and (C) reduced PEPC activity correlates with reduced fibre length in 10-d fibre of Li1 mutant, as compared with its parental line TM-1. YL, young leaves; ML, mature leaves. Each value is the mean ±SE of three replicates.
To provide further genetic evidence on the role of PEPC in fibre elongation, a monogenic, dominant cotton mutant Ligon lintless (Li1) was chosen for comparison with its parent, TM-1. The Li1 mutant displayed normal fibre initiation but much shorter fibre length compared with TM-1 (Karaca et al., 2002; Zhu et al., 2003). Figure 2C shows that, at the rapid elongation phase of 10 DAA, the PEPC activity in the mutant was only 25% of the wild type, which corresponded to ∼55% reduction of fibre length.
Inhibition of PEPC activity reduces cotton fibre elongation
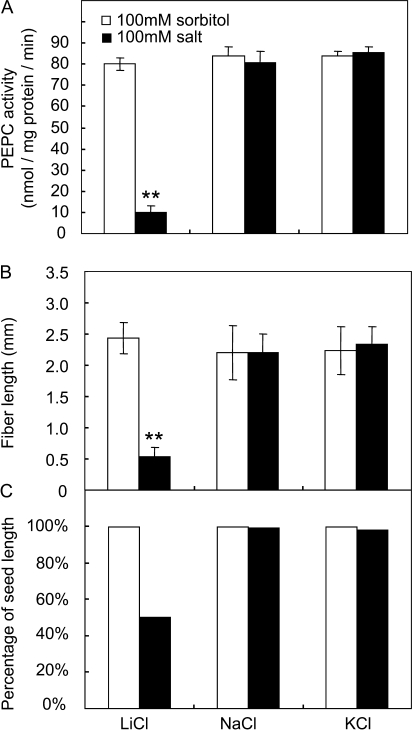
To examine whether high PEPC activity (Fig. 2B) is required for cotton fibre elongation, a potent inhibitor against the activity of this enzyme, LiCl (Monreal et al., 2007), was applied to cultured cotton seed and fibre (Beasley and Ting, 1973; Shi et al., 2006). In this context, the application of 100 mM LiCl reduced fibre PEPC activity by 88% (Fig. 3A), which led to 80% and 50% reduction in fibre length and seed size (Fig. 3B, C), respectively, compared with the control samples treated with sorbitol. The effect appeared to be attributable specifically to LiCl, since treatment with NaCl or KCl at the same concentration elicited no such inhibitory response (Fig. 3).
Fig. 3.
Effect of a high concentration of LiCL on fibre elongation and seed size. (A) PEPC activity in cotton fibre. Note PEPC activity was significantly reduced by 100 LiCl but not by NaCl and KCl at the same concentration. (B) Fibre length is significantly reduced by LiCl but not by NaCl and KCl. (C) Seed size (measured as% of seed length) was also reduced by LiCl. Each value is the mean ±SE of three replicates. An asterisk indicates a significant difference (t test, *P <0.05; **P <0.01).
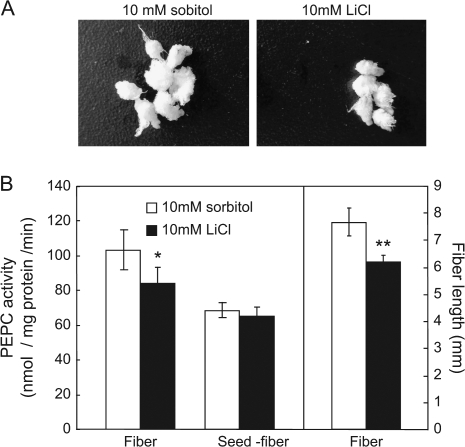
To determine unequivocally the effect of LiCl on fibre elongation without parallel impact on seed development, LiCl at various concentrations was applied to cultured seeds. The analyses revealed that treatment with 10 mM LiCl has no effect on seed size (Fig. 4A) but significantly inhibited fibre PEPC activity and, hence, fibre length (Fig. 4B). It is of note that PEPC activity was not reduced by LiCl in seed with the fibre removed (Fig. 4B), indicating that the observed effect of 10 mM LiCl is fibre-specific.
