Abstract
Background
Laboratory systems to study bacterial transmission and mucosal colonization leading to infection have not been utilized.
Methods
We determined if transmission of various strains of P. aeruginosa among individual mice could occur and the properties of such strains in establishing GI mucosal colonization as well as disseminating systemically following induction of neutropenia.
Results
Isolates of P. aeruginosa associated with epidemic spread among CF patients readily established GI colonization at higher levels than strains associated with systemic or chronic infection and out-competed these strains. Colonization was associated with resistance to bile salts. However, the epidemic CF isolates did not disseminate following induction of neutropenia and did not induce as much mucosal pathology as did strains capable of disseminating.
Conclusion
Murine models can be used to study P. aeruginosa transmission and early colonization, and the properties of these strains associated with their known clinical behaviors are mimicked in this setting.
Keywords: Pseudomonas aeruginosa, colonization, horizontal transmission, translocation, murine model, epidemic strain, cystic fibrosis, nosocomial infection, neutropenia, immunocompromised
Background
Pseudomonas aeruginosa is a major cause of nosocomial infections in intensive care unit (ICU) [1, 2], cancer, and bone marrow transplant (BMT) patients [3]. Infection leads to significant morbidity and mortality [4-6]. P. aeruginosa also causes chronic lung infections in patients with bronchiectasis or cystic fibrosis (CF) and is associated with poorer prognosis [7].
Since P. aeruginosa is at best a transient inhabitant of the normal human microbiome, acquisition and mucosal colonization is an initial and crucial step of pathogenesis. In the ICU, acquisition of exogenous P. aeruginosa via cross-transmission accounts for the majority of the colonization or infectious episodes [8, 9]. For CF patients, P. aeruginosa strains are often acquired from diverse environmental sources outside the hospital [10]. However, well-documented outbreaks in a number of CF clinics involving highly transmissible “epidemic” strains of P. aeruginosa have occurred [11-14].
Little is known about these early steps of acquisition, mucosal colonization, and transmission of P. aeruginosa, specifically with respect to the “epidemic” strains. In a murine model of oropharyngeal colonization, chronic P. aeruginosa infection has been achieved in transgenic CF mice, however, the colonization levels are too low for quantitative analysis [15, 16]. Here we used a murine model [17] to study P. aeruginosa gastrointestinal (GI) colonization, competitive co-colonization between different strains, and horizontal transmission in the setting of antibiotic-induced depletion of the indigenous GI flora. We also evaluated bacterial dissemination following neutropenia. These findings identified and validated a suitable animal model for studying acquisition of P. aeruginosa that can be used to define determinants of transmission and colonization relevant to person-to-person transmission.
Materials and Methods
Bacterial strains
The strains used are listed in table 1. P. aeruginosa “epidemic” strains LES and C3719 are LPS rough, non-mucoid CF respiratory isolates whose genomes have recently been sequenced [18, 19]. Strain PA2192nm is a non-mucoid variant of mucoid strain PA2192 obtained from a CF patient with 8 years of chronic infection, also with a recently sequenced genome [18]. Strains PAO1 (wound isolate) and PA14 (isolate from burn patient) are well-studied, sequenced strains [20, 21] and, along with strain PA2192nm, are referred as “non-epidemic” strains. All the strains had virtually identical in-vitro growth rates, except the epidemic strains took a slightly longer time to reach log phase growth (not shown). Strains were tested for swimming [22] and twitching motility [23] as well as in-vitro cytotoxicity on Caco-2 cells (CytoTox 96, Promega). All of the strains had intact genes for hslV (PA5053), hslU (PA5054), hslO (PA1468610), pqsA (PA0996), pqsB (PA0997), pqsC (PA0998), pqsD (PA0999), and pqsE (PA1000) as determined by BLAST search (not shown). The presence of exoS (PA3841) or exoU (PA14 51530) genes was also determined by BLAST search. LPS glycoforms were analyzed by SDS-PAGE [24].
Table 1.
Bacterial strains used
| Strain | Genotype and Phenotype | LPS structure | Motility | Cytotoxicity | exoS or exoU | Origin and reference | |
|---|---|---|---|---|---|---|---|
| Swimming | Twitching | ||||||
| PAO1 | Wild-type wound isolate, Cm sensitive | LPS smooth, complete core | Yes | Yes | No [48] | exoS | M. Vasil [49] |
| PAO1 ΔgalU | ΔgalU mutant | LPS rough, incomplete outer core | n/a | n/a | No [48] | n/a | [26] |
| PA14 | Wild-type isolate from burn patient | LPS smooth, complete core | Yes | Yes | Yes | exoU | F. Ausubel [50] |
| PA2192 non-mucoid (nm) | non-mucoid variant of mucoid CF isolate PA2192 | LPS rough, complete core | No | Yes | No | exoS | Our collection [18] |
| LES | Epidemic CF isolate (Liverpool clone) | LPS rough, complete core | No | No | No | exoS | S. Lory [13] |
| C3719 | Epidemic CF isolate (Manchester clone) | LPS rough, complete core | No | No | No | exoS | S. Lory [12] |
Abbreviation: Cm, Chloramphenicol
Murine Model of GI Tract Colonization by P. aeruginosa
As described [17], C3H/HeN mice (6- to 8-week-old females) were housed in groups of 4 in sterilized cages with sterile filter hoods and maintained under specific pathogen-free conditions in compliance with the Harvard Medical Area Institutional Animal Care and Use Committee guidelines. Mice were fed sterile water with 2 mg streptomycin/ml and 1,500 U penicillin G/ml for 4 days to deplete indigenous GI flora (confirmed by bacterial stool cultures [17]). Stable GI colonization by P. aeruginosa is not achieved in the presence of indigenous bowel flora [25]. Next, mice were fed sterile water with 1,500 U penicillin G/mL and P. aeruginosa strains (approximately 107 CFU/ml for 5 days). Water containing P. aeruginosa was changed after 2 to 3 days to maintain bacterial levels. Stool samples were collected from individual mice daily starting 24 h after the initiation of P. aeruginosa water, weighed, homogenized, diluted in 1 ml 1% protease peptone, and plated on cetrimide agar to quantify bacterial levels. The presence of green oxidase-positive colonies was used to confirm P. aeruginosa GI colonization.
