Abstract
Recent studies indicate that the small heat shock protein αB-crystallin is expressed in poor prognosis basal-like breast tumors and likely contributes to their aggressive phenotype. However, the mechanisms underlying the deregulated expression of αB-crystallin in basal-like tumors are poorly understood. Using a bioinformatics approach, we identified a putative DNA binding motif in the human αB-crystallin promoter for the proto-oncogene Ets1, a member of the ETS transcription factor family that bind to DNA at palindromic ETS-binding sites (EBS). Here we demonstrate that ectopic expression of Ets1 activates the αB-crystallin promoter by an EBS-dependent mechanism and increases αB-crystallin protein levels, while silencing Ets1 reduces αB-crystallin promoter activity and protein levels. Chromatin immunoprecipitation analyses showed that endogenous Ets1 binds to the αB-crystallin promoter in basal-like breast cancer cells in vivo. Interrogation of publically available gene expression data revealed that Ets1 is expressed in human basal-like breast tumors and is associated with poor survival. Collectively, our results point to a previously unrecognized link between the oncogenic transcription factor Ets1 and αB-crystallin in basal-like breast cancer.
Keywords: αB-crystallin, Ets1, molecular chaperone, basal-like breast cancer, gene regulation
Introduction
The small heat shock protein αB-crystallin is a molecular chaperone that enhances survival in response to cellular stress by inhibiting protein aggregation and reducing intracellular reactive oxygen species levels [1–3]. In addition, αB-crystallin directly interacts with the cell death machinery to suppress apoptosis by inhibiting caspase-3 activation and preventing the mitochondrial translocation of proapoptotic Bcl-2 family members Bax and Bcl-xs [4–6]. αB-crystallin is commonly expressed in many cancers, and its expression correlates with poor clinical outcomes in breast and head and neck carcinomas [7–9]. Consistent with its antiapoptotic function, αB-crystallin expression in breast cancer is associated with resistance to neoadjuvant chemotherapy [10]. Moreover, αB-crystallin is predominantly expressed in a subset of poor prognosis, triple (ER/PR/HER2) negative breast tumors with a basal epithelial gene expression profile (basal-like breast cancer) and likely contributes to the aggressive phenotype of these tumors [9–11].
Despite its emerging pathogenic significance in cancer, very little is known about the regulation of αB-crystallin gene (CRYAB) expression in breast cancer or other malignancies. The mammalian αB-crystallin gene and the adjacent small heat shock protein HspB2/MKBP gene are arranged head-to-head and share a conserved intergenic promoter that is differentially transcribed in an orientation-specific manner [12, 13]. Unlike in cancer, the transcriptional regulation of the murine αB-crystallin gene in normal tissue, such as muscle and lens, has been studied extensively. Multiple tissue-specific regulatory elements activated by distinct transcription factors, including MyoD, Pax-6 and HSF, have been identified [14–16].
In the present study, we used a bioinformatics approach to identify putative transcriptional regulators of the human αB-crystallin gene in breast cancer cells. One such candidate we identified was the oncogenic transcription factor Ets1. Ets1 is a cellular homologue of the avian erythroblastosis E26 viral oncogene that contains a conserved 85 amino acid ETS DNA binding domain that forms a winged helix-turn-helix motif [17, 18]. ETS family members bind to palindromic ETS-binding sites (EBS) composed of a 5’-GGA(A/T)-3’ consensus core sequence and regulate expression of genes involved in proliferation (Myc), invasion (matrix metalloproteinase (MMP)-1, MMP-3, MMP-9 and urokinase plasminogen activator), and angiogenesis (VEGF receptor 1). Here we report that Ets1 binds to the αB-crystallin promoter and regulates its expression by an EBS-dependent mechanism. We also show that overexpression of Ets1 in breast cancer cells increases αB-crystallin protein levels, while silencing Ets1 reduces αB-crystallin levels. In addition, we demonstrate that Ets1 is expressed in basal-like tumors from patients and is associated with poor survival. Taken together, our results point to a previously unrecognized and direct link between the proto-oncogene Ets1 and αB-crystallin in basal-like breast cancer.
Materials and Methods
Bioinformatics analysis of the shared human αB-crystallin/HspB2 promoter
The human αB-crystallin promoter, the 1111 base pair intergenic region between the ATG translational initiation sites of the adjacent αB-crystallin and HspB2 genes [12], was queried for DNA motifs within the TRANSFAC library using an internet-based MOTIF search (http://motif.genome.jp/). A cut-off score of 80 was used to identify putative transcription factor DNA binding elements.
Cell Culture
Human MCF-10A breast epithelial cells and MDA-MB-231 breast carcinoma cells were purchased from ATCC. MCF-10A cells were cultured in DMEM/F12 medium (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml of EGF (Sigma-Aldrich), 10 µg/ml insulin (Sigma-Aldrich), 0.5 mg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml cholera toxin (Sigma-Aldrich), and 1X Penicillin-Streptomycin-Glutamine (Invitrogen). MDA-MB-231 cells were grown in MEM medium with Earl’s salts plus L-glutamine (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 1X non-essential amino acids (Mediatech), 10 mM HEPES buffer (Mediatech) and 1X Penicillin-Streptomycin-Glutamine.