Fig. 4.
Effect of 10 mM LiCl treatment on fibre elongation. (A) Seed and fibre morphology after in vitro culture with 10 mM LiCl. (B) Fibre length and the PEPC activity in fibre and seeds (with fibre removed) after salt-treatment. Each value is the mean ±SE of three replicates. *P <0.05, **P <0.01.
Cloning of major PEPC cDNAs that are highly expressed in elongating fibres
The strong correlation between PEPC activity and fibre elongation prompted us to explore the molecular basis of PEPC activity in cotton fibre. Extensive database searches identified two major cDNAs, GhPEPC1 and GhPEPC2 (Genbank accession numbers AF008939 and EU032328, respectively) that accounted for ∼75% of GhPEPC ESTs expressed in cotton fibre (http://www.cottondb.org/).
The GhPEPC1 and GhPEPC2 encoded 966 and 971 amino acids, respectively, exhibiting the typical size of plant PEPCs (Izui et al., 2004). Alignment of their deduced amino acid sequences with those from other species (see Supplementary Fig. S1 at JXB online) revealed that GhPEPC1 shared 89.2%, 86%, and 79% sequence identity with that of AtPPC1, maize C3 PEPC, and maize C4 PEPC, respectively. Similarly, GhPEPC2 shared 87% identity with that of AtPPC1 and maize C3 PEPC and 77% to maize C4 PEPC. GhPEPC1 shared 87.8% identity with GhPEPC2.
As highlighted in Supplementary Fig. 1 at JXB online, GhPEPC1 and 2 had the typical features of plant PEPCs. Here, a Ser residue located in the N terminus forming part of the sequence acid base, XXSIDAQLR, is invariably present in all plant PEPCs and is the target for phosphorylation (Chollet et al., 1996; Vidal and Chollet, 1997; Nimmo, 2000). The signature C3 Ala was present in GhPEPC1 and 2, but was absent in Zm PEPC-C4 (Izui et al., 2004). Furthermore, GhPEPC1 and 2 contained four loops and two motifs, which are highly conserved domains in plant PEPCs (Izui et al., 2004). Among them, loops I and II were the PEP and HCO3− binding sites, respectively, whereas loops III and IV were regulatory domains. A conserved Arg in GhPEPC1-379 and GhPEPC2-372 may account for subunit interaction (Chollet et al., 1996; Izui et al., 2004). Finally, five Cys, present in all plant PEPCs, were also present in GhPEPC1 and 2 (Chollet et al., 1996).
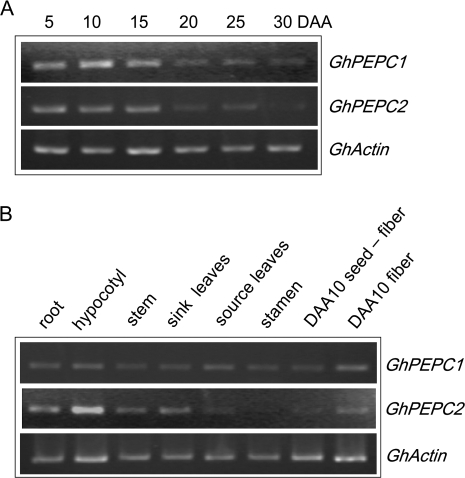
The expression patterns of GhPEPC1 and 2 in fibres and other tissues were examined by using semi-quantitative reverse transcriptase (RT)-PCR with gene-specific primers. The transcript levels of GhPEPC1 and GhPEPC2 were high during the rapid elongation phase of 5–15 DAA but decreased sharply at the slow elongation phase at 15 DAA onwards and became almost undetectable by 30 DAA when fibre elongation has terminated (Fig. 5A; see also Fig. 2A). GhPEPC2 was also highly expressed in elongating hypocotyls and roots as well as in expanding leaves (Fig. 5B). Within cotton seed, both PEPC genes were mainly expressed in elongating fibres with much weaker signals detected from the remaining seed tissue (Fig. 5B), corresponding to high and low PEPC activities in the two tissues, respectively (Fig. 4B).