Murine Model of P. aeruginosa Co-Colonization
To evaluate P. aeruginosa strain competition during murine GI colonization, one epidemic strain and one non-epidemic strain were mixed together in sterile water with 1,500 U penicillin G/mL at ~107 CFU/ml for each strain and fed to 8 antibiotic-treated mice. GI colonization levels were measured daily for 5 days. Cetrimide agar with or without 0.060 mg gentamicin/ml was used to grow colonies of the epidemic strains for 48 h at 37°C. We could not find antibiotics to select for growth of the non-epidemic strains and against the epidemic strains. To quantify the non-epidemic strains, we utilized a significant colony growth differential on cetrimide agar between these and the epidemic strains. All colonies of the non-epidemic strains were macroscopically visible by 24 h of incubation, whereas colonies of the epidemic strains were not visualized until 48 h after incubation. Therefore, we counted the macroscopically visible colonies on cetrimide agar at 24 h as those of the non-epidemic strains, and further confirmed that the colonies that emerged between 24 and 48 hrs of incubation were equivalent in number to those found on cetrimide with gentamicin used to select for the epidemic strains.
Murine Model of Horizontal Transmission of P. aeruginosa
Horizontal transmission among mice within a cage was analyzed by placing one mouse previously colonized with P. aeruginosa into a cage with 3 other antibiotic-treated mice not given P. aeruginosa. To prevent P. aeruginosa transmission via the drinking water, acidified water (0.2 M sodium acetate, pH 4.0) was used and monitored daily by culture on cetrimide agar. Cages and bedding were changed daily. Stool was collected daily from each mouse for 5 consecutive days and processed to quantify colonization levels after introduction of the colonized mouse.
Murine Model of P. aeruginosa GI Translocation
After establishing P. aeruginosa colonization, one 200 μg dose of monoclonal antibody (MAb) RB6-8C5 specific for the Ly6 antigen on PMNs was administered intraperitoneally [17]. The drinking water was replaced with sterile water containing 1500 U penicillin G/ml. Mice were monitored twice a day for morbidity and mortality for 7 days. Prior experience has documented no deaths occur after 7 days in this experimental setting. Moribund mice were euthanized and along with mice found dead between observation periods, the spleens were resected, homogenized in 1 ml of 1% proteose peptone, serially diluted, and plated on TSA (1.5% agar in TSB), MacConkey and cetrimide agar. The presence of green oxidase-positive colonies on cetrimide agar was used for confirmation of dissemination of P. aeruginosa. In prior work [17], we showed that when P. aeruginosa translocates from the GI tract into the systemic circulation the organism can be recovered not only from the blood and liver but also from the spleen.
Bile-salt Sensitivity Assay
P. aeruginosa was grown on TSA at 37°C overnight. Colonies were suspended in PBS to a concentration of ~107 CFU/ml. 100 μl of bacterial suspension was placed, in triplicate, in a sterile 96-well microtiter plate. 100 μl of 1% sodium deoxycholate (Sigma) in PBS was added to individual wells. 100 μl of 2% BSA in PBS was added to other wells to determine survival during the incubation period. The plate was incubated at 37°C for 4 h with gentle shaking, samples serially diluted, plated on TSA, and incubated at 37°C for bacterial enumeration.
Serum Sensitivity Assay
100-μl aliquots of TSB-grown P. aeruginosa that were diluted in PBS to contain ~106 CFU were placed, in triplicate, in individual wells of sterile 96-well plates. Serum from a healthy human was diluted in PBS to 10%. Heat inactivated serum (56°C for 30 min) and PBS without serum served as controls. Equal volumes (100 μl) of sera and bacterial suspensions were mixed and incubated at 37°C for 1 h with gentle shaking. Samples were serially diluted, plated on TSA, and incubated overnight at 37°C for enumeration.
Histological Analysis of Murine GI Tract
Neutropenic C3H/HeN mice colonized with P. aeruginosa strains PA14 or LES were sacrificed at 0, 24, or 42 h after neutropenia was induced, while mice colonized with strain LES were additionally sacrificed at 72 or 96 h. After sacrifice, ceca were resected, fixed in Bouin's solution and sections stained with hematoxylin and eosin then reviewed by a veterinary pathologist. Gross histopathology of the GI tract was also recorded.
Measurement of Cytokine Responses in Murine GI Tissue
Mice colonized with either strain PA14 or LES, and non-colonized controls, were made neutropenic then sacrificed 42 h later. After sacrifice, ceca were resected, contents gently flushed out, cecal tissue washed in Modified Eagles Medium (Gibco) then homogenized in 1 ml of ice-cold PBS with protease inhibitors (complete Mini, Roche Diagnostics) and 0.5 % Triton-X100 (Amresco). The sample was centrifuged (5 min at 5000 x g at 4°C), Tumor Necrosis Factor-alpha (TNF-α) and Interleukin-one beta (IL-1ß) levels in supernatant were measured by ELISA assays (eBioscience) and protein concentrations by BCA Protein Assay Kit (Pierce).
