Abstract
Redox-responsive polyplexes represent a promising class of non-viral gene delivery vectors. The reducible disulfide bonds in the polyplexes undergo intracellular reduction owing to the presence of high concentrations of reduced glutathione (GSH). Available evidence suggests improved transfection activity of redox-sensitive polyplexes upon artificial modulation of intracellular GSH. This study investigates the effect of innate differences in GSH concentration in a panel of human pancreatic cancer cell lines on activity of reducible polyplexes of the four major classes of nucleic acid therapeutics: plasmid DNA (pDNA), messenger RNA (mRNA), antisense oligodeoxynucleotides (AON) and siRNA. In general, reducible polyplexes of linear poly(amido amines) (PAA) show improved activity compared to non-reducible polyplexes of PAA. Results demonstrate that increased GSH levels are associated with improved transfection of mRNA polyplexes but no clear trend is observed for pDNA, AON and siRNA polyplexes.
Keywords: non-viral gene delivery, reducible polyplexes, glutathione, transfection, pancreatic cancer
1. INTRODUCTION
Polyplexes are investigated as promising delivery vectors for a variety of nucleic acid therapeutics [1]. Polyplexes capable of responding to environmental changes or stimuli by altering their properties and behavior seem to promise a significant improvement in delivery efficacy [2, 3]. One of several stimuli used for improving efficiency of nucleic acid delivery is the redox potential gradient existing between extracellular and intracellular environments [4-8]. The redox potential gradient has been exploited by incorporating disulfide bonds into the structure of the delivery vectors, which provide the vectors with the capability to release the nucleic acids selectively in the subcellular reducing space [4, 9-12].
The intracellular cleavage of disulfide bonds in polyplexes is mediated by thiol/disulfide exchange reactions with small redox molecules like GSH and thioredoxin; either alone or with the help of redox enzymes [13]. The reduction results in enhanced rates of disassembly of the polycation-nucleic acid complexes, which is believed to increase intracellular bioavailability of the delivered nucleic acids [14-16] and decrease polycation cytotoxicity [11, 17-19].
Available evidence suggests that artificially changing cellular GSH levels leads to changes in biological activity of polyplexes [3, 4, 7, 20]. Modulation of GSH levels using either buthionine sulfoximine (BSO), an inhibitor of cytoplasmic GSH synthesis or enhancement of GSH levels using GSH monoethyl ester demonstrated a small, but significant effect on the transfection activity of pDNA polyplexes. We previously tested the effect of increased GSH levels associated with overexpression of Bcl-2 (an anti-apoptotic protein overexpressed in multiple human cancers). The results demonstrated that Bcl-2 overexpression can be utilized as a proxy redox stimulus to selectively enhance the activity of all four major classes of potential nucleic acid therapeutics, when delivered by redox-sensitive vectors [21].
Cancer cells often exhibit elevated levels of GSH. The published evidence on the improved activity of reducible polyplexes associated with artificial modulation of GSH levels and with increased GSH related to Bcl-2 overexpression in cancers motivated us to study if similar improvement would also be observed in cancer cells with innate differences in GSH levels. The present study tests the hypothesis in a panel of human pancreatic cancer cell lines.