Fig. 5.
Transcript levels of GhPEPC1 and 2 in cotton fibre during development and in other tissues. (A) Expression of GhPEPC1and 2 during fibre development. Note their transcript abundance was evidently higher at the elongation phase (5–15 DAA) than that at the slow-to-terminal elongation stage (20–30 DAA). (B) Expression of GhPEPC1and 2 in other tissues. Note the high transcript level in fibres as compared to the seed with fibre removed. GhActin was used as an internal control.
GhPEPC1 and GhPEPC2 are expressed in fibres early in elongation but not in non-differentiating ovular epidermis
Cotton fibre elongation starts from 0 DAA, protruding above the ovule epidermis (Ruan and Chourey, 1998). However, genes involving in fibre elongation may be expressed in ovule epidermis prior to 0 DAA (Ruan, 2005; Wu et al., 2006). Thus, further studies were conducted by using in situ hybridization to determine if GhPEPC1 and GhPEPC2 were expressed in fibre cells early in elongation or in ovules prior to fibre elongation.
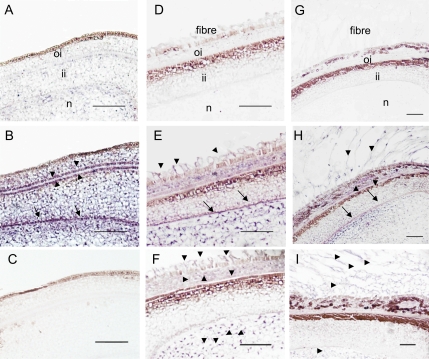
For GhPEPC1 mRNA, the signal was not detected in the ovule epidermis at 2 d before anthesis (–2 DAA; Fig. 6B), compared with its sense control (Fig. 6A). Instead, the GhPEPC1 transcripts were detected in the integument and nucellus, particularly, at the interface between the outer and inner integument and at the innermost cell layer of the inner integument (Fig. 6B). By contrast, no GhPEPC2 transcript signal was detected in ovule sections at –2 DAA (Fig. 6C). By 1 DAA, however, GhPEPC1 transcript became evident in the young elongating fibre cells (Fig. 6E), compared with its sense control (Fig. 6D). The hybridization signal was also detected in the nucellus and, to a less extent, in the outer and inner integuments (Fig. 6E). For GhPEPC2 mRNA, its signal was observed in fibre cells, the outer integument, and in the nucellus, but was absent in the inner integument (Fig. 6F). Hybridization of 5 d seed sections with the GhPEPC1 antisense probe yielded similar spatial expression patterns (Fig. 6H) as that in 1 d sections (Fig. 6E). It showed strong signals of the GhPEPC1 transcript in rapidly elongating fibres and at the inmost cell layer of the seed coat (Fig. 6H) compared with the control (Fig. 6G). At 5 DAA, the transcript of GhPEPC2 became restricted to fibre cells and the nucellus (Fig. 6I). Throughout the experiments, the sense controls for GhPEPC2 showed the same background as that for GhPEPC1.
Fig. 6.