Statistical Analyses
Pair-wise comparisons between strains were analyzed by Mann-Whitney U tests except competitive co-colonization data were analyzed by Wilcoxon Signed Rank test. Survival data were analyzed by the Log rank test. Bonferroni's correction was used when multiple comparisons were performed. One-way ANOVA with Dunnett's multiple comparison was used for analysis of results from the bile salt sensitivity and serum sensitivity assays. As appropriate, non-normally distributed data were log transformed. Kruskal-Wallis tests with Dunn's multiple comparison were used for analyzing proinflammatory cytokine levels. Correlations between TNF-α and IL-1β levels in each mouse were analyzed by Spearman correlation test.
Results
Comparative GI colonization levels of different strains
P. aeruginosa strains PAO1, PA14, LES, C3719, and PA2192nm all colonized the murine GI tract. One day after the start of oral feeding of bacteria, the cfu/g stool of the epidemic strains LES and C3719 were higher than those of the other strains (figure 1A; P<0.01 by Mann Whitney U test with Bonferonni's correction). By day 5, strains PAO1, PA14 and PA2192nm reached colonization levels comparable to strain C3719, but all still colonized at a lower level than strain LES (figure 1B; P<0.01).
Figure 1.
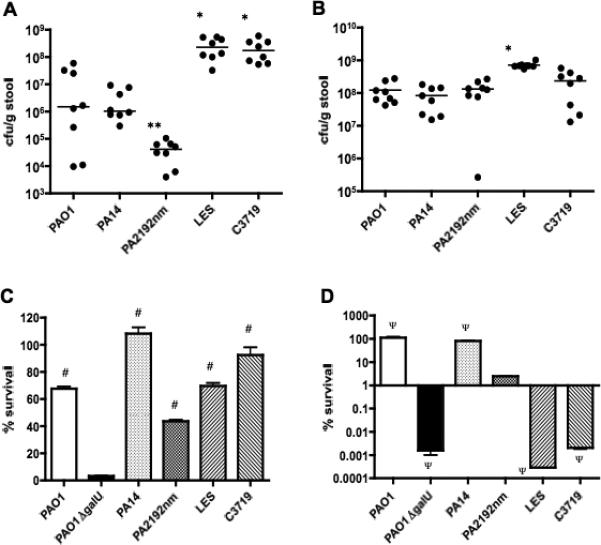
Gastrointestinal colonization levels in C3H/HeN mice by individual P. aeruginosa strains PAO1, PA14, PA2192nm, LES, or C3719 on day 1 (A) and day 5 (B) after the initiation of oral bacterial administration. Points represent results from individual animals, and horizontal lines represent the medians (n = 8 per strain). * P<0.01 compared to the non-epidemic strains of PAO1, PA14 and PA2192nm, ** P<0.01 compared to strain PA14. P values: Mann-Whitney test with Bonferroni's correction.
(C), Bile salt sensitivities of P. aeruginosa strains PAO1, PAO1ΔgalU, PA14, PA2192nm, LES and C3719. Percent survival was defined as the number of CFUs counted after exposure to 1% sodium deoxycholate for 4 h at 37°C divided by CFUs after exposure to 2% BSA (control) in the same condition X 100. The bars show the average of triplicate samples for each strain, and the error bars represent standard error of the means. # P<0.01 compared to strain PAO1ΔgalU by one-way ANOVA with Dunnett's multiple comparison.
(D), Serum sensitivities of P. aeruginosa strains PAO1, PAO1ΔgalU, PA14, PA2192nm, LES and C3719. Percent survival was defined as the number of CFUs counted after exposure to 10% human serum for 1 h at 37°C divided by CFUs after exposure to heat inactivated serum in the same condition X 100. The bars show the average of triplicate samples for each strain, and the error bars represent standard error of the means. Note Y axis is a log scale. Ψ P<0.01 compared to strain PA2192nm by one-way ANOVA with Dunnett's multiple comparison.
Relationship of resistance to bile salts with GI colonizing capacity
P. aeruginosa strain PAO1ΔgalU, which lacks the outer core oligosaccharide of lipopolysaccharide [26], is unable to colonize murine GI tracts [17]. Since strains C3719 and PA2192nm have a full LPS outer core but lack LPS O-side chains [18, 27] as does strain LES (S. Lory, unpublished data), we hypothesized that their LPS structure conferred resistance to bile salts and potentially the ability to colonize the GI tract. We found strain PAO1ΔgalU was significantly more sensitive to bile salts compared to all the other strains tested (figure 1C; P<0.01 by ANOVA with Dunnett's multiple comparison), indicating that possession of an intact LPS outer core, even in the absence of LPS O-side chains, conferred sufficient resistance to bile salts to allow for GI colonization by P. aeruginosa.
Serum Sensitivity Assay
LPS smooth strains PAO1 and PA14 were more serum resistant than the LPS rough strain PA2192nm, whereas epidemic strains LES and C3719 and strain PAO1ΔgalU, also LPS rough strains, were significantly more serum-sensitive than strain PA2192nm (figure 1D; P<0.01 by one-way ANOVA with Dunnett's multiple comparison).
Competitive colonization between the epidemic and systemic strains
When the epidemic strains LES or C3719 were administered concurrently with the non-epidemic strains PA14 or PA2191nm, the epidemic strains colonized 38 of 40 mice at higher levels than the non-epidemic strains throughout the observation period of 5 days (figure 2 A-E for day 5, data not shown for days 1 to 4; P<0.05 by Wilcoxon Signed Rank test). Co-colonization by strains C3719 and PAO1 on day 5 showed only 4 of 8 mice with higher levels of the epidemic strain and the difference was not significant (figure 2F). Competitive indices between epidemic and non-epidemic strains are also shown (figure 2G). When comparing the cfu/g stool of mice mono-colonized with strains PAO1, PA14 and 2192 nm with the cfu/g stool of these strains in mice co-colonized with the epidemic strains, the levels of the non-epidemic strains were significantly lower (P<0.01, Mann Whitney U test) in the co-colonized mice except for strain PAO1 in mice co-colonized by epidemic strain C3719. Notably, the two epidemic strains achieved comparable GI colonization levels regardless of whether the animals were mono-colonized with these two strains (figure 1) or co-colonized along with non-epidemic strains (figure 2).