2. MATERIALS AND METHODS
2.1 Materials
N,N’-hexamethylenebisacrylamide (HMBA) and N,N’-cystaminebisacrylamide (CBA) were obtained from MP Biomedicals (Solon, OH). 1-(2-Aminoethyl) piperazine (AEPZ) was obtained from Acros Organics (Fisher Scientific, Pittsburgh, PA). CellTracker™ Green CMFDA (5-chloromethylfluorescein diacetate), Hoechst 33342, phosphate buffered saline (PBS), Dulbecco’s Modified Eagle Medium (DMEM), Roswell Park Memorial Medium (RPMI) and fetal bovine serum (FBS) were from Invitrogen (Carlsbad, CA). pDNA vectors, gWiz™ high-expression luciferase (gWiz-Luc) containing luciferase reporter gene, and gWiz™ high-expression GFP (gWiz-GFP) containing enhanced green fluorescent protein reporter gene were from Aldevron (Fargo, ND). Luciferase SP6 pDNA template for synthesis of mRNA was obtained from Promega (Madison, WI) and pDNA template for synthesis of eGFP mRNA, pGEM4Z/GFP/A64 was a kind gift from Dr. David Boczkowski, Duke University (Durham, NC). mRNA encoding for luciferase and eGFP were synthesized using mMESSAGE mMACHINE® kit obtained from Applied Biosystems (Foster City, CA). Firefly luciferase (GL3) and control siRNA were obtained from Ambion (Austin, TX). Luciferase antisense oligodeoxynucleotides (AON) (5′-AACCGCTTCCCCGACTTCC-3′) and negative control AON (5′-CCAATGTCAAGCACTTCCGTT-3′) with phosphorothioate linkages were custom synthesized by Midland Certified Reagent Company (Midland, TX). All other reagents were from Fisher Scientific, unless otherwise mentioned.
2.2 Synthesis of linear poly(amido amines) (PAA)
Non-reducible PAA and reducible PAA (rPAA) were synthesized by following a previously published method [22] with minor changes. Briefly, calculated quantity of HMBA or CBA was added into a small vial containing AEPZ in methanol/water mixture (7/3 v/v). The polymerization was carried out in dark at 37 °C to inhibit reactivity of the secondary amines in the intermediate product and thus avoid formation of hyperbranched structures [23]. The reaction was allowed to proceed for three days to yield a viscous solution. Subsequently, a small amount (1 mol% of starting material) of 1-methylpiperazine was added to consume any unreacted acrylamide groups, and reaction was continued for 12 h at 37 °C. The resulting solution was diluted with water, acidified with 6 M HCl to pH ~4, and then purified using centrifugal filters with molecular weight cut-off 3000 (Vivaspin, Sartorius). The concentrate was then lyophilized and dry polymers were stored at −20 °C. The composition and linear nature of polymers was established by NMR (D2O, 400 MHz) (Figure S2). Molecular weights were measured by size exclusion chromatography (SEC) using Shimadzu LC-10ADVP liquid chromatograph using Polymer Labs PL gel 5 μm mixed C column. The system was equipped with miniDAWN multiangle light scattering detector and OPTILAB DSP interferometric refractometer (Wyatt Technology). Sodium acetate buffer (300 mM, pH 4.5) was used as an eluent at a flow rate of 1.0 mL/min and temperature of 35 °C.
2.3 Cell lines
Human pancreatic cancer cell lines (Panc-1, MiaPaCa, AsPC-1, and Panc-28) were a kind gift from Dr. Fazlul H. Sarkar (Karmanos Cancer Institute). Panc-1, MiaPaCa and Panc-28 are human ductal pancreatic adenocarcinoma cell lines and AsPC-1 is human pancreatic adenocarcinoma derived from ascites metastatic site. The cells were maintained in DMEM supplemented with 4 mM L-alanyl-L-glutamine and 10% FBS at 37 °C in 5% CO2 atmosphere.
2.4 Characterization of pancreatic cancer cell lines
Cellular GSH levels [21, 24], subcellular distribution of GSH [21], concentration of cell surface protein thiols [25] and population doubling time [21] were measured for all the cell lines. Intracellular GSH concentration was measured on reverse-phase ion-exchange Waters HPLC system and normalized to cell volume determined using Packed-Cell-Volume tubes using the protocol described by the manufacturer (MidSci). Subcellular distribution of GSH was studied by fluorescence microscopy. Cells in log phase were stained with 7.5 μM CMFDA to label intracellular thiols (GSH) and 10 μM Hoechst 33342 to label the nucleus. MetaVue software was used to quantitate the average fluorescence intensity of the nuclear and cytoplasmic GSH staining by creating distinct regions encompassing the nucleus and entire intracellular space excluding the nucleus (cytoplasm). Results are expressed as a ratio of average fluorescence intensity of the nuclear to the cytoplasmic regions. At least 100 cells were counted per data point. To determine the concentration of redox-active cell surface protein thiols, the cells were incubated with 5,5′-dithiobis-2-nitrobenzoic acid at a final concentration of 190 μM for 30 min at room temperature, centrifuged at 3000 rpm for 5 min and absorbance was measured at 412 nm using an extinction coefficient of 14,150 M-1cm-1. Growth curves were constructed for each cell line depicting total number of cells vs. time since seeding and the doubling period was calculated from a linear portion of the log phase. GSH levels following BSO treatment was measured using a previously described enzymatic recycling method [21].