Reporter Assays
The full-length human αB-crystallin gene promoter (−1081/+30 relative to the transcription start site) and 5’-truncated promoter constructs were PCR amplified from genomic DNA isolated from human MCF-10A breast epithelial cells using KOD Hot Start DNA Polymerase (Novagen) according to manufacturer’s protocol using a sense primer containing a SacI restriction enzyme cleavage site (5’-CGAGCTCCATGGCTGCAGATGCAGC-3’ (full-length promoter), 5’-CGAGCTCTGGTGCTGACATGTTGACC-3’ (−516 truncation) or 5’-CGAGCTCACACTACGCCGGCTCCCATC-3’ (−356 truncation)) and an antisense primer containing a BglII site (5’-GGAAGATCTCATGGTGGCTAGGTGAGTGTGGGG-3’). PCR products were subcloned into the SacI and BglII sites of the pGL3-Basic luciferase reporter plasmid (Promega). The putative EBS was mutated using the QuickChange Site-Directed Mutagenesis kit (Stratagene) and the following primers: 5’-CCTGGGGCTCAGCCTAAAAAGATTTTAGTCCC-3’ and 5’-GGGACTAAAATGTTTTTAGGCTGAGCCCCAGG-3’. All constructs were verified by DNA sequencing. For ectopic expression experiments, MCF-10A cells were transiently co-transfected with 700 ng of pcDNA3.1-Ets1 (kindly provided by Dr. M. Zhou, Emory University) or pcDNA3.1 vector, 100 ng of pGL3 firefly luciferase reporter, and 1 ng of control pRL-TK Renilla luciferase reporter using Lipofectamine 2000 (Invitrogen). A reporter pGL3-MMP9 (kindly provided by Dr. M. Sharon Stack, University of Missouri) was used as a positive control. Lysates were assayed for luciferase activity 48 h later with the Dual Luciferase Reporter Assay system (Promega) using a Clarity Luminescence Microplate Reader (Bio-Tek). Firefly luciferase activity was normalized to Renilla luciferase activity and expressed as fold induction relative to the activity in empty vector-transfected cells. For gene silencing experiments, MDA-MB-231 cells were transiently co-transfected with 300 ng of pGL3 reporter, 3 ng of pRL-TK reporter and 25 µM of siRNA targeting Ets1 (Dharmacon ON-TARGETplus SMARTpool) or lamin A/C (control, Dharmacon) using Lipofectamine 2000. Luciferase activity was measured 72 h later as described above and expressed as fold induction relative to the activity in cells transfected with control lamin A/C siRNA.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using the LightShift Chemiluminescent EMSA kit (Pierce). The human αB-crystallin promoter region containing the putative EBS (WT: 5’-TCAGCCTAGGAAGATTCCAGTCCCTGC-3’) or a mutant EBS (5’-TCAGCCTAAAAAGATTTTAGTCCCTGC-3’) was duplexed with the corresponding antisense oligonucleotide and used as probes. Twenty fmol of biotinylated WT or mutant probe was added to 10 µg of nuclear extract (Santa Cruz Biotechnology) for 20 minutes at RT. For competition experiments, 20 fmol of biotinylated WT probe was incubated in the presence of 4 pmol of unbiotinylated WT probe. For antibody abrogation experiments, 5 µg of Ets1 antibody (SantaCruz Biotechnologies; sc-111) was added to the binding reaction for 20 minutes at RT before adding the WT probe; the reaction was then incubated for an additional 20 minutes at RT. Protein-DNA complexes were resolved by 5% native-PAGE using 0.5 × Tris-Borate EDTA buffer, transferred to a Biodyne B membrane (Pall Life Sciences) and UV cross-linked to the membrane. Biotin-labeled DNA was detected by chemiluminescence following the manufacturer’s protocol.
Chromatin immunoprecipitation (ChIP)
ChIP was performed using the EZ ChIP kit (Upstate) according to the manufacturer's protocol. Briefly, ~2 × 107 MDA-MB-231 cells were fixed with 1% formaldehyde for 10 min at RT. Crosslinked chromatin was sonicated using the Bioruptor 200 (Diagenode) for 30 min at full power with 30 second on/off cycles. Sheared chromatin was pre-cleared and incubated with 2 µg of antibody (Ets1 or control rabbit IgG; SantaCruz Biotechnologies, catalogue no. sc-350 and sc-2027, respectively). Immunoprecipitated complexes were washed and eluted, crosslinks reversed, and samples RNase-treated according to the manufacturer’s protocol. DNA was isolated using the QIAquick PCR Purification kit (QIAGEN) and PCR amplified with the following primers for the αB-crystallin promoter: 5’-AGATGGCTGGTGCTGACATGTTGA-3’; 5’-AATCAGGCCAGCAACTATCTTGGG-3. PCR products were resolved on a 2% agarose gel.
Creation of breast cancer cells stably overexpressing Ets1
Retroviral supernatants were generated by transfecting the Phoenix amphotrophic retrovirus packaging cell line (ATCC) with pBABE-Ets1 and pBABE vector as described previously [9]. MDA-MB-231 cells were retrovirally transduced as described [9] and puromycin-resistant pools were selected by growth in 2 µg/ml puromycin. The expression of Ets1 and αB-crystallin in puromycin-resistant pools was determined by immunoblotting as described using the following primary antibodies: αB-crystallin (Stressgen Biotechnologies; SPA-222), Ets1 (Abcam, ab10936) or actin (Sigma-Aldrich; # A4700).
Analysis of Ets1 gene expression data from human breast tumors
The expression of the human Ets1 gene in human breast tumors was determined from publically available gene profiling datasets by Oncomine analysis (http://www.oncomine.org/) as described [19].
Statistical Analyses
The statistical significance of differences for reporter assays was determined by ANOVA with a Bonferroni posttest using Prism 4 sofware (GraphPad).