Figure 2.
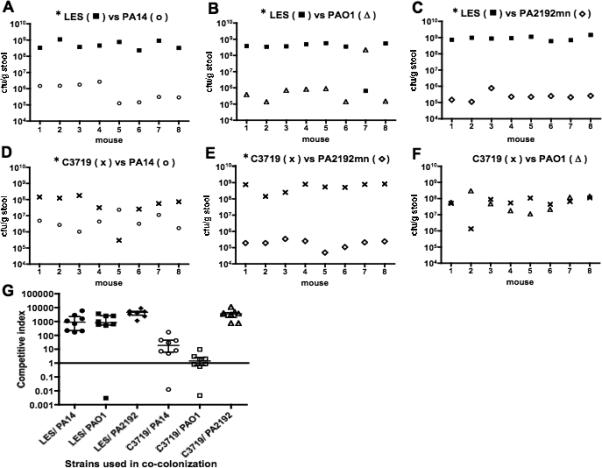
(A), (B), (C), (D), (E), (F); Gastrointestinal (GI) co-colonization levels by P. aeruginosa strains in individual mice 5 days after being given a mixture of one epidemic strain (LES or C3719) and one non-epidemic strain (PAO1, PA14 or PA2192nm) (n = 8 mice per each combination of strains). Colonization levels achieved by the epidemic strains were higher than those by the non-epidemic strains ((A), (B), (C), (D), (E), * P<0.05 by Wilcoxon signed rank test) except for one combination of strains C3719 and PAO1 (F).
(G), Competitive Indices (C.I.) of GI co-colonization levels by epidemic versus non-epidemic strains in each mouse in the different groups. C.I. was defined as the colonization level of an epidemic strain divided by the level of a non-epidemic strain. A C.I. above 1.0 indicates the epidemic strain out-competed the non-epidemic strain in the co-colonization level. Horizontal bars indicate median, error bars indicate interquartile range.
Horizontal transmission of P. aeruginosa strains
All the P. aeruginosa strains could be transmitted from a single colonized mouse placed into a cage with 3 uncolonized, antibiotic-treated mice (figure 3). For all strains, colonization levels in the GI tract established an equilibrium, increasing until days 2 or 3 (levels ranging from 107 to 109 cfu/g stool) and then decreasing by day 5 (levels ranging from 105 to 106 cfu/g stool). P. aeruginosa was never detected in the acidified drinking water.
Figure 3.
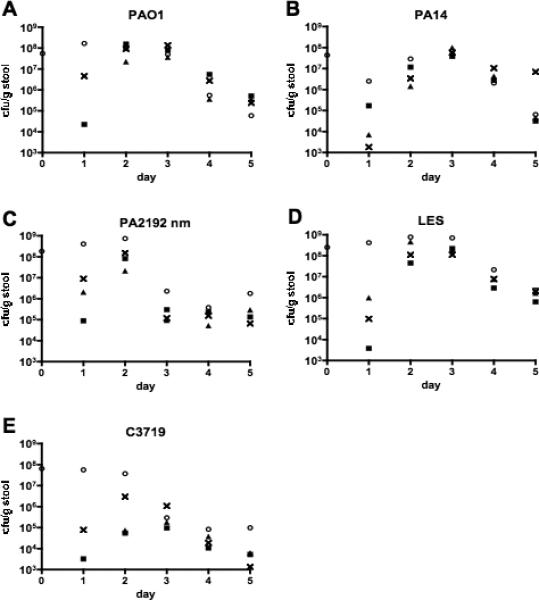
Horizontal transmission of P. aeruginosa strains PAO1 (A), PA14 (B), PA2129nm (C), LES (D), and C3719 (E) from a previously colonized C3H/HeN mouse (○) to antibiotic-treated non-colonized mice (△, ■, ×) within a single cage. Points represent results from individual animals. All the P. aeruginosa strains were transmitted from a single colonized mouse to 3 antibiotic-treated uncolonized cage mates. Colonization levels in the gastrointestinal tract established an equilibrium, increasing until days 2 or 3 and then decreasing by day 5 for all the strains. P. aeruginosa was not detected in the acidified drinking water provided in the cages.
Systemic spread of P. aeruginosa following induction of neutropenia
Seven out of 8 mice mono-colonized with strain PAO1 and all 8 mice mono-colonized with strain PA14 died or became moribund after neutropenia was induced (figure 4A). Five out of eight mice mono-colonized with strain PA2192nm died, but the overall survival was significantly longer than for mice mono-colonized with strains PAO1 or PA14 (figure 4A; P<0.05 by log rank test with Bonferonni's correction). None of 8 mice died mono-colonized with epidemic strains LES or C3719 died (figure 4A). Strain PA2192nm was isolated from the spleen of the dead or moribund mice but at a significantly lower level than were strains PAO1 or PA14 (figure 4B; P<0.05 by Mann Whitney test with Bonferonni's correction). GI colonization levels in all the surviving mice were comparable to those achieved before induction of neutropenia (data not shown).
Figure 4.