2.5 Transfection experiments
All transfections were conducted in 48-well plates with cells seeded at a density of 30,000 (Panc-1), 75,000 (MiaPaCa and AsPC-1) and 40,000 (Panc-28) cells/well 24 h before transfection, such that cells were 70–80% confluent (confirmed by visual examination) at the time of transfection. Differences in cell volume among the selected cell lines were taken into account in estimating the seeding density. Previously published protocols were followed to conduct all transfections [7, 14, 21]. 2 μg of polycation/well was used in all transfection studies. pDNA polyplexes (as indicated) and polyplexes of mRNA, AON and siRNA were formed as ternary complexes (DOTAP/polycation/nucleic acid) to improve their endosomal escape. For AON and siRNA transfections, the cells were co-transfected with complexes containing 0.5 μg pDNA and the indicated amounts of AON or siRNA. Complexes were diluted either in medium lacking FBS (−FBS) or medium with 10% FBS (+FBS). All transfection experiments were repeated at least three times on different occasions. Raw transfection activity of luciferase pDNA and mRNA complexes was expressed as RLU/mg cellular protein ± standard deviation (SD) of quadruplicate samples. Raw transfection activity of eGFP pDNA and mRNA was expressed as % eGFP positive (+ve) cells ± SD of quadruplicate samples. Fluorescence intensity of eGFP expression (mean fluorescence intensity per cell) was obtained from dot plot analyses of flow cytometry data. Transfection activity of AON and siRNA complexes was expressed as % luciferase knockdown, relative to cells transfected with control pDNA polyplexes. Effect of GSH levels on transfection activity was expressed as relative transfection activity, i.e. the ratio of gene expression mediated by reducible (rPAA) polyplexes to that mediated by control non-reducible (PAA) polyplexes. In GSH inhibition experiments, cells were incubated with 5 mM BSO for 24 h in complete culture medium, washed once with warm PBS prior to adding complexes and transfection was carried out as described above.
2.6 Biophysical characterization of polyplexes
The following biophysical properties of pDNA polyplexes were characterized using previously reported methods: hydrodynamic size and ζ potential [17], redox sensitivity [5], and serum stability [21].
2.7 Statistical analysis
Student’s t-test was used to determine if differences between means of two groups were significant and ANalysis Of Variance (ANOVA) was used to determine if differences among multiple groups was significant. A P-value < 0.05 was considered statistically significant in all cases.
3. RESULTS
3.1 Biological characterization of the pancreatic cancer cell lines
3.1.1 GSH levels
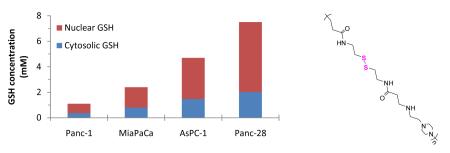
Several human pancreatic cancer cell lines were screened and a panel of four cell lines was identified that exhibited a range of intracellular GSH concentrations. GSH levels depend on the cell growth phase and were measured in log and stationary phases (Table 1). The GSH concentration ranged from 1 to 7.5 mM and 0.3 to 3 mM, in the log and stationary phases, respectively. The GSH levels were 50–75% lower in the stationary phase than in the log phase, which is in agreement with published observations showing decreased GSH levels with increasing cell density [26, 27].
Table 1.
