Summary
Newcastle disease virus (NDV) is an avian paramyxovirus that exists as hundreds of strains with widely different virulence properties. The NDV V protein exhibits interferon (IFN) antagonistic activity, which contributes to the virulence of the virus. The IFN antagonistic activities of the V proteins from the avirulent strain La Sota and the moderately virulent strain Beaudette C (BC) were compared in an assay for the rescue of a recombinant NDV expressing the green fluorescent protein (NDV-GFP). Consistent with the virulence properties of the two viruses, the BC V protein exhibits a 4-fold greater ability to rescue replication of NDV-GFP than the La Sota V protein. Four amino acid differences in the C-terminal region of V, as well as the N-terminal region, contribute to the difference in IFN antagonistic activity between the two V proteins.
Newcastle disease virus (NDV) is a member of the Paramyxoviridae family of enveloped negative-stranded RNA viruses, which also includes Sendai virus, the various parainfluenza viruses, measles virus, respiratory syncytial virus and the henipaviruses. The NDV genome contains six genes, which encode six major proteins: nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase protein (HN) and the large (L) polymerase (Lamb and Parks, 2007). NDV also produces the V and W proteins by RNA editing during P gene transcription. The P gene mRNA is edited by insertion of one or two additional G residues into a run of G's within the conserved editing site, thus generating the V- and W-encoding mRNAs, respectively (Steward et al., 1993).
NDV causes respiratory, neurological or enteric disease in birds. Strains are classified into three pathotypes. Avirulent (lentogenic) strains cause mild or asymptomatic infections, whereas virulent (velogenic) strains cause high mortality. Strains of intermediate virulence are called mesogenic (Alexander, 1997). NDV is also being used as a vaccine vector (Huang et al., 2003a; Dinapoli et al., 2009) and oncolytic agent due to its ability to kill tumor cells (Elankumaran et al., 2006; Freeman et al., 2006).
Cleavage of the F protein precursor (F0) produces the active fusion protein (Scheid and Choppin, 1974) and is the primary determinant of virulence as determined by the number of basic residues in the cleavage site (Glickman et al., 1988; Nagai et al., 1976; Toyoda et al., 1989). However, other viral proteins contribute to virulence (Panda et al., 2004; Peeters et al., 1999). Recombinant viruses lacking V have impaired growth in cell culture and embryonated chicken eggs and are highly attenuated in young chickens (Huang et al., 2003b; Mebatsion et al., 2001). These mutant viruses also exhibit increased sensitivity to exogenous interferon (IFN) (Elankumaran et al., 2006; Huang et al., 2003b). Using an IFN-sensitive NDV-GFP-based assay, it was demonstrated that the NDV V protein possesses IFN antagonistic activity, defined by the C-terminal region of the protein (Park et al., 2003). This is consistent with the IFN-antagonistic activity of the NDV V protein contributing to virulence. However, the role of V in the differential virulence patterns exhibited by NDV pathotypes has not been examined.
Here, the NDV-GFP-based assay (Park et al., 2003) was used to compare the relative IFN antagonistic activities of the V proteins from mesogenic strain Beaudette C (BC) and lentogenic strain LaSota. DF1 cells (chicken embryo fibroblast cell line) (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco's Modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 2 mM L-glutamine, 4 U/ml penicillin and 4 μg/ml streptomycin. The enhanced GFP gene was inserted between the P and M genes of the BC cDNA and the virus was rescued from cDNA (Elankumaran et al., 2006). It was demonstrated that the NDV-BC-GFP virus is susceptible to IFN by inhibition of growth following treatment with 1000 U/ml of chicken IFN-α (AbD serotec, Kidlington, Oxford, UK) prior to infection (data not shown).
The P genes of the La Sota and BC viruses were cloned into pBluescript SK(+) (pBSK) (Stratagene, La Jolla, CA) within a few egg passages of the original stock (Veterinary Services Laboratory, Ames, IA). Each V gene was generated from the respective P gene by insertion of a G nucleotide into the editing site as described previously (Corey and Iorio, 2007). Mutated V genes were prepared using the same protocol. The presence of all mutations was confirmed by DNA sequencing. The wild type (wt) or mutated V genes were subcloned into pCAGGS by blunt-end ligation.