Results
Identification of a putative Ets1 binding site in the shared human αB-Crystallin/HspB2 promoter
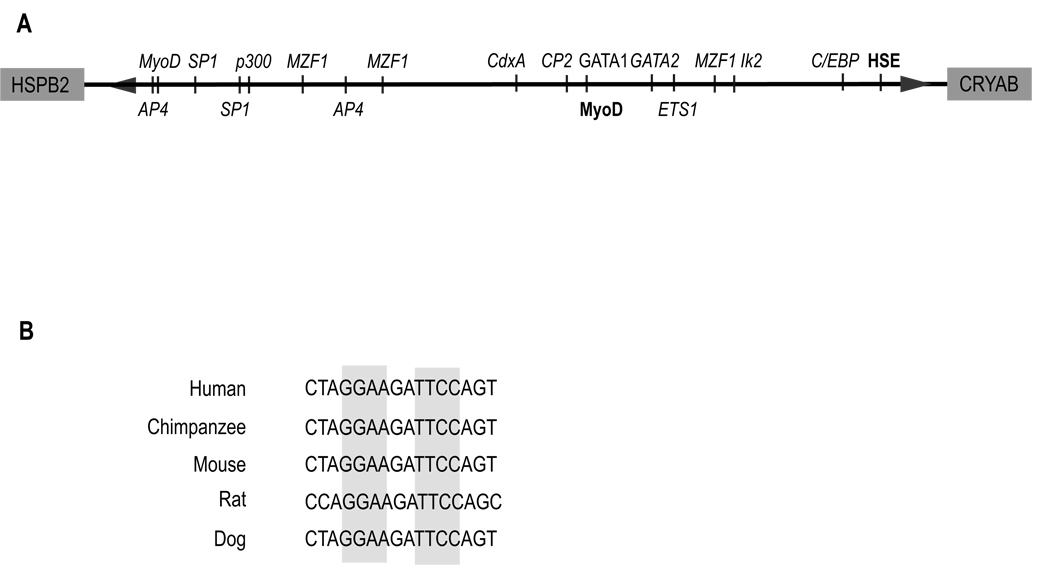
We examined the shared human αB-crystallin/HspB2 promoter for putative transcription factor DNA binding elements using an internet-based MOTIF search. Our analysis identified previously published MyoD and heat shock elements [14, 16], as well as a putative binding site for Ets1, a member of the ETS family of transcription factors (Fig. 1A). The putative palindromic EBS at −441 to −432 in the human αB-crystallin promoter (Fig. 1B, highlighted in gray) is absolutely conserved across many mammalian species.
Figure 1. Schematic representation of the shared human αB-crystallin/HspB2 promoter and conservation of the putative ETS-binding site (EBS).
(A) Using the MOTIF-search platform, we performed a bioinformatics analysis of the shared human αB-crystallin/HspB2 promoter spanning the region between the start ATG for each gene. The search identified published (bold) transcriptional regulators of αB-crystallin, as well as additional putative (italics) regulators, including Ets1. (B) Alignment of the putative EBS in the human αB-crystallin promoter with the corresponding region from other species.
Transcriptional regulation of the human αB-crystallin promoter by Ets1
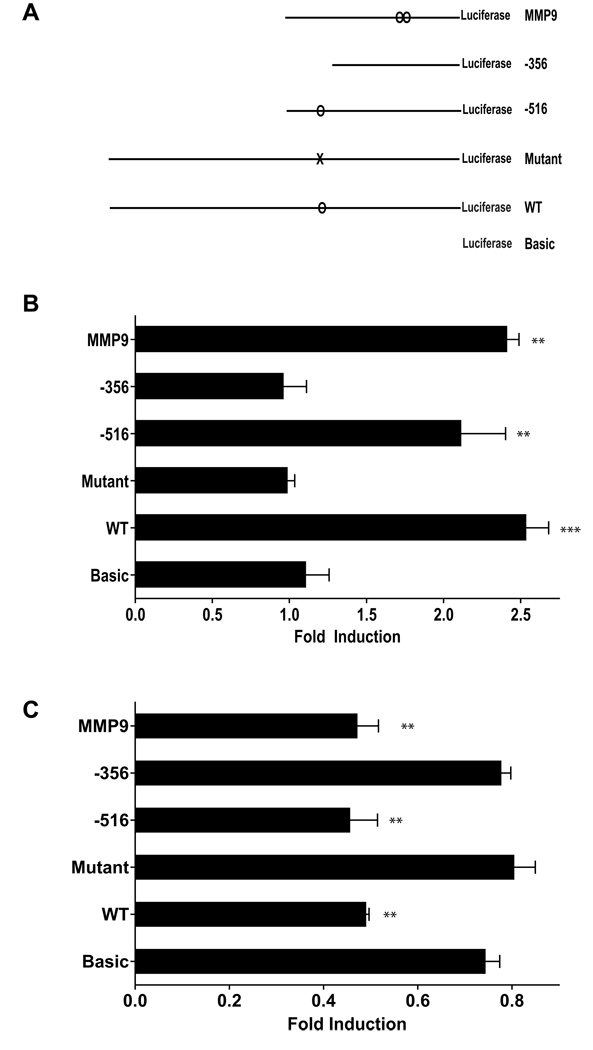
To determine whether the putative EBS in the αB-crystallin promoter is activated by Ets1, we co-transfected human MCF-10A breast epithelial cells (which express low levels of Ets1 and αB-crystallin) with cDNAs encoding Ets1 and a luciferase reporter under the control of the αB-crystallin promoter (−1081/+30 for the full-length WT promoter) (Fig. 2A). Ectopic expression of Ets1 resulted in a ~2.5 fold increase in luciferase activity in cells co-transfected with the WT αB-crystallin promoter reporter (Fig. 2B). Importantly, the activation of the WT αB-crystallin promoter by Ets1 was similar in magnitude to the Ets1-induced activation of the MMP-9 promoter, a well-established Ets1 transcriptional target [20]. In contrast, the related ETS family member ESX did not significantly activate the WT αB-crystallin promoter (data not shown), indicating specificity within the ETS family. Moreover, Ets1 activated a truncated αB-crystallin promoter (−516/+30) that contains the putative EBS at −441 to −432, but not a truncated αB-crystallin promoter (−356/+30) that lacks the EBS. Mutation of both core elements within the putative EBS (AAAAGATTTT, nucleotide alterations in bold) in the αB-crystallin promoter abrogated Ets1 activation. These results indicate that the αB-crystallin promoter contains a functional EBS, which is required for its activation by Ets1.