In situ mRNA localization of GhPEPC1 and 2 in cotton ovules and seeds. (A) Cross-section of ovules at –2 DAA hybridized with a GhPEPC1 sense probe as a negative control. (B) Cross-section of ovules at –2 DAA hybridized with a GhPEPC1 anti-sense probe. Note the absence of GhPEPC1 mRNA signals in the ovule epidermis but strong mRNA signals, indicated by the purple colour, in integument and nucellus, particularly at the interface between outer- and inner-integument (arrow heads) and at the innermost layer of inner integument (arrows). (C) Cross section of ovules at –2 DAA hybridized with a GhPEPC2 anti-sense probe. Note, no GhPEPC2 mRNA signals were detected. (D) Cross-section of seed at 1 DAA, hybridized with a sense probe of GhPEPC1. (E) Cross-section of seed at 1 DAA, hybridized with an antisense probe of GhPEPC1, showing the presence of GhPEPC1 mRNA in elongating fibres (arrows) in addition to that in the seed coat and nucellus. (F) Cross-section of seed at 1 DAA, hybridized with an antisense probe of GhPEPC2, showing the presence of its mRNA in elongating fibres in addition to that in the outer seed coat and nucellus (arrow heads). (G) Cross-section of seed at 5 DAA, hybridized with a sense probe of GhPEPC1. (H) Cross-section of seed at 5 DAA, hybridized with an anti-sense probe GhPEPC1. (I) Cross-section of seed at 5 DAA, hybridized with an antisense probe of GhPEPC2. oi, Outer integument; ii, inner integument; n, nucellus. Bars=50 μm.
Discussion
Several lines of evidence are presented here that high activity of PEPC is required for the rapid and extensive expansion of cotton fibre cells. Developmentally, PEPC activities correlated positively with fibre elongation rate (Fig. 2A, B; see Supplementary Fig. 2 at JXB online). Consistently, the transcript levels of GhPEPC 1 and 2, two major PEPC genes expressed in cotton fibre, were higher in the rapid elongation phase than in the slow-to-termination phase (Fig. 5A). Genotypically, fibres with the highest PEPC activity from Gb elongated the fastest and had the highest regression coefficient (R2=0.8305) between fibre elongation rate and PEPC activity among the three genotypes (see Supplementary Fig. S2 at JXB online). By contrast, the genotype, Ga, with the lowest PEPC activity displayed the slowest fibre elongation rate, leading to short fibres (Fig. 2A , B). Furthermore, PEPC activity from a short fibre mutant, Li, was only about 25% of its wild-type level (Fig. 2C). Consistent with the notion that fibre elongation requires high PEPC activity is the observation that GhPEPC1 and 2 were highly expressed in elongating fibres as compared to the remaining seed tissues (Fig. 5B) and PEPC activity in elongation fibres was higher than that in developing leaves (Fig. 2B). Finally, the casual relationship between PEPC activity and fibre elongation was shown pharmacologically, in which inhibition of fibre PEPC activity by LiCl reduced fibre elongation (Fig. 4). Collectively, the data show that PEPC probably plays an important role in rapid fibre elongation.
The role of PEPC in plant cell expansion has been implicated in stomatal guard cell movement. To this end, drought-induced stomata closure is associated with a guard cell-specific reduction of PEPC gene expression in potato (Kopka et al., 1997). Similarly, a decrease in PEPC activity reduced stomatal conductance and slowed its opening in response to increased light or reduced CO2 partial pressure in a PEPC-deficient mutant of Amaranthus edulis (Cousins et al., 2007). It remains difficult, however, to establish a relationship between temporal changes in PEPC expression or activities and the kinetics of guard cell expansion. In tomato, the expression of two PEPC genes has been shown to correlate with the increase in PEPC activity and the accumulation of malate and citrate during fruit expansion from 8–20 d post-anthesis (Guillet et al., 2002). However, the multicellular nature of tomato fruit renders it difficult to evaluate the contribution of PEPC to the expansion of given cell type(s) within the fruit.
In view of the above analyses, our findings on the role of PEPC in cotton fibre and seed are of significance for two reasons. First, the developmental and genotypic correlation between PEPC activity and cotton fibre elongation provides compelling correlative evidence on the role of this enzyme in plant cell expansion at the single-celled level during the course of its normal development. The role of PEPC in fibre elongation is further supported by the observation that in vitro-cultured cotton fibre grew faster under high concentrations of CO2 (Dhindsa et al., 1975), most likely through its refixation by PEPC in the fibre cells (Ruan, 2005). Second, in addition to its role in fibre elongation, the differential in situ expression patterns of GhPEPC1 and 2 in cotton seed (Fig. 6) indicate the potential function of PEPC in ovule and seed early in development. To our knowledge, there has been no report thus far regarding the expression of PEPC in seed development before and immediately after pollination.