The IFN antagonistic activities of the V proteins were tested by their ability to rescue growth of NDV-GFP virus. DF1 cells were seeded in 6-well plates and transfected in triplicate at 80% confluence using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were washed with PBS and infected with NDV-GFP (moi of 0.001). Virus growth was monitored at 24 h post-infection (Fig. 1A) and quantitated by counting fluorescent cells in 3-5 fields (approximately 3000 cells) (Fig. 1B). It should be noted that, although the NDV-GFP virus is a BC virus and has an intact V open reading frame, the IFN-induced inhibition of NDV-GFP growth occurs prior to infection. Thus, within the time frame of the assay, an antiviral state has already been established before the V protein is expressed from the virus rendering it a non-factor.
Fig 1.
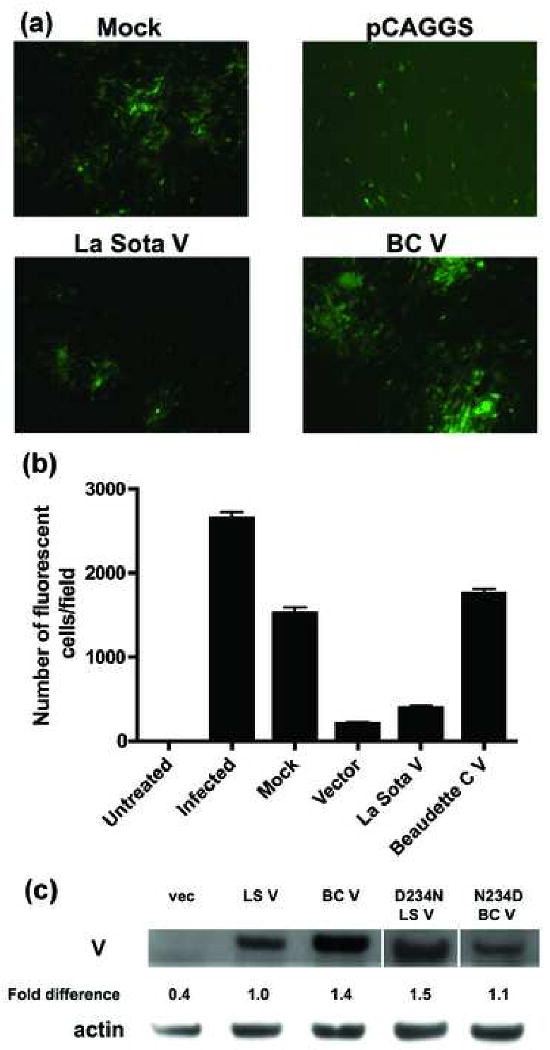
BC V exhibits a four-fold greater ability than La Sota V to rescue growth of NDV-GFP. (A) DF1 cells were mock-transfected, or transfected with empty pCAGGS vector, La Sota V or BC V. After 24 h, the cells were infected with NDV-GFP virus and examined for fluorescence 24 h later. (B) The growth of NDV-GFP virus was quantitated by counting the number of fluorescent cells from three different fields. The average values and standard deviations (error bars) are shown for one experiment out of a total of four. The absolute numbers of fluorescent cells vary from one experiment to another but the relative activities are consistent. (C) DF1 cells were transfected with wt or mutant V plasmids. After 24 h, lysates were prepared and Western blot was performed using V18 antiserum. V protein levels standardized to actin were determined by densitometry and are expressed relative to wt (set at 100%). These data represent one out of at least two experiments.
Although the virus grows well in mock-transfected cells, transfection with the empty pCAGGS vector inhibits growth by inducing IFN and the subsequent antiviral state, as shown previously by Park et al. (2003). Transfection with empty vector results in a significant decrease (211 ± 15 infected cells) in virus growth as compared to mock (1522 ± 70 infected cells) (Fig. 1B). Transcription from a plasmid may lead to generation of double-stranded RNA or plasmid DNA may be recognized by a cytosolic DNA sensor, triggering IFN production. Most importantly, expression of either V protein rescues virus growth (Fig. 1A), but with BC V (1753 ± 58 infected cells) doing so four times more efficiently than La Sota V (394 ± 29 infected cells) (Fig. 1B). This correlates with the difference in virulence between the two strains of the virus.