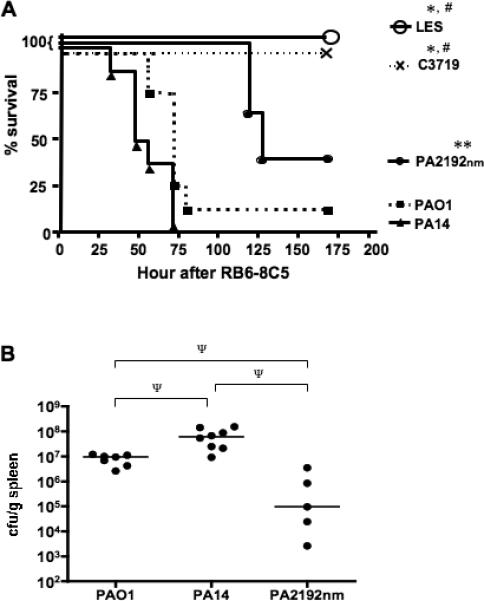
(A), Survival curves of C3H/HeN mice mono-colonized with P. aeruginosa strain PAO1, PA14, PA2192nm, LES, or C3719 followed by MAb RB6-8C5 induced neutropenia (n = 8 per strain). * P< 0.01, ** P<0.05 compared to mice colonized with strain PAO1 or PA14, # P<0.05 compared to those with strain PA2192nm by log rank test with Bonferroni's correction. (B), Bacterial levels of P. aeruginosa strains isolated from the spleens of moribund or dead mice after gastrointestinal colonization followed by the induction of neutropenia (n = 7, 8, and 5 for strain PAO1, PA14, and PA2192nm respectively). Ψ P<0.05 by Mann Whitney test with Bonferonni's correction.
Cecal histopathology in neutropenic mice
Forty-two h after induction of neutropenia, the GI tracts of mice colonized by strain PA14 was edematous and hyperemic (figure 5A). The GI tracts of mice that had been neutropenic for 48 h and colonized by strain LES (figure 5B) or mice colonized with strain PA14 prior to the induction of neutropenia appeared normal and comparable to uncolonized mice (not shown). Histopathologic analysis of the ceca from neutropenic mice colonized with strain PA14 revealed shortening and flattening of the villi along with marked sloughing of the epithelial cells (figure 5C, E). No abnormalities were observed in mice colonized with strain PA14 just prior to induction of neutropenia (not shown), nor were any observed in mice colonized with strain LES at 42 h (figure 5 D, F) or up to 96 h after induction of neutropenia (not shown).
Figure 5.
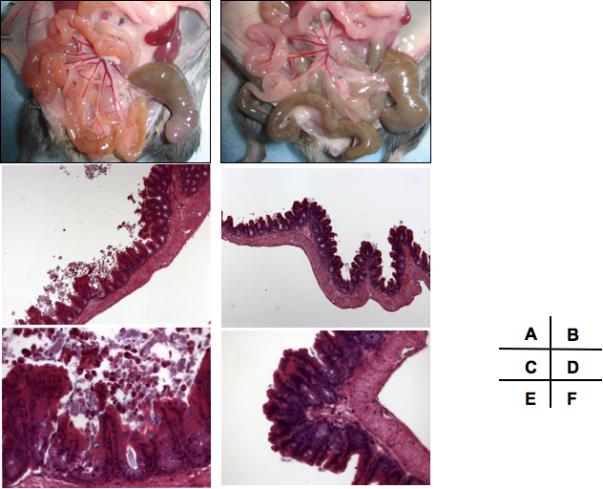
Gross anatomy and microscopic histology of the murine (C3H/HeN) gastrointestinal (GI) tract after prior mono-colonization with P. aeruginosa strain PA14 ((A), (C), (E)) or LES ((B), (D), (F)) and subsequent MAb RB6-8C5 induced neutropenia. Mice were sacrificed and sections were obtained 42 h after induction of neutropenia.
(A), Diffuse erythema in noncontiguous areas of the GI tract, notably in the small intestine as well as the tip of the cecum. (C), (E); Shortening and flattening of the villi along with marked sloughing of the epithelial cells are clearly seen. (B), (D), (F); Normal macro- and microscopic appearances. Magnification: (C), (D) 100X, (E), (F) 400X.
Cecal TNF-α and IL-1β concentrations in neutropenic mice
Animals colonized with strain PA14 had significantly higher tissue levels of TNF-α and IL-1β compared with levels in neutropenic mice colonized with strain LES (figure 6; P<0.05 by Kruskal-Wallis tests with Dunn's multiple comparison test). Tissue levels of TNF-α and IL-1β in each neutropenic mouse colonized with strain PA14 were significantly correlated (P<0.05, Spearman correlation test, data not shown).
Figure 6.
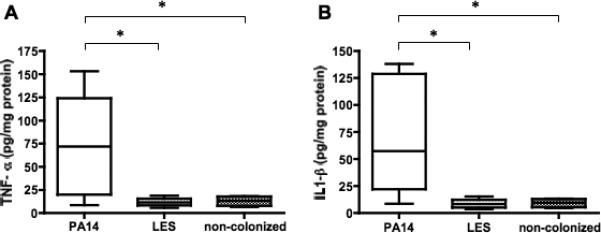
Levels of Tumor Necrosis Factor alpha (TNF-α) (A), and Interleukin one beta (IL-1ß) (B) in the ceca of C3H/HeN mice whose gastrointestinal tract were mono-colonized by either P. aeruginosa strain PA14 (n = 8) or strain LES (n = 6), followed by MAb RB6-8C5 induced neutropenia. Mice that were antibiotic-treated but not given any P. aeruginosa were also induced neutropenic to serve as non-colonized controls (n = 4). Mice were sacrificed 42 h after MAb administration and ceca harvested to measure the tissue cytokine levels. Lines indicate medians, boxes 25th and 75th percentiles, error bars 10th and 90th percentiles. * P<0.05 by Kruskal-Wallis test with Dunn's multiple comparison post-hoc test.