Pancreatic cancer cell characterization
| Cell line | GSH concentration (mM) | GSH distribution (nuc/cyt)a |
Surface thiol content (nmol/106 cells) |
Doubling time (h) |
|
|---|---|---|---|---|---|
| log | stationary | ||||
| Panc-1 | 1.1 ± 0.2 | 0.3 ± 0.2 | 2.0 ± 0.5 | 30 ± 2 | 31 |
| MiaPaCa | 2.4 ± 0.4 | 1.3 ± 0.5 | 2.0 ± 0.2 | 44 ± 4 | 17 |
| AsPC-1 | 4.7 ± 0.4 | 1.4 ± 0.6 | 2.2 ± 0.3 | 30 ± 2 | 27 |
| Panc-28 | 7.5 ± 0.4 | 3.0 ± 1.5 | 2.7 ± 0.4 | 66 ± 5 | 38 |
from average fluorescence intensities in nuclear and cytoplasmic compartments of cells
3.1.2 Subcellular distribution of GSH
Cellular GSH is predominantly present in the cytoplasm (1–11 mM), which is also the principal site of GSH biosynthesis [28-30]. The nuclear GSH concentrations are typically greater (although the total amount is lower) than those found in the cytoplasm and can reach up to 20 mM [31-33]. GSH distribution between the cytoplasm and nucleus was studied in log phase by staining intracellular GSH and cell nuclei (not shown). The nuc/cyt GSH ratio ranged from 2.0 to 2.7 (Table 1), in support of a published observation that nuc/cyt ratios are higher than 2.0 in the log phase of cell growth [34].
3.1.3 Population doubling time
To account for a possible effect of cell growth rate on transfection activity, population doubling times for each cell line were determined (Table 1). MiaPaCa cells showed the fastest growth rate, followed by AsPC-1, Panc-1 and Panc-28. The doubling time of pancreatic cancer cells is related to their differentiation grade [35]. Grade 2 and grade 3 cell lines have doubling times ranging from 22 to 41 h and 19 to 32 h, respectively. Our data show that AsPC-1 (grade 2) and MiaPaCa and Panc-1 (grade 3) have doubling times of 27, 17 and 31 h, respectively, while the doubling time of Panc-28 was 38 h [36].
3.1.4 Cell surface thiol content
Exofacial surface of cellular plasma membrane contains redox active thiol groups [25, 37-40] that could potentially participate in interactions of reducible polyplexes with cells during transfection. As shown in Table 1, surface thiol content in Panc-1 and AsPC-1 (30 nmol/106 cells) falls within the general range of thiols (~ 4 to ~ 30 nmol/106 cells) reported previously [25, 41]. The highest surface thiol content was observed in Panc-28, also the cell line with the highest intracellular GSH concentration. The surface thiol content in Panc-28 was 2-fold higher than that in Panc-1 and AsPC-1 (P=0.0003) and 1.5-fold higher than that in MiaPaCa (P=0.0004).