PEPC may control cotton fibre elongation in several ways. Previous studies have indicated that the generation and maintenance of cell turgor is essential for cotton fibre elongation (reviewed by Ruan, 2007). In cotton fibre, malate, along with K+ and hexoses, is a major osmotic solute. The in vivo content of malate is about 0.5–0.7 mmol g−1 DW in the rapidly elongating fibres from 5–10 DAA (Fig. 1), which is equivalent to about 50–70 mM. Thus, it is reasonable to argue that malate accumulation probably plays an important role in fibre elongation through its osmotic effect to attract the influx of water. The high level of PEPC expression (Figs 5, 6) and activity (Figs 2, 3, 4) in fibres provides a biochemical basis for the generation of malate, and hence an osmotic potential and turgor that drives fibre elongation.
High PEPC activity may also be required for the biosynthesis of membrane lipids during fibre elongation where the plasma membrane and tonoplast are enlarged at an extraordinary rate (Ruan et al., 2000, 2001). Indeed, genes involved in fatty acid biosynthesis, chain elongation, and lipid transfer are highly up-regulated during early fibre development (Ji et al., 2003; Qin et al., 2005, 2007; Shi et al., 2006; Gou et al., 2007). In plant cells, fatty acids are synthesized in leucoplasts using acetate, pyruvate, and malate as substrates (Smith et al., 1992). However, the rate of fatty acid synthesis was approximately 4.5 and 120 times higher when malate was the precursor compared with that when pyruvate and acetate served as the precursors, respectively (Smith et al., 1992). Thus, PEPC may play a major role in the enlargement of the plasma membrane and tonoplast during fibre elongation by providing malate for lipid biosynthesis.
Our in situ hybridization analyses further revealed several important findings. First, the observation that GhPEPC 1 and 2 were expressed in young fibres as early as 1 DAA (Fig. 6) suggests that PEPC plays a role in fibre elongation from the very beginning. Interestingly, their transcripts were undetectable in ovule epidermis at –2 DAA (Fig. 6), when fibres are undergoing initiation at the molecular level but remain morphologically indistinguishable from the adjacent ovule epidermal cells (Ruan, 2005). These findings, together with expression analyses of GhPEPC1 and 2 in the later stages of fibre development (Fig. 5) point to the involvement of these genes in fibre elongation but not in the initiation process.
It is also of significance to note the differential expression patterns of GhPEPC 1 and 2 in cotton ovules and seed. While GhPEPC2 was not expressed in ovules before anthesis (Fig. 6C) in this work, GhPEPC1 mRNA was abundantly detected in this tissue (Fig. 6B). This indicates a specific role that GhPEPC1 may play in ovule development. After anthesis, the two PEPC genes exhibited similar expression patterns. Their mRNAs were detected in the nucellus, the outer integument, and fibres, although the signal strength of GhPEPC2 appeared to be weaker than that of GhPEPC1 (Fig. 6).
The role of GhPEPC 1 and 2 in early cotton seed development is not clear at this stage. However, seed development typically requires the local synthesis of amino acids from ammonia-N and the carbon skeleton (Sodek, 1980). Thus, it is possible that activities of GhPEPC1 and 2 may yield oxalacetate, which can serve as a carbon acceptor for amino acid biosynthesis (Radchuk et al., 2007) in cotton ovules or seed before and immediately after fertilization. Noteworthy is the strong mRNA signals of GhPEPC1 at the interface between the outer and inner integuments and at the innermost layer of inner integument in ovules before anthesis (Fig. 6B). These tissue boundaries may be active cellular sites for the synthesis of amino acids from the PEPC-derived oxalacetate, which may then be transported inwards to support the young embryonic tissues after fertilization.
In summary, data presented here show that (i) high activity of PEPC is probably required for rapid cotton fibre cell elongation; and (ii) GhPEPC1 and 2 may play temporally different roles in cotton ovule and seed development in addition to their roles in fibre elongation. The findings provide new insights into the roles of PEPC in plant cell expansion and ovule and seed development.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Alignment analyses of GhPEPC1 and 2 with PEPCs from other plant species.