Discussion
We conducted this study to validate a murine model related to host acquisition and infection by P. aeruginosa using GI colonization and horizontal transmission to study these early steps in the infectious process. Antibiotic-treated mice could be rapidly colonized in their GI tract by all of the P. aeruginosa strains tested and could facilely transmit them to antibiotic-treated, non-colonized cagemates. The epidemic strains achieved higher GI colonization levels than the non-epidemic strains and out-competed them in the co-colonization setting. We also used PMN depletion, a risk factor for systemic nosocomial infections by P. aeruginosa, to study the association of colonization and dissemination, and found the epidemic strains failed to disseminate from the GI tract, which was an interesting corollary to the localized, non-disseminating nature of lung infections that develop in CF patients infected with these strains. This murine model of acquisition, colonization, transmission and dissemination of P. aeruginosa can now provide further opportunities to gain insights into why certain P. aeruginosa strains may achieve a competitive advantage and/or have a predilection for specific clinical manifestations of infection.
We utilized a GI model of colonization to study P. aeruginosa epidemic strains that are responsible for chronic lung infections because the GI tract is an ideal mucosal surface for serial, quantitative analysis of P. aeruginosa colonization. It is also an important source of this organism that can ultimately lead to colonization of the respiratory tract [28]. It is plausible that a fecal-oral transmission route contributes for spreading strains of P. aeruginosa among CF patients (e.g. via oropharyngeal or upper GI tract colonization followed by aspiration to the lungs), as it has been shown that such patients have high fecal burdens of P. aeruginosa [29]. In non-CF populations, a significant portion of P. aeruginosa ventilator associated pneumonia in the ICU has been shown to be caused by the endogenous strains that originally existed in the patients’ GI flora [30]. Based on these observations, P. aeruginosa GI colonization appears highly relevant to P. aeruginosa colonization/infection in the respiratory tract.
While our animal model is not representative of the hygienic conditions in most hospitals, we do demonstrate that in a laboratory setting P. aeruginosa can be transmitted from a carrier to an antibiotic-treated, non-colonized host without contaminated water serving as a source – most likely via a fecal-oral route, an important mode of transmission readily achieved even in a modern hygienic environment [31]. In fact, although water is a well-established environmental source of P. aeruginosa in hospitals [32-35], several recent studies on outbreaks of P. aeruginosa nosocomial infections reported a lack of a persistent environmental source [11, 36, 37], suggesting some other method of horizontal transmission between infected and non-infected individuals.
It is not clear why the epidemic strains spread so well among patients in CF clinics in England and Wales [36-38]. Using a laminar flow model of survival of bacteria in aerosols, Clifton et al. [39] did not find a difference among epidemic and non-epidemic P. aeruginosa isolates, but did find mucoid strains of P. aeruginosa had better overall survival in the aerosols, with survival affected by temperature and humidity. Our finding that the epidemic strains colonized the GI mucosal surface better than the non-epidemic strains may partly explain the “epidemic” capacity of the strains. Moreover, these epidemic strains appear to be genetically different from other strains [18, 40], with strain LES harboring a large number of phage-encoded pathogenicity islands [40].
Our findings in the co-colonization experiments also correlated with the clinical observation that strain LES can “superinfect” a CF lung previously colonized by other P. aeruginosa strains and out-compete the original strains [41]. The inability of the epidemic strains to disseminate, even in the setting of neutropenia, fits with the chronic and localized nature of lung infection by these strains in CF patients. These in-vivo characteristics are associated with the serum-sensitivity of the strains, a well-known property of CF isolates obtained from chronically-infected patients [42-44]. Thus it appears that strains of P. aeruginosa do not need to synthesize O-antigens in order to initiate mucosal colonization or resist host innate immunity in the GI tract and lung.
Our findings are also somewhat comparable to those obtained with strain LES in a rat model of chronic lung infection [19]. In this setting, strain LES was recovered from co-infected mice at levels lower than strain PAO1 but at higher levels than strain PA14 [19]. Also, the distribution of these non-epidemic strains in the lungs was different from that of strain LES. Overall, both models showed that the epidemic isolates can be highly successful in establishing tissue colonization and can out-compete some non-epidemic isolates.
Antibiotic depletion of the indigenous murine GI microbial flora that is needed to establish P. aeruginosa GI colonization is likely analogous to what occurs in non-CF hospitalized patients exposed to broad-spectrum antibiotics. Whether antibiotic treatment in CF affects acquisition of P. aeruginosa colonization of their lungs is difficult to ascertain due to the highly variable nature of clinical care of these patients early in life. Studies have shown that treatment of CF patients with antibiotics directed against Staphylococcus aureus leads to earlier acquisition of P. aeruginosa [45, 46] and while the increasing use of early, intensive and routine antibiotic treatments in young CF patients following initial acquisition of P. aeruginosa does lead to temporary eradication, the vast majority of these patients acquire new strains of P. aeruginosa after the conclusion of therapy [47]. Thus it appears that use of antibiotics to condition a mucosal surface for P. aeruginosa colonization may reflect a common situation encountered in clinical medicine.
In conclusion, we developed and validated a murine model of P. aeruginosa acquisition, GI colonization and horizontal transmission. Both epidemic and non-epidemic strains were readily acquired and horizontally transmitted to non-infected mice, and epidemic strains showed a superior ability to mono-colonize and co-colonize the GI mucosa when compared to the other strains. The epidemic strains failed to disseminate, even in the setting of neutropenia, consistent with the actual clinical properties of these strains. Further use of this model may provide critical insights into these early steps in bacterial pathogenesis and possibly help to develop novel means for therapy and infection control.
Acknowledgements
We thank Charles Czuprynski for provision of the RB6-8C5 hybridoma, and Roderick Bronson for murine pathology analysis.