Figure 2. Transcriptional regulation of the human αB-crystallin promoter by Ets1.
(A) Schematic representation of the reporter constructs used. The full-length WT human αB-crystallin promoter (−1081/+30) and 5’ truncations (−516/+30, and −356/+30) were subcloned into the pGL3-Basic luciferase vector (Basic). The mutant EBS (AAAAGATTTT) was generated by site-directed mutagenesis. A pGL3-MMP9 reporter was used as a positive control. (B) MCF-10A cells were transiently co-transfected with 700 ng of pcDNA3.1-Ets1 or vector, 100 ng of pGL3 firefly luciferase reporter, and 1 ng of control pRL-TK Renilla luciferase reporter. Firefly luciferase activity was normalized to Renilla luciferase activity and expressed as fold induction relative to the activity in empty vector-transfected cells. **p < 0.01 ***p < 0.001 versus pGL3-Basic. (C) MDA-MB-231 cells were transiently co-transfected with 300 ng pGL3 reporter, 3 ng of pRL-TK reporter, and 25 µM of siRNA targeting Ets1 or lamin A/C (control). Normalized firefly luciferase activity was expressed as fold induction relative to the activity in cells transfected with control lamin A/C siRNA. **p < 0.01 versus pGL3-Basic.
We next examined whether silencing Ets1 inhibited αB-crystallin promoter activity. To this end, we co-transfected human MDA-MB-231 basal-like breast cancer cells (which express moderate levels of Ets1) with an Ets1 siRNA (or control lamin A/C siRNA) and each of the αB-crystallin promoter reporter constructs. The activity of the WT αB-crystallin promoter was inhibited in cells co-transfected with the Ets1 siRNA compared to cells co-transfected with a control siRNA (Fig. 2C). Similarly, silencing Ets1 inhibited the activity of the αB-crystallin promoter truncated at −516 and the MMP-9 promoter, but did not inhibit activity of αB-crystallin promoter constructs lacking a functional EBS and shown to be unresponsive to Ets1 overexpression (Fig. 2B). Collectively, these findings demonstrate that the αB-crystallin promoter is regulated by the expression levels of Ets1, which activates transcription by an EBS-dependent mechanism.
Ets1 binds to the αB-crystallin promoter in vitro and in vivo
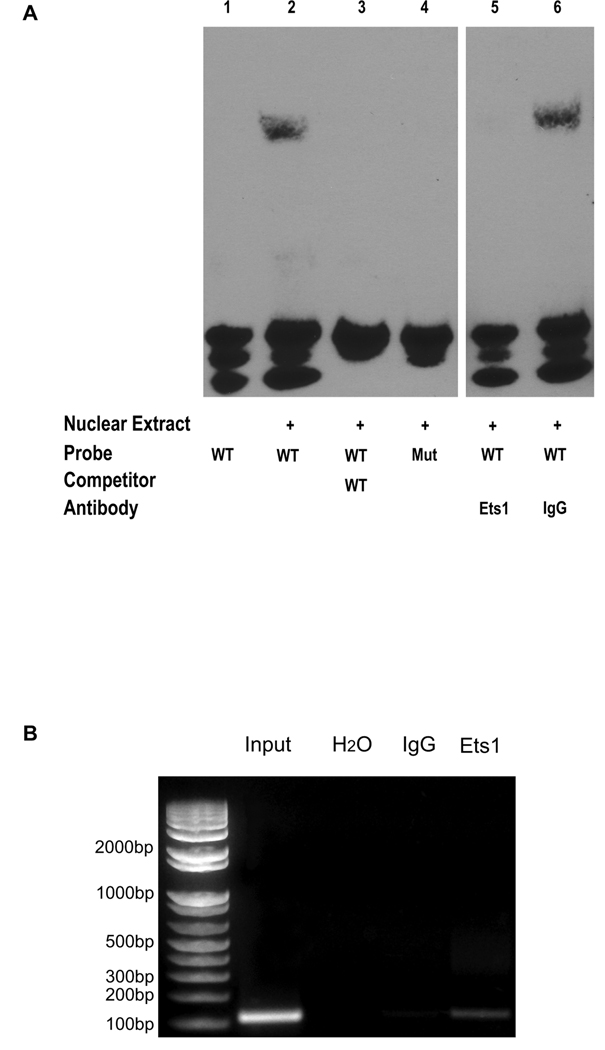
To determine whether Ets1 binds to the human αB-crystallin promoter, we performed electrophoretic mobility shift assays (EMSA) and chromatin immunoprecipitation (ChIP) analyses. For EMSA experiments, nuclear extracts were incubated with a biotinylated probe containing the putative αB-crystallin promoter EBS (WT or mutant) in the absence or presence of excess unbiotinylated probe. Incubation of the nuclear extract with the WT probe led to a gel shift of the protein-DNA complex (Fig. 3A, lane 2) that was not observed when excess unbiotinylated probe was added (Fig. 3A, lane 3) or when a probe containing a mutant EBS was used (Fig. 3A, lane 4). Moreover, preincubating the nuclear extract with an Ets1 specific antibody (but not IgG control) prior to the addition of the biotinylated WT probe prevented the formation of the DNA-protein complex (Fig. 3A, lanes 5 and 6), confirming that Ets1 was responsible for the observed gel shift. To determine whether endogenous Ets1 binds to the human αB-crystallin promoter in vivo, we performed a ChIP assay using human MDA-MB-231 basal-like breast cancer cells. PCR amplification of Ets1-immunoprecipitated DNA with primers flanking the αB-crystallin promoter EBS revealed a band of the expected size that was also observed when input DNA was amplified (Fig. 3B). These results indicate that a putative EBS in the αB-crystallin promoter is both necessary and sufficient for Ets1 binding in vitro and that endogenous Ets1 binds to this EBS in the αB-crystallin promoter in vivo.