Supplementary Fig. S2. Correlation analyses between PEPC activity and fibre elongation rate.
Supplementary Material
Acknowledgments
This work was funded, in part, by the University of Newcastle (Grants, 1021404 and 1030156) and the National Science Foundation of China (Grant, 30425043) to YLR. We thank Professor Guixian Xia for providing the Li cotton mutant.
References
- Basra AS, Malik CP. Development of the cotton fibre. International Review of Cytology. 1984;87:65–113. [Google Scholar]
- Beasley CA, Ting IP. Effects of plant growth substances on in vitro fibre development from unfertilized cotton ovules. American Journal of Botany. 1973;60:130–139. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. Biochemistry and molecular biology of plants. The American Society of Plant Physiologists; 2000. pp. 549–556. [Google Scholar]
- Chollet R, Vidal J, O'Leary MH. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Plant Molecular Biology. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Coursol S, Pierre JN, Vidal J. Role of the phosphoinositide pathway in the light-dependent C4 phosphoenolpyruvate carboxylase phosphorylation cascade in Digitaria sanguinalis protoplasts. Biochemical Society Transactions. 2000;28:821–833. [PubMed] [Google Scholar]
- Cousins AB, Baroli I, Badger MR, Ivakov A, Lea PJ, Leegood RC, Caemmerer SV. The role of phosphoenolpyruvate carboxylase during C4 photosynthetic isotope exchange and stomatal conductance. Plant Physiology. 2007;145:1006–1017. doi: 10.1104/pp.107.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Goldberg RB. Analysis of gene expression. In: Show CH, editor. Plant molecular biology: a practical approach. Oxford: IRL Press; 1988. pp. 1–35. [Google Scholar]
- Dhindsa RS, Beasley CA, Ting IP. Osmoregulation in cotton fibre. Plant Physiology. 1975;56:394–398. doi: 10.1104/pp.56.3.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famiani F, Walker RP, Técsi L, Chen ZH, Proietti P, Leegood RC. An immunohistochemical study of the compartmentation of metabolism during the development of grape (Vitis vinifera L.) berries. Journal of Experimental Botany. 2000;345:675–683. [PubMed] [Google Scholar]
- Gehrig H, Heute V, Kluge M. New partial sequences of phosphoenolpyruvate carboxylase as molecular phylogenetic markers. Molecular Phylogenetics and Evolution. 2001;20:262–274. doi: 10.1006/mpev.2001.0973. [DOI] [PubMed] [Google Scholar]
- Giglioli-Guivarc'h N, Pierre JN, Brown S, Chollet R, Vidal J, Gadal P. The light-dependent transduction pathway controlling the regulatory phosphorylation of C4-phosphoenolpyruvate carboxylase in protoplasts from Digitaria sanguinalis. The Plant Cell. 1996;8:573–586. doi: 10.1105/tpc.8.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Wang LJ, Chen SP, Hu WL, Chen XY. Gene expression and metabolite profiles of cotton fibre during cell elongation and secondary cell wall synthesis. Cell Research. 2007;17:422–434. doi: 10.1038/sj.cr.7310150. [DOI] [PubMed] [Google Scholar]
- Guillet C, Just D, Bénard N, Destrac-Irvine A, Baldet P, Hernould M, Causse M, Hatch MD, Oliver IR. Activation and inactivation of phosphoenolpyruvate carboxylase in leaf extracts from C4 species. Australian Journal of Plant Physiology. 1978 5, 571–580. [Google Scholar]
- Hatch MD, Oliver IR. Activation and inactivation of phosphoenolpyruvate carboxylase in leaf extracts from C4 species. Australian Journal of Plant Physiology. 1978;5:571–580. [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y. PHOSPHOENOLPYRUVATE CARBOXYLASE: a new era of structural biology. Annual Review of Plant Biology. 2004;55:69–84. doi: 10.1146/annurev.arplant.55.031903.141619. [DOI] [PubMed] [Google Scholar]
- Jiao J, Chollet R. Light/dark regulation of maize leaf phosphoenolpyruvate carboxylase by in vivo phosphorylation. Archives of Biochemistry and Biophysics. 1988;261:409–417. doi: 10.1016/0003-9861(88)90357-8. [DOI] [PubMed] [Google Scholar]
- Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH, Fu Q, Liu D, Luo JC, Zhu YX. Isolation and analyses of genes preferentially expressed during early cotton fibre development by subtractive PCR and cDNA array. Nucleic Acids Research. 2003;31:2534–2543. doi: 10.1093/nar/gkg358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ni DA, Ruan YL. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. The Plant Cell. 2009;21:2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JF, Vance CP, Allan DL. Phosphorus deficiency in Lupinus albus. Altered lateral root development and enhanced expression of phosphoenolpyruvate carboxylase. Plant Physiology. 1996;112:31–41. doi: 10.1104/pp.112.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca M, Saha S, Jenkins JN, Zipf A, Kohel R, Stelly DM. Simple sequence repeat (SSR) markers linked to the Ligon Lintless (Li1) mutant in cotton. The Journal of Heredity. 2002;93:221–224. doi: 10.1093/jhered/93.3.221. [DOI] [PubMed] [Google Scholar]
- Kopka J, Provart NJ, Müller-Röber B. Potato guard cells respond to drying soil by a complex change in the expression of genes related to carbon metabolism and turgor regulation. The Plant Journal. 1997;11:871–882. doi: 10.1046/j.1365-313x.1997.11040871.x. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development. Annals of Botany. 2007;100:1391–1401. doi: 10.1093/aob/mcm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC. C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. Journal of Experimental Botany. 2002;53:581–590. doi: 10.1093/jexbot/53.369.581. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Vidal J, Chollet R, Gadal P, Crétin C. Phosphoenolpyruvate carboxylase: structure, regulation and evolution. Plant Science. 1994;99:111–124. [Google Scholar]
- Lowry OH, Passonneau JV. A flexible system of enzymatic analysis. New York: Academic Press; 1972. [Google Scholar]
- Magnin NC, Cooley BA, Reiskind JB, Bowes G. Regulation and localization of key enzymes during the induction of Kranz-less, C4-type photosynthesis in Hydrilla verticillata. Plant Physiology. 1997;115:1681–1689. doi: 10.1104/pp.115.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Bhatt K, Baumann K. Shaping in plant cells. Current Opinion in Plant Biology. 2001;4:540–549. doi: 10.1016/s1369-5266(00)00213-2. [DOI] [PubMed] [Google Scholar]
- Monreal JA, López-Baena FJ, Vidal J, Echevarría C, García-Mauriño S. Effect of LiCl on phosphoenolpyruvate carboxylase kinase and the phosphorylation of phosphoenolpyruvate carboxylase in leaf disks and leaves of Sorghum vulgare. Planta. 2007;225:801–812. doi: 10.1007/s00425-006-0391-0. [DOI] [PubMed] [Google Scholar]
- Nimmo HG. The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends in Plant Science. 2000;5:75–80. doi: 10.1016/s1360-1385(99)01543-5. [DOI] [PubMed] [Google Scholar]
- Pfluger J, Zambryski PC. Cell growth: the power of symplastic isolation. Current Biology. 2001;11:R436–R439. doi: 10.1016/s0960-9822(01)00254-8. [DOI] [PubMed] [Google Scholar]
- Qin YM, Pujol FM, Hu CY, Feng JX, Kastaniotis AJ, Hiltunen JK, Zhu YX. Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid elongation. Journal of Experimental Botany. 2007;58:473–481. doi: 10.1093/jxb/erl218. [DOI] [PubMed] [Google Scholar]
- Qin YM, Pujol FMA, Shi YH, Feng JX, Liu YM, Kastaniotis AJ, Hiltunen JK, Zhu YX. Cloning and functional characterization of two cDNAs encoding NADPH-dependent 3-ketoacyl-CoA reductase from developing cotton fibres. Cell Reseach. 2005;15:465–473. doi: 10.1038/sj.cr.7290315. [DOI] [PubMed] [Google Scholar]
- Radchuk R, Radchuk V, Götz KP, Weichert H, Richter A, Emery RJ, Weschke W, Weber H. Ectopic expression of phosphoenolpyruvate carboxylase in Vicia narbonensis seeds: effects of improved nutrient status on seed maturation and transcriptional regulatory networks. The Plant Journal. 2007;51:819–839. doi: 10.1111/j.1365-313X.2007.03196.x. [DOI] [PubMed] [Google Scholar]