Financial support: National Institute of Health grants RO1 AI22535 (GBP), AI48917 (GBP), AI62983 (AYK) and AI 50036 (GPP).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Vincent JL, Bihari DJ, Suter PM, et al. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee. JAMA. 1995;274:639–44. [PubMed] [Google Scholar]
- 2.Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in pediatric intensive care units in the United States. National Nosocomial Infections Surveillance System. Pediatrics. 1999;103:e39. doi: 10.1542/peds.103.4.e39. [DOI] [PubMed] [Google Scholar]
- 3.Elting LS, Rubenstein EB, Rolston KV, Bodey GP. Outcomes of bacteremia in patients with cancer and neutropenia: observations from two decades of epidemiological and clinical trials. Clin Infect Dis. 1997;25:247–59. doi: 10.1086/514550. [DOI] [PubMed] [Google Scholar]
- 4.Crouch Brewer S, Wunderink RG, Jones CB, Leeper KV., Jr Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest. 1996;109:1019–29. doi: 10.1378/chest.109.4.1019. [DOI] [PubMed] [Google Scholar]
- 5.Siegman-Igra Y, Ravona R, Primerman H, Giladi M. Pseudomonas aeruginosa bacteremia: an analysis of 123 episodes, with particular emphasis on the effect of antibiotic therapy. Int J Infect Dis. 1998;2:211–5. doi: 10.1016/s1201-9712(98)90055-8. [DOI] [PubMed] [Google Scholar]
- 6.Rello J, Rue M, Jubert P, et al. Survival in patients with nosocomial pneumonia: impact of the severity of illness and the etiologic agent. Crit Care Med. 1997;25:1862–7. doi: 10.1097/00003246-199711000-00026. [DOI] [PubMed] [Google Scholar]
- 7.Doring G, Conway SP, Heijerman HG, et al. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: a European consensus. Eur Respir J. 2000;16:749–67. doi: 10.1034/j.1399-3003.2000.16d30.x. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand X, Thouverez M, Talon D, et al. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med. 2001;27:1263–8. doi: 10.1007/s001340100979. [DOI] [PubMed] [Google Scholar]
- 9.Agodi A, Barchitta M, Cipresso R, Giaquinta L, Romeo MA, Denaro C. Pseudomonas aeruginosa carriage, colonization, and infection in ICU patients. Intensive Care Med. 2007;33:1155–61. doi: 10.1007/s00134-007-0671-6. [DOI] [PubMed] [Google Scholar]
- 10.Geddes DM. Of isolates and isolation: Pseudomonas aeruginosa in adults with cystic fibrosis. Lancet. 2001;358:522–3. doi: 10.1016/S0140-6736(01)05742-7. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong DS, Nixon GM, Carzino R, et al. Detection of a widespread clone of Pseudomonas aeruginosa in a pediatric cystic fibrosis clinic. Am J Respir Crit Care Med. 2002;166:983–7. doi: 10.1164/rccm.200204-269OC. [DOI] [PubMed] [Google Scholar]
- 12.Jones AM, Govan JR, Doherty CJ, et al. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet. 2001;358:557–8. doi: 10.1016/s0140-6736(01)05714-2. [DOI] [PubMed] [Google Scholar]
- 13.Cheng K, Smyth RL, Govan JR, et al. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet. 1996;348:639–42. doi: 10.1016/S0140-6736(96)05169-0. [DOI] [PubMed] [Google Scholar]
- 14.Scott FW, Pitt TL. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J Med Microbiol. 2004;53:609–15. doi: 10.1099/jmm.0.45620-0. [DOI] [PubMed] [Google Scholar]
- 15.Coleman FT, Mueschenborn S, Meluleni G, et al. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc. Natl. Acad. Sci. U S A. 2003;100:1949–1954. doi: 10.1073/pnas.0437901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mena A, Macia MD, Borrell N, et al. Inactivation of the mismatch repair system in Pseudomonas aeruginosa attenuates virulence but favors persistence of oropharyngeal colonization in cystic fibrosis mice. J. Bacteriol. 2007;189:3665–3668. doi: 10.1128/JB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh AY, Priebe GP, Pier GB. Virulence of Pseudomonas aeruginosa in a murine model of gastrointestinal colonization and dissemination in neutropenia. Infect Immun. 2005;73:2262–72. doi: 10.1128/IAI.73.4.2262-2272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathee K, Narasimhan G, Valdes C, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U S A. 2008;105:3100–5. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kukavica-Ibrulj I, Bragonzi A, Paroni M, et al. In vivo growth of Pseudomonas aeruginosa strains PAO1 and PA14 and the hypervirulent strain LESB58 in a rat model of chronic lung infection. J Bacteriol. 2008;190:2804–13. doi: 10.1128/JB.01572-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 21.Liberati NT, Urbach JM, Miyata S, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U S A. 2006;103:2833–8. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arora SK, Dasgupta N, Lory S, Ramphal R. Identification of two distinct types of flagellar cap proteins, FliD, in Pseudomonas aeruginosa. Infect. Immun. 2000;68:1474–1479. doi: 10.1128/iai.68.3.1474-1479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradley DE. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 1980;26:146–54. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 24.Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 25.Pier GB, Meluleni G, Neuger E. A murine model of chronic mucosal colonization by Pseudomonas aeruginosa. Infect. Immun. 1992;60:4768–4776. doi: 10.1128/iai.60.11.4768-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhury B, Carlson RW, Goldberg JB. The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohydr. Res. 2005;340:2761–72. doi: 10.1016/j.carres.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Knirel YA, Bystrova OV, Shashkov AS, et al. Structural analysis of the lipopolysaccharide core of a rough, cystic fibrosis isolate of Pseudomonas aeruginosa. Eur J Biochem. 2001;268:4708–19. doi: 10.1046/j.1432-1327.2001.02396.x. [DOI] [PubMed] [Google Scholar]