Figure 3. Ets1 binds to the αB-crystallin promoter in vitro and in vivo.
(A) EMSA analysis of Ets1 binding to the putative EBS in the human αB-crystallin promoter in vitro. The human αB-crystallin promoter region containing the putative WT EBS or a mutant (Mut) EBS was duplexed with the corresponding antisense oligonucleotide and used as probes. Twenty fmol of biotinylated WT or mutant probe was added to nuclear extract in the absence or presence of 4 pmol of unbiotinylated WT probe. For antibody abrogation experiments, the binding reaction was preincubated with an Ets1 antibody or control IgG before adding the WT probe. Protein-DNA complexes were resolved by native-PAGE, transferred to a membrane, and detected by chemiluminescence. (B) ChIP analysis of endogenous Ets1 binding to the human αB-crystallin promoter in vivo. Input DNA-protein complexes or DNA-protein complexes immunoprecipitated with water, IgG or Ets1 antibody were PCR amplified using primers flanking the αB-crystallin promoter EBS.
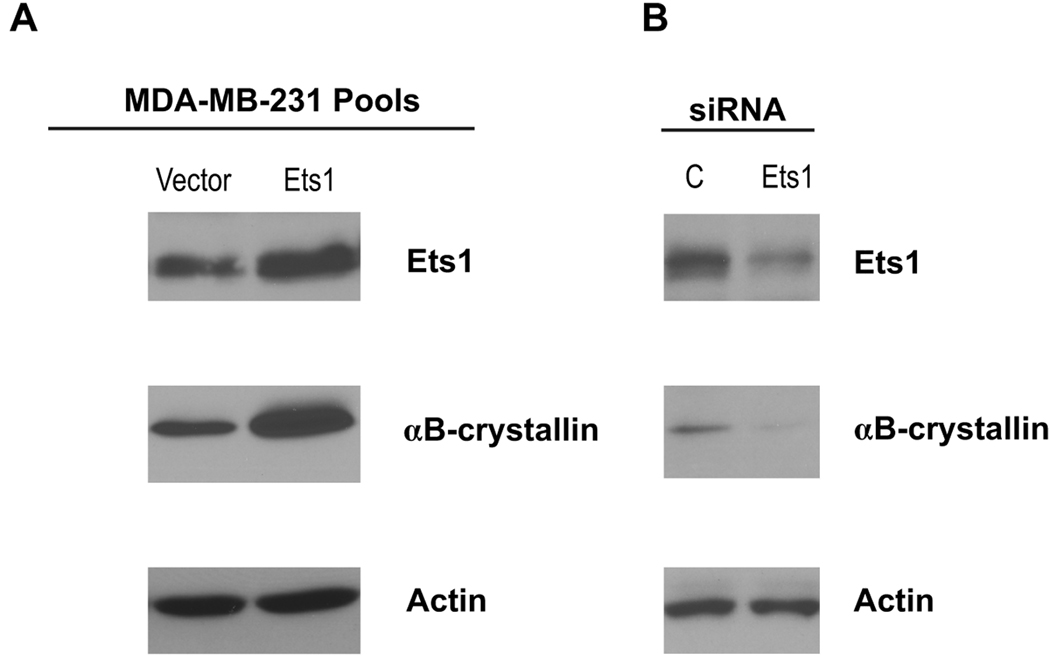
To determine whether Ets1 regulates αB-crystallin protein levels in breast cancer cells, we generated MDA-MB-231 pools stably expressing Ets1 by retroviral transduction. Stable overexpression of Ets1 resulted in a 2.4-fold increase in Ets1 protein levels and a 3.1-fold increase in αB-crystallin protein levels (Fig. 4A). Conversely, transfection of MDA-MB-231 cells with an Ets1 siRNA reduced Ets1 protein levels 2.3-fold and decreased αB-crystallin protein levels 6.0-fold compared to the levels in MDA-MB-231 cells transfected with a control lamin A/C siRNA (Fig. 4B). These latter findings indicate that the expression of αB-crystallin protein in these breast cancer cells is regulated by the endogenous levels of Ets1.
Figure 4. Regulation of endogenous αB-crystallin protein levels by Ets1 in basal-like breast cancer cells.