One possibility is that the difference in IFN antagonistic activity of the two V proteins is due merely to differences in expression. Western blots were performed at 24 h post-transfection to quantitate V protein expression. Briefly, the cells were scraped into cold phosphate buffered saline (PBS), pelleted for 5 min at 16,000×g and lysed for 30 minutes using 50 μl NP-40 cell lysis buffer (Biosource International, Inc., Camarillo, CA) supplemented with 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Sigma Chemical Co., St. Louis, MO). Twenty μl of the cleared lysate were electrophoresed on a NuPAGE 4-12% Bis-Tris gel (Invitrogen) under reducing conditions. Proteins were transferred onto an Immobilon-P membrane (Millipore, Bedford, MA) and blocked overnight at 4°C with Detector Block (Kirkegaard and Perry Laboratories, Gaithersburg, MD). A polyclonal antibody (V18) raised in rabbits against the C-terminal 18 amino acids of the BC V protein was used followed by horseradish peroxidase-conjugated goat anti-rabbit antibody (Kirkegaard and Perry Laboratories). Proteins were visualized using the Amersham ECL Western Blotting Analysis System (GE Healthcare, Buckinghamshire, UK). The membrane was stripped using Re-Blot Plus Strong Solution (Chemicon International, Temecula, CA), blocked overnight and re-probed with anti-actin (Sigma).
Both V proteins are expressed, although the BC V protein is detected 1.4-fold more efficiently than LaSota V (Fig. 1C). There are two possible explanations for this result. One is that the BC protein is actually expressed more efficiently. Alternatively, the V18 antiserum may recognize La Sota V less efficiently than it does BC V. The antiserum is directed against the C-terminal sequence of BC V and there is a difference between La Sota (aspartic acid) and BC (asparagine) at amino acid 234 in this region.
To determine whether this amino acid difference modulates antiserum reactivity, both genes were mutated at position 234 to the amino acid in the other protein. D234N-mutated La Sota V is recognized by the V18 antiserum at a level similar to wt BC V and N234D-mutated BC V is recognized at a reduced level similar to wt La Sota V (Fig. 1C). Hence, the amino acid difference at position 234 appears to account for the differential recognition of the two V proteins by the V18 antiserum. Still, we cannot definitively rule out the possibility that the residue at position 234 modulates expression except to note that, if it does completely account for the observed difference in IFN antagonism between the two V proteins, then a 40% difference in expression would have to account for a four-fold difference in IFN antagonism.
Previously, the C-terminal region of NDV V was demonstrated to be necessary and sufficient for its IFN antagonistic activity (Park et al. 2003). The C-terminal regions of the two proteins have four amino acid differences at positions 144, 153, 161 and 234. These residues are S, E, S and D, respectively, in La Sota V and P, K, P and N, respectively, in BC V (Fig. 2). To test the contribution of these residues to IFN antagonistic activity, both proteins were mutated at each position to the amino acid in the other protein and the IFN antagonistic activities of the mutated proteins were determined (Fig. 3A). All of the La Sota V proteins carrying single mutations have an increased ability to rescue growth of the NDV-GFP virus, ranging from between 512 ± 13 and 761 ± 27 compared to 394 ± 29 for La Sota V. Conversely, all of the individually mutated BC V proteins have decreased ability to rescue growth compared to wt ranging from 569 ± 40 to 1329 ± 138, relative to BC V (1753 ± 58) (Fig. 3B). Thus, each residue contributes to IFN antagonism.
Fig. 2.