- 28.Bonten MJ, Bergmans DC, Speijer H, Stobberingh EE. Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units. Implications for infection control. Am J Respir Crit Care Med. 1999;160:1212–9. doi: 10.1164/ajrccm.160.4.9809031. [DOI] [PubMed] [Google Scholar]
- 29.Speert DP, Campbell ME, Davidson AGF, Wong LTK. Pseudomonas aeruginosa colonization of the gastrointestinal tract in patients with cystic fibrosis. J. Infect. Dis. 1993;167:226–229. doi: 10.1093/infdis/167.1.226. [DOI] [PubMed] [Google Scholar]
- 30.Kropec A, Huebner J, Riffel M, et al. Exogenous or endogenous reservoirs of nosocomial Pseudomonas aeruginosa and Staphylococcus aureus infections in a surgical intensive care unit. Intensive Care Med. 1993;19:161–165. doi: 10.1007/BF01720533. [DOI] [PubMed] [Google Scholar]
- 31.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bert F, Maubec E, Bruneau B, Berry P, Lambert-Zechovsky N. Multi-resistant Pseudomonas aeruginosa outbreak associated with contaminated tap water in a neurosurgery intensive care unit. J Hosp Infect. 1998;39:53–62. doi: 10.1016/s0195-6701(98)90243-2. [DOI] [PubMed] [Google Scholar]
- 33.Cobben NA, Drent M, Jonkers M, Wouters EF, Vaneechoutte M, Stobberingh EE. Outbreak of severe Pseudomonas aeruginosa respiratory infections due to contaminated nebulizers. J Hosp Infect. 1996;33:63–70. doi: 10.1016/s0195-6701(96)90030-4. [DOI] [PubMed] [Google Scholar]
- 34.Trautmann M, Lepper PM, Haller M. Ecology of Pseudomonas aeruginosa in the intensive care unit and the evolving role of water outlets as a reservoir of the organism. Am J Infect Control. 2005;33:S41–9. doi: 10.1016/j.ajic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Rogues AM, Boulestreau H, Lasheras A, et al. Contribution of tap water to patient colonisation with Pseudomonas aeruginosa in a medical intensive care unit. J Hosp Infect. 2007;67:72–8. doi: 10.1016/j.jhin.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 36.Jones AM, Govan JR, Doherty CJ, et al. Identification of airborne dissemination of epidemic multiresistant strains of Pseudomonas aeruginosa at a CF centre during a cross infection outbreak. Thorax. 2003;58:525–7. doi: 10.1136/thorax.58.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panagea S, Winstanley C, Walshaw MJ, Ledson MJ, Hart CA. Environmental contamination with an epidemic strain of Pseudomonas aeruginosa in a Liverpool cystic fibrosis centre, and study of its survival on dry surfaces. J Hosp Infect. 2005;59:102–7. doi: 10.1016/j.jhin.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Al-Aloul M, Crawley J, Winstanley C, Hart CA, Ledson MJ, Walshaw MJ. Increased morbidity associated with chronic infection by an epidemic Pseudomonas aeruginosa strain in CF patients. Thorax. 2004;59:334–6. doi: 10.1136/thx.2003.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clifton IJ, Fletcher LA, Beggs CB, Denton M, Peckham DG. A laminar flow model of aerosol survival of epidemic and non-epidemic strains of Pseudomonas aeruginosa isolated from people with cystic fibrosis. BMC Microbiol. 2008;8:105. doi: 10.1186/1471-2180-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winstanley C, Langille MG, Fothergill JL, et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2008 doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCallum SJ, Corkill J, Gallagher M, Ledson MJ, Hart CA, Walshaw MJ. Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet. 2001;358:558–60. doi: 10.1016/s0140-6736(01)05715-4. [DOI] [PubMed] [Google Scholar]
- 42.Pier GB, Ames P. Mediation of the killing of rough, mucoid isolates of Pseudomonas aeruginosa from patients with cystic fibrosis by the alternative pathway of complement. J. Infect. Dis. 1984;150:223–228. doi: 10.1093/infdis/150.2.223. [DOI] [PubMed] [Google Scholar]
- 43.Schiller NL, Hatch RA, Joiner KA. Complement activation and C3 binding by serum-sensitive and serum-resistant strains of Pseudomonas aeruginosa. Infect. Immun. 1989;57:1707–1713. doi: 10.1128/iai.57.6.1707-1713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hancock REW, Mutharia LM, Chan L, Darveau RP, Speert DP, Pier GB. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: A class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side-chains. Infect. Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stutman HR, Lieberman JM, Nussbaum E, Marks MI. Antibiotic prophylaxis in infants and young children with cystic fibrosis: A randomized controlled trial. J Pediatr. 2002;140:299–305. doi: 10.1067/mpd.2002.121930. [DOI] [PubMed] [Google Scholar]
- 46.Ratjen F, Comes G, Paul K, Posselt HG, Wagner TO, Harms K. Effect of continuous antistaphylococcal therapy on the rate of P. aeruginosa acquisition in patients with cystic fibrosis. Pediatr. Pulmonol. 2001;31:13–16. doi: 10.1002/1099-0496(200101)31:1<13::aid-ppul1001>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 47.Taccetti G, Campana S, Festini F, Mascherini M, Doring G. Early eradication therapy against Pseudomonas aeruginosa in cystic fibrosis patients. Eur. Respir. J. 2005;26:458–61. doi: 10.1183/09031936.05.00009605. [DOI] [PubMed] [Google Scholar]
- 48.Priebe GP, Dean CR, Zaidi T, et al. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect Immun. 2004;72:4224–32. doi: 10.1128/IAI.72.7.4224-4232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preston MJ, Fleiszig SM, Zaidi TS, et al. Rapid and sensitive method for evaluating Pseudomonas aeruginosa virulence factors during corneal infections in mice. Infect Immun. 1995;63:3497–501. doi: 10.1128/iai.63.9.3497-3501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]