(A) MDA-MB-231 pools stably expressing pBABE vector or pBABE-Ets1 were created by retroviral transduction. Ets1, αB-crystallin and actin levels were determined by immunoblotting. (B) MDA-MB-231 cells were transiently transfected with 25 µM Ets1 siRNA or a control (C) lamin A/C siRNA. Ets1, αB-crystallin and actin levels were determined by immunoblotting 72 h later.
Ets1 is expressed in basal-like breast tumors and is associated with poor survival
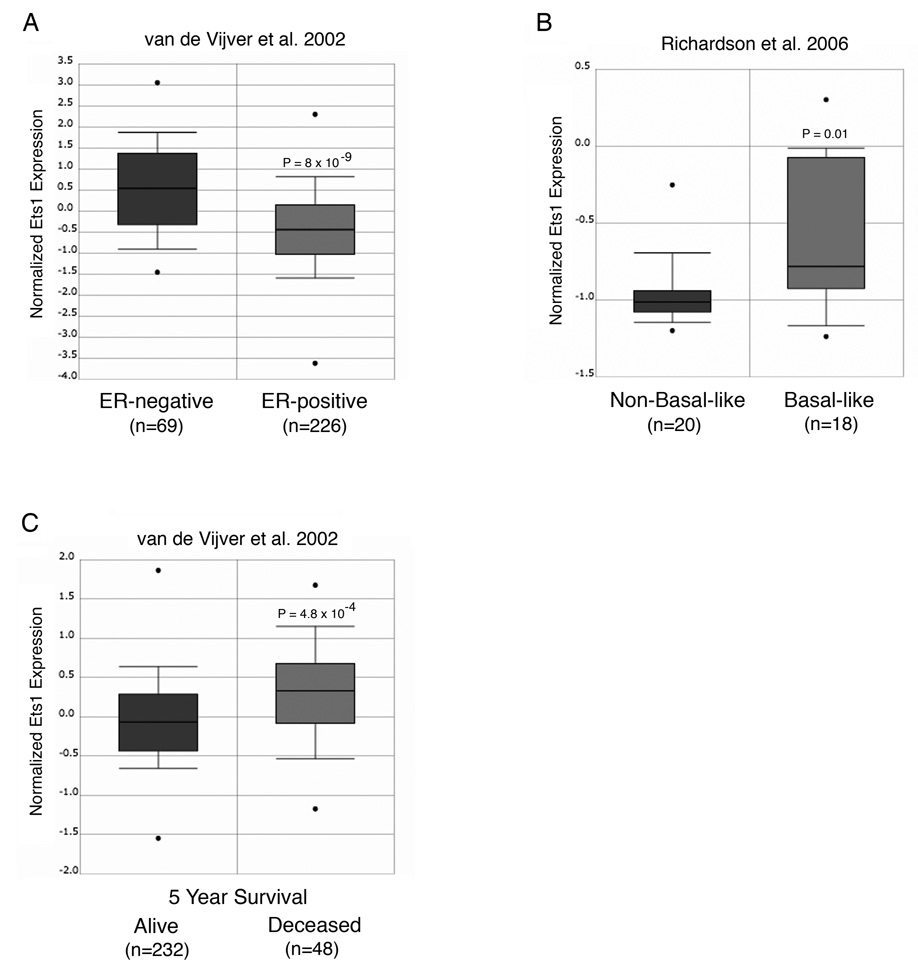
Intriguingly, Ets1 mRNA and protein are expressed in the stroma, endothelial cells and/or epithelium of poor prognosis breast carcinomas and in basal-like breast cancer cell lines [21–25], suggesting that Ets1 and αB-crystallin may be co-expressed in human basal-like breast tumors. To determine whether Ets1 is differentially expressed in human breast tumor molecular subtypes, we interrogated publically available gene expression datasets by Oncomine analysis [19]. The Ets1 gene was predominantly expressed in estrogen receptor (ER)-negative breast tumors (Fig. 5A) with a basal-like gene signature (Fig. 5B). Furthermore, Ets1 gene expression in human breast carcinomas was associated with poor survival at five years (Fig. 5C). Taken together, our results indicate that Ets1 is expressed in poor prognosis basal-like breast tumors, a distinctive gene expression pattern similar to that previously reported for αB-crystallin [9–11].
Figure 5. Ets1 is expressed in basal-like breast tumors and is associated with poor survival.
Publically available gene expression datasets were interrogated by Oncomine analysis. (A) Ets1 expression as a function of ER-status in breast cancer [35]. (B) Ets1 expression in non-basal-like and basal-like breast tumors [36]. (C) Ets1 expression and five-year survival in breast cancer [35].
Discussion
We have described a previously unrecognized and direct link between the oncogenic transcription factor Ets1 and αB-crystallin in breast cancer: Ets1 is a novel transcriptional activator of the αB-crystallin gene. Several lines of experimental evidence support this conclusion. First, ectopic expression of Ets1, but not the structurally related ETS family member ESX, activates the αB-crystallin promoter by an EBS-dependent mechanism. Conversely, silencing endogenous Ets1 reduces the activity of the αB-crystallin promoter. Second, Ets1 binds to the EBS in the αB-crystallin promoter in vitro. Third, endogenous Ets1 binds to the αB-crystallin promoter in basal-like breast cancer cells in vivo. Fourth, overexpression of Ets1 in breast cancer cells increases αB-crystallin protein levels, while silencing Ets1 reduces αB-crystallin levels. Although our results do not rule out the potential role of other ETS family members in regulating αB-crystallin gene expression, they point to a functionally important and specific role of Ets1 in this process. Such specificity may be conferred by the nucleotides flanking the conserved EBS or by coregulatory proteins that interact with ETS family members and cooperatively bind DNA [26, 27]. Taken together, our results demonstrate unequivocally that endogenous Ets1 binds to the αB-crystallin promoter in vivo, directly regulating αB-crystallin gene and protein expression levels.