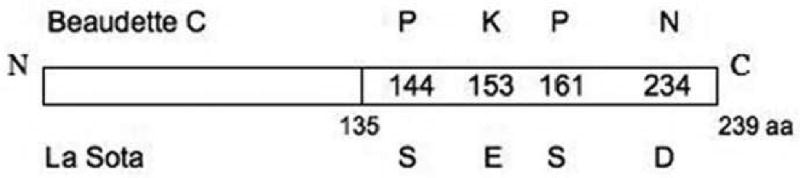
Schematic representation of the amino acid differences in the BC and LaSota V proteins. The figure shows the residues at the four positions in the C-terminal region that differ between the two proteins.
Fig 3.
Effect of wt and mutated V proteins on the growth of NDV-GFP virus. DF1 cells were mock-transfected (mock), or transfected with empty vector (vector), La Sota wt or mutated V plasmids (panel A) or BC wt or mutated V plasmids (panel B). After 24 h, the cells were infected with NDV-GFP at a moi of 0.001 and examined for fluorescence 24 h later. In panels C and D, the percent expression relative to wt (open bars) is shown for mutated LaSota (panel C) and BC V proteins (panel D), along with the percent of wt rescue of growth of NDV-GFP virus (filled bars), based on the data in panels A and B. Expression levels were determined as described in the legend to Fig. 1C. These data represent one experiment out of at least two.
To determine the possible contribution of differences in expression to the differences in IFN antagonism, the plasmid constructs were transfected into DF1 cells and Western blots were performed using the V18 antibody. With the exception of the D234N-mutated La Sota V protein, the increased IFN antagonistic activities (filled bars) of the individually mutated proteins cannot be accounted for by increased expression (open bars) (Fig. 3C). This exception is consistent with the results in Fig. 1C. Conversely, none of the individually mutated BC V proteins exhibits an expression level reduced enough to account for its reduced IFN antagonism (Fig. 3D).
To test whether the mutations in the C-terminal region of V have an additive effect, double, triple and quadruple mutants of both wt V proteins were evaluated. The double mutated proteins of La Sota V are better than the single mutated proteins in rescue of viral growth (Fig. 3A). The number of fluorescent cells for La Sota V S144P/E153K and S161P/D234N mutated proteins was 1144 ± 17 and 897 ± 28, respectively. However, the triple and quadruple mutated proteins of La Sota V are not significantly more effective than the double mutated proteins.
Fig. 3B shows that the BC V P144S/K153E double mutated protein (645 ± 47 fluorescent cells) has a decreased ability to rescue the virus as compared to the proteins carrying individual P144S and K153E mutations. The BC V P161S/N234D double mutated protein rescues viral growth at almost the same level as the P161S and N234D mutated proteins. Surprisingly, the triple and quadruple mutated proteins of BC V are better at rescuing the NDV-GFP virus than the double mutated proteins. In no case can increased IFN antagonistic activity be accounted for by increased expression (Fig. 3D). For the BC V protein carrying multiple mutations, the results are less clear. For the two proteins carrying N234D mutations (P161S/N234D and the quadruple mutated protein), reduced rescue can be accounted for by reduced expression. This correlates with the presence of the mutation at position 234, though the single mutant does not exhibit this problem.
Overall, the ability of the mutated V proteins to rescue growth of NDV-GFP does not correlate with the expression levels. For example, even though the V18 antibody detects the BC V quadruple mutated protein at only 20% of BC V, it rescues growth of the virus at a greater level than that of other mutated proteins that are recognized to a greater extent by the antibody (i.e., BC V P161S-mutated protein which is recognized at 100% of BC V). Most importantly, neither the quadruple-mutated LaSota V nor BC V protein exhibits rescue activity commensurate with the other wt V protein. Thus, other differences between the two proteins located in the N-terminal region of V contribute to their differences in IFN antagonistic activity.
The V proteins of paramyxoviruses show 44% overall homology, with the region of highest homology being the C-terminus, which contains seven conserved cysteines capable of binding two zinc atoms (Paterson et al., 1995; Steward et al., 1995). Several studies point to the importance of the C-terminus of the paramyxovirus V protein to IFN antagonism and virulence (Fukuhara et al., 2002; He et al., 2002; Huang et al., 2000; Kawano et al., 2001; Nishio et al., 2001; Park et al., 2003). Our results indicate that residues 144, 153, 161 and 234 in the C-terminal region of the NDV V protein all contribute to IFN antagonistic activity.