- Rao SK, Magnin NC, Reiskind JB, Bowes G. Photosynthetic and other phosphoenolpyruvate carboxylase isoforms in the single-cell, facultative C4 system of Hydrilla verticillata. Plant Physiology. 2002;130:876–886. doi: 10.1104/pp.008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL. Recent advances in understanding cotton fibre and seed development. Seed Science Research. 2005;15:269–280. [Google Scholar]
- Ruan YL. Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Functional Plant Biology. 2007;34:1–10. doi: 10.1071/FP06234. [DOI] [PubMed] [Google Scholar]
- Ruan YL, Chourey PS. A fibreless seed mutation in cotton is associated with lack of fibre cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiology. 1998;118:399–406. doi: 10.1104/pp.118.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Chourey PS, Delmer PD, Perez-Grau L. The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiology. 1997;115:375–385. doi: 10.1104/pp.115.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. Pathway and control of sucrose import into initiating cotton fibres. Australian Journal of Plant Physiology. 2000;27:795–800. [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. The control of single-celled cotton fibre elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. The Plant Cell. 2001;13:47–63. doi: 10.1105/tpc.13.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. Suppression of sucrose synthase gene expression represses cotton fibre cell initiation, elongation, and seed development. The Plant Cell. 2003;15:952–964. doi: 10.1105/tpc.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Xu SM, White R, Furbank RT. Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fibre elongation mediated by callose turnover. Plant Physiology. 2004;136:4104–4113. doi: 10.1104/pp.104.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert AM, Benedict CR, Berlin JD, Kohel RJ. Cotton fibre development: kinetics of cell elongation and secondary wall thickening. Crop Science. 1973;13:704–709. [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fibre cell elongation. The Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RG, Gauthier DA, Dennis DT, Turpin DH. Malate- and pyruvate-dependent fatty acid synthesis in leucoplasts from developing castor endosperm. Plant Physiology. 1992;98:1233–1238. doi: 10.1104/pp.98.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek L. Distribution and properties of a potassium-dependent asparaginase isolated from developing seeds of Pisum sativum and other plants. Plant Physiology. 1980;65:22–26. doi: 10.1104/pp.65.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD. Brassinosteroid regulates fibre development on cultured cotton ovules. Plant and Cell Physiology. 2005;46:1384–1391. doi: 10.1093/pcp/pci150. [DOI] [PubMed] [Google Scholar]
- Tarczynski MC, Outlaw WH., Jr Partial characterization of guard-cell phosphoenolpyruvate carboxylase: kinetic datum collection in real time from single-cell activities. Archives of Biochemistry and Biophysics. 1990;280:153–158. doi: 10.1016/0003-9861(90)90530-c. [DOI] [PubMed] [Google Scholar]
- Vidal J, Chollet R. Regulatory phosphorylation of C4 PEP carboxylase. Trends in Plant Science. 1997;2:230–237. [Google Scholar]
- Wu YR, Machado AC, White RW, Llewellyn DJ, Dennis ES. Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant and Cell Physiology. 2006;47:107–127. doi: 10.1093/pcp/pci228. [DOI] [PubMed] [Google Scholar]
- Zhu YQ, Xu KX, Chen XY. The reduction of auxin polarity transportations in the Ligon Lintless (Li1) mutant of cotton. Journal of Plant Physiology and Molecular Biology. 2003;29:15–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.