αB-crystallin, then, can be added to the growing network of cancer-related genes activated by Ets1 and related family members. Like Ets1, αB-crystallin has been implicated in angiogenesis, migration and invasion, and apoptosis-resistance [4, 9, 17, 18, 28–30]. For example, both Ets1 and αB-crystallin are selectively expressed in endothelial cells during developmental and tumor angiogenesis [28, 29]. Recent studies indicate that αB-crystallin is required for endothelial cell survival during tube morphogenesis [29]. Moreover, the coordinated regulation of several MMPs (including 1, 3, and 9) and αB-crystallin by Ets1 likely promotes metastasis by initiating invasion and suppressing apoptosis. These findings suggest that αB-crystallin may be an important downstream target of Ets1 in promoting tumor progression, an hypothesis we will explore in future studies.
We have also shown that the Ets1 gene is expressed in clinically aggressive basal-like breast tumors, the same molecular subtype which expresses αB-crystallin [9–11]. Basal-like breast tumors are associated with a poor prognosis because they are highly proliferative and invasive, and they metastasize rapidly to the lungs and brain [31]. Given the well established role of ETS family members in promoting proliferation, invasion and angiogenesis [17, 18], it is tempting to speculate that Ets1 may contribute to the aggressive phenotype of basal-like tumors. Consistent with our findings, Ets1 was one of ten proteins recently reported to define an immunohistochemistry phenotype capable of identifying basal-like breast cancer cell lines [24]. Although one clinical study suggested a correlation between Ets1 and HER2 expression [25], Oncomine analysis indicated that Ets1 was highly expressed in hereditary Brca1-associated breast tumors, which often have a basal-like gene expression profile [31], providing additional evidence linking Ets1 expression to basal-like tumors (data not shown). Intriguingly, the closely related ETS family member Ets2 has been shown to transcriptionally repress the Brca1 gene [32]. Because reduced expression of BRCA1 and/or BRCA1 dysfunction is likely to play a key role in the pathogenesis of basal-like tumors [31], ETS family members may contribute to the molecular etiology of these tumors via their effects on multiple downstream targets. We also observed that Ets1 gene expression was associated with poor survival in breast cancer, consistent with the results of two earlier studies, one using RT-PCR to measure Ets1 levels, and the second using immunohistochemistry [22, 25]. These findings from patient tumors are consistent with the preclinical data implicating Ets1 in tumor progression [33, 34]. In future studies, it will be important to examine whether Ets1 and αB-crystallin are co-expressed in basal-like tumors and to evaluate their prognostic value in these tumors.
Acknowledgements
We thank Drs. M. Zhou and M. Sharon Stack for plasmids. This work was supported by NIH grants R01CA097198 (VLC), R21CA125181 (VLC) and T32GM08061 (JDB), and by the Breast Cancer Research Foundation (VLC).
References
- 1.Horwitz J. α-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehlen P, Kretz-Remy C, Preville X, Arrigo AP. Human hsp27, Drosophila hsp27 and human αB-crystallin expression-mediated increase in glutathione is essential for the protective activity of these proteins against TNFα-induced cell death. EMBO J. 1996;15:2695–2706. [PMC free article] [PubMed] [Google Scholar]
- 3.Clark JI, Muchowski PJ. Small heat-shock proteins and their potential role in human disease. Curr Opin Struct Biol. 2000;10:52–59. doi: 10.1016/s0959-440x(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 4.Kamradt MC, Chen F, Cryns VL. The small heat shock protein αB-crystallin negatively regulates cytochrome c- and caspase-8-dependent activation of caspase-3 by inhibiting its autoproteolytic maturation. J Biol Chem. 2001;276:16059–16063. doi: 10.1074/jbc.C100107200. [DOI] [PubMed] [Google Scholar]
- 5.Kamradt MC, Lu M, Werner ME, et al. The small heat shock protein αB-crystallin is a novel inhibitor of TRAIL-induced apoptosis that suppresses the activation of caspase-3. J Biol Chem. 2005;280:11059–11066. doi: 10.1074/jbc.M413382200. [DOI] [PubMed] [Google Scholar]
- 6.Mao YW, Liu JP, Xiang H, Li DW. Human αA- and αB-crystallins bind to Bax and Bcl-XS to sequester their translocation during staurosporine-induced apoptosis. Cell Death Differ. 2004;11:512–526. doi: 10.1038/sj.cdd.4401384. [DOI] [PubMed] [Google Scholar]
- 7.Chelouche-Lev D, Kluger HM, Berger AJ, Rimm DL, Price JE. αB-crystallin as a marker of lymph node involvement in breast carcinoma. Cancer. 2004;100:2543–2548. doi: 10.1002/cncr.20304. [DOI] [PubMed] [Google Scholar]
- 8.Chin D, Boyle GM, Williams RM, et al. αB-crystallin, a new independent marker for poor prognosis in head and neck cancer. Laryngoscope. 2005;115:1239–1242. doi: 10.1097/01.MLG.0000164715.86240.55. [DOI] [PubMed] [Google Scholar]
- 9.Moyano JV, Evans JR, Chen F, et al. αB-crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J Clin Invest. 2006;116:261–270. doi: 10.1172/JCI25888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov O, Chen F, Wiley EL, et al. αB-crystallin is a novel predictor of resistance to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2008;111:411–417. doi: 10.1007/s10549-007-9796-0. [DOI] [PubMed] [Google Scholar]