However, the quadruple mutated La Sota and BC V proteins fail to convert rescue to the level of the other protein. This indicates that residues in the N-terminal region of V also contribute to IFN antagonistic activity. This is consistent with earlier results obtained with other paramyxoviruses (Chatziandreou et al., 2002; Fontana et al., 2008). It is conceivable that N-terminal changes could affect the structure of the protein making the C-terminus less accessible to interacting proteins. There are five amino acid differences between La Sota and BC in the N-terminal region at residues 30, 41, 46, 65 and 82. It will be interesting to determine how the identity of the residues at these positions affects IFN antagonistic activity. This is the first evidence that the N-terminus of NDV V contributes to its IFN antagonistic activity.
Though we unable to examine the IFN antagonistic activity of a V protein from a velogenic strain due to the requirement for BSL-3 containment for these viruses, we have compared the sequence of velogenic strains (Locke et al., 2000) to that of La Sota. Surprisingly, lentogenic strains are more similar to velogenic strains than they are to mesogenic strains at positions 144, 153 and 234 and there is no pattern for residue 161 for these strains. In addition, there are no amino acids anywhere in the V protein that are specific for any of the three pathotypes (data not shown).
There are hundreds of NDV isolates, which exhibit widely different virulence patterns. The attenuation of virulence in a recombinant NDV that fails to express the V protein has been attributed to its inability to antagonize IFN-α (Huang et al., 2003b). To better understand the role of the V protein in virulence, we compared the IFN antagonistic activities of the V proteins from a lentogenic and a mesogenic strain. We showed that the V protein from a mesogenic strain has a four-fold greater ability to rescue the growth of an NDV-GFP virus than that from an avirulent, lentogenic strain. This is consistent with the two proteins having significantly different IFN antagonistic activities. These differences correlate with their virulence and suggests that the IFN antagonistic activity of the NDV V protein may have an important role in the virulence of NDV. This possibility can be tested by evaluation of recombinant viruses in which the two V proteins have been exchanged.
Acknowledgments
We thank Matteo Porotto and Anne Moscona for the pCAGGS vector. We thank Katherine Fitzgerald, Nadege Goutagny, Paul Rota and Madelyn Schmidt for helpful suggestions. We also thank Daniel Rockemann for technical assistance. We are grateful to Paul Mahon for his help in the preparation of the manuscript. This work was supported by grant AI-49268 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander DJ. Newcastle disease and other avian Paramyxoviridae infections. In: Calnek BW, editor. Diseases of poultry. 10th. Iowa State University Press; Ames: 1997. pp. 541–569. [Google Scholar]
- Chatziandreou N, Young D, Andrejeva J, Goodbourn S, Randall RE. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology. 2002;293:234–42. doi: 10.1006/viro.2001.1302. [DOI] [PubMed] [Google Scholar]
- Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol. 2007;81:9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinapoli JM, Ward JM, Chen L, yang L, Elankumaran S, Murphy BR, Samal SK, Collins PL, Bukreyev A. Delivery to the lower respiratory tract is required for effective immunization with Newcastle disease virus-vectored vaccines intended for humans. Vaccine. 2009;27:1530–1539. doi: 10.1016/j.vaccine.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elankumaran S, Rockemann D, Samal SK. Newcastle disease virus exerts oncolysis by both intrinsic and extrinsic caspase-dependent pathways of cell death. J Virol. 2006;80:7522–34. doi: 10.1128/JVI.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana JM, Bankamp B, Bellini WJ, Rota PA. Regulation of interferon signaling by the C and V proteins from attenuated and wild-type strains of measles virus. Virology. 2008;374:71–81. doi: 10.1016/j.virol.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Freeman AI, Zakay-Rones Z, Gomori JM, Linetsky E, Rasooly L, Greenbaum E, Rozenman-Yair S, Panet A, Libson E, Irving CS, Galun E, Siegal T. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol Ther. 2006;13:221–8. doi: 10.1016/j.ymthe.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Huang C, Kiyotani K, Yoshida T, Sakaguchi T. Mutational analysis of the Sendai virus V protein: importance of the conserved residues for Zn binding, virus pathogenesis, and efficient RNA editing. Virology. 2002;299:172–81. doi: 10.1006/viro.2002.1516. [DOI] [PubMed] [Google Scholar]