- 11.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 12.Swamynathan SK, Piatigorsky J. Orientation-dependent influence of an intergenic enhancer on the promoter activity of the divergently transcribed mouse Shsp/αB-crystallin and Mkbp/HspB2 genes. J Biol Chem. 2002;277:49700–49706. doi: 10.1074/jbc.M209700200. [DOI] [PubMed] [Google Scholar]
- 13.Doerwald L, van Rheede T, Dirks RP, et al. Sequence and functional conservation of the intergenic region between the head-to-head genes encoding the small heat shock proteins alphaB-crystallin and HspB2 in the mammalian lineage. J Mol Evol. 2004;59:674–686. doi: 10.1007/s00239-004-2659-y. [DOI] [PubMed] [Google Scholar]
- 14.Gopal-Srivastava R, Piatigorsky J. The murine αB-crystallin/small heat shock protein enhancer: identification of αBE-1, αBE-2, αBE-3, and MRF control elements. Mol Cell Biol. 1993;13:7144–7152. doi: 10.1128/mcb.13.11.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopal-Srivastava R, Cvekl A, Piatigorsky J. Pax-6 and αB-crystallin/small heat shock protein gene regulation in the murine lens. Interaction with the lens-specific regions, LSR1 and LSR2. J Biol Chem. 1996;271:23029–23036. doi: 10.1074/jbc.271.38.23029. [DOI] [PubMed] [Google Scholar]
- 16.Somasundaram T, Bhat SP. Canonical heat shock element in the αB-crystallin gene shows tissue-specific and developmentally controlled interactions with heat shock factor. J Biol Chem. 2000;275:17154–17159. doi: 10.1074/jbc.M000304200. [DOI] [PubMed] [Google Scholar]
- 17.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Lincoln DW, 2nd, Bove K. The transcription factor Ets-1 in breast cancer. Frontiers in Bioscience. 2005;10:506–511. doi: 10.2741/1546. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York) 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watabe T, Yoshida K, Shindoh M, et al. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer. 1998;77:128–137. doi: 10.1002/(sici)1097-0215(19980703)77:1<128::aid-ijc20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Behrens P, Rothe M, Wellmann A, Krischler J, Wernert N. The Ets-1 transcription factor is up-regulated together with MMP 1 and MMP 9 in the stroma of pre-invasive breast cancer. J Pathol. 2001;194:43–50. doi: 10.1002/path.844. [DOI] [PubMed] [Google Scholar]
- 22.Span PN, Manders P, Heuvel JJ, et al. Expression of the transcription factor Ets-1 is an independent prognostic marker for relapse-free survival in breast cancer. Oncogene. 2002;21:8506–8509. doi: 10.1038/sj.onc.1206040. [DOI] [PubMed] [Google Scholar]
- 23.Buggy Y, Maguire TM, McGreal G, et al. Overexpression of the Ets-1 transcription factor in human breast cancer. Br J Cancer. 2004;91:1308–1315. doi: 10.1038/sj.bjc.6602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 25.Myers E, Hill AD, Kelly G, et al. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin Cancer Res. 2005;11:2111–2122. doi: 10.1158/1078-0432.CCR-04-1192. [DOI] [PubMed] [Google Scholar]
- 26.Woods DB, Ghysdael J, Owen MJ. Identification of nucleotide preferences in DNA sequences recognised specifically by c-Ets-1 protein. Nucleic Acids Res. 1992;20:699–704. doi: 10.1093/nar/20.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R, Pei H, Watson DK. Regulation of Ets function by protein-protein interactions. Oncogene. 2000;19:6514–6523. doi: 10.1038/sj.onc.1204035. [DOI] [PubMed] [Google Scholar]
- 28.Wernert N, Raes MB, Lassalle P, et al. c-ets1 proto-oncogene is a transcription factor expressed in endothelial cells during tumor vascularization and other forms of angiogenesis in humans. Am J Pathol. 1992;140:119–127. [PMC free article] [PubMed] [Google Scholar]
- 29.Dimberg A, Rylova S, Dieterich LC, et al. αB-crystallin promotes tumor angiogenesis by increasing vascular survival during tube morphogenesis. Blood. 2008;111:2015–2023. doi: 10.1182/blood-2007-04-087841. [DOI] [PubMed] [Google Scholar]
- 30.Maddala R, Rao VP. α-crystallin localizes to the leading edges of migrating lens epithelial cells. Exp Cell Res. 2005;306:203–215. doi: 10.1016/j.yexcr.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 31.Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med. 2006;12:537–544. doi: 10.1016/j.molmed.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Baker KM, Wei G, Schaffner AE, Ostrowski MC. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J Biol Chem. 2003;278:17876–17884. doi: 10.1074/jbc.M209480200. [DOI] [PubMed] [Google Scholar]
- 33.Delannoy-Courdent A, Mattot V, Fafeur V, et al. The expression of an Ets1 transcription factor lacking its activation domain decreases uPA proteolytic activity and cell motility, and impairs normal tubulogenesis and cancerous scattering in mammary epithelial cells. J Cell Sci. 1998;111:1521–1534. doi: 10.1242/jcs.111.11.1521. [DOI] [PubMed] [Google Scholar]
- 34.Hahne JC, Okuducu AF, Kaminski A, Florin A, Soncin F, Wernert N. Ets-1 expression promotes epithelial cell transformation by inducing migration, invasion and anchorage-independent growth. Oncogene. 2005;24:5384–5388. doi: 10.1038/sj.onc.1208761. [DOI] [PubMed] [Google Scholar]
- 35.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 36.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]