- Glickman RL, Syddall RJ, Iorio RM, Sheehan JP, Bratt MA. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J Virol. 1988;62:354–6. doi: 10.1128/jvi.62.1.354-356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Paterson RG, Stock N, Durbin JE, Durbin RK, Goodbourn S, Randall RE, Lamb RA. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology. 2002;303:15–32. doi: 10.1006/viro.2002.1738. [DOI] [PubMed] [Google Scholar]
- Huang C, Kiyotani K, Fujii Y, Fukuhara N, Kato A, Nagai Y, Yoshida T, Sakaguchi T. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J Virol. 2000;74:7834–7841. doi: 10.1128/jvi.74.17.7834-7841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Elankumaran S, Panda A, Samal SK. Recombinant Newcastle disease virus as a vaccine vector. Poult Sci. 2003a;82:899–906. doi: 10.1093/ps/82.6.899. [DOI] [PubMed] [Google Scholar]
- Huang Z, Krishnamurthy S, Panda A, Samal SK. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J Virol. 2003b;77:8676–85. doi: 10.1128/JVI.77.16.8676-8685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Kaito M, Kozuka Y, Komada H, Noda N, Nanba K, Tsurudome M, Ito M, Nishio M, Ito Y. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology. 2001;284:99–112. doi: 10.1006/viro.2001.0864. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th. Vol. 1. Woltus Kluwer/Lippincott Williams and Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- Locke DP, Selleres HS, Crawford JM, Schultz-Cherry S, King DJ, Meinersmann RJ, Seal BS. Newcastle disease virus phosphoproein gene analysis and transcriptional editing in avian cells. Virus Res. 2000;69:55–68. doi: 10.1016/s0168-1702(00)00175-1. [DOI] [PubMed] [Google Scholar]
- Mebatsion T, Verstegen S, De Vaan LT, Romer-Oberdorfer A, Schrier CC. A recombinant newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J Virol. 2001;75:420–8. doi: 10.1128/JVI.75.1.420-428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y, Klenk HD, Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72:494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- Nishio M, Tsurudome M, Ito M, Kawano M, Komada H, Ito Y. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J Virol. 2001;75:9165–76. doi: 10.1128/JVI.75.19.9165-9176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda A, Huang Z, Elankumaran S, Rockemann DD, Samal SK. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb Pathog. 2004;36:1–10. doi: 10.1016/j.micpath.2003.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003;77:1501–11. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson RG, Leser GP, Shaughnessy MA, Lamb RA. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology. 1995;208:121–31. doi: 10.1006/viro.1995.1135. [DOI] [PubMed] [Google Scholar]
- Peeters BP, de Leeuw OS, Koch G, Gielkens AL. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J Virol. 1999;73:5001–9. doi: 10.1128/jvi.73.6.5001-5009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid A, Choppin PW. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology. 1974;57:475–90. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- Steward M, Samson AC, Errington W, Emmerson PT. The Newcastle disease virus V protein binds zinc. Arch Virol. 1995;140:1321–8. doi: 10.1007/BF01322759. [DOI] [PubMed] [Google Scholar]
- Steward M, Vipond IB, Millar NS, Emmerson PT. RNA editing in Newcastle disease virus. J Gen Virol. 1993;74(Pt 12):2539–47. doi: 10.1099/0022-1317-74-12-2539. [DOI] [PubMed] [Google Scholar]
- Toyoda T, Sakaguchi T, Hirota H, Gotoh B, Kuma K, Miyata T, Nagai Y. Newcastle disease virus evolution. II. Lack of gene recombination in generating virulent and avirulent strains. Virology. 1989;169:273–82. doi: 10.1016/0042-6822(89)90152-9. [DOI] [PubMed] [Google Scholar]