3.2 Polycations and biophysical characterization of polyplexes
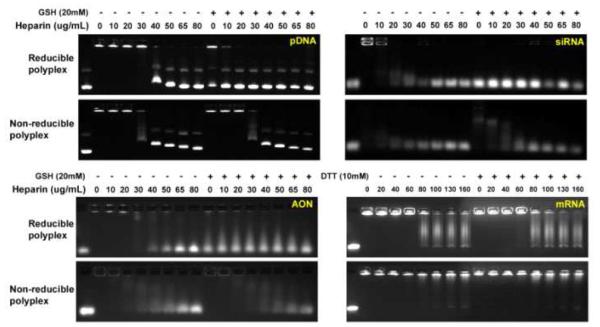
Reducible PAAs are a versatile class of polycations characterized by easy synthesis, good hydrolytic stability, and adjustable endosomal buffering capability [22]. Non-reducible and reducible PAAs (Scheme 1) were synthesized by Michael addition polymerization. Weight-average molecular weight and polydispersity index were found to be Mw = 8300 and Mw/Mn = 1.05 for PAA and Mw = 6200 and Mw/Mn = 1.12 for rPAA. Both synthesized polymers could condense pDNA in an ethidium bromide exclusion assay. The stability and redox sensitivity of the polyplexes was evaluated by resistance to polyelectrolyte exchange reactions with heparin using agarose gel electrophoresis (Fig. 1). The results show that siRNA forms the least stable polyplexes, with as low as 20 μg/mL heparin being able to destabilize the polyplexes. In comparison, both pDNA and AON polyplexes are destabilized at higher heparin concentrations; ≥40 g/mL for pDNA and ≥30 g/mL for AON. A notable difference between pDNA and siRNA and AON polyplexes is the gradually increasing release with increasing heparin concentration for the short nucleotides. pDNA polyplexes, on the other hand, are released in a more abrupt manner. Polyplexes of mRNA exhibited strikingly higher stability against heparin exchange with signs of mRNA release observed at 80 μg/mL heparin but no complete release even at 160 μg/mL heparin. No significant differences were observed between reducible and non-reducible polyplexes in the absence of GSH. Treatment of pDNA, siRNA, and AON polyplexes with 20 mM GSH resulted in substantial destabilization of reducible polyplexes and nucleic acid release observed even in the absence of heparin, while no effect was observed in non-reducible polyplexes. Treatment with GSH had no effect on mRNA polyplexes and it was observed that GSH itself is capable of retarding the movement of mRNA (not shown). Alternative treatment with 10 mM DTT, which had no effect on electrophoretic migration of mRNA, produced a small destabilizing effect on reducible polyplexes, with signs of release observed at 60 μg/mL heparin. The possibility of premature release of pDNA from reducible polyplexes as a result of thiol-disulfide reactions mediated by free thiols present in serum (protein thiols, cysteine, homocysteine, cysteinylglycine) was studied as described earlier [21]. In our earlier study, we observed no effect of serum thiols on the stability of reducible polyplexes based on reducible poly-L-lysine (rPLL) [21]. However, reduction of rPLL resulted in peptide residues with 11 positive charges, whereas complete reduction of rPAA results in oligocation residues with only 3 positive charges. To study if the lower number of charges in degradation products of rPAA would influence stability of their polyplexes in serum, polyplexes were incubated with 10% FBS for 3 h, simulating transfection studies (Fig. S1). The results indicated that serum did not affect the stability of reducible polyplexes. Biophysical properties, such as size and surface charge, influence transfection activity of polyplexes [17]. Size and charge of polyplexes were measured under conditions simulating their use in transfection experiments (Table 2). The results confirm that PAA and rPAA form polyplexes of similar sizes and zeta potentials with each of the four studied nucleic acids. mRNA which forms the most stable polyplexes also forms polyplexes of the largest size.
Scheme 1.
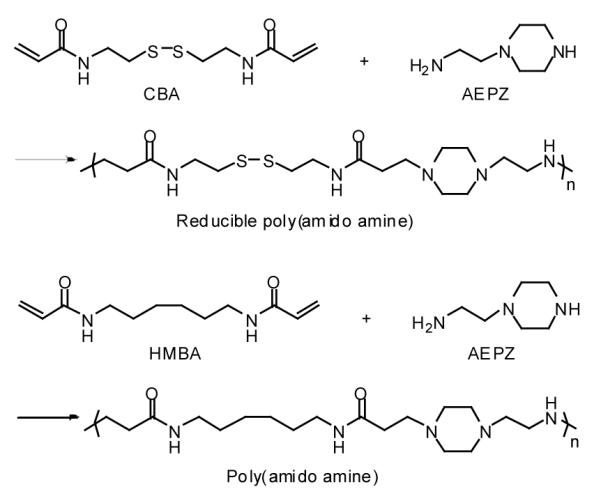
Synthesis of linear reducible poly(amido amine)s.
Figure 1. Polyplex stability against exchange reactions with heparin in non-reducing and reducing conditions.
(a) pDNA polyplexes; (b) AS ODN polyplexes; (c) siRNA polyplexes; and (d) mRNA polyplexes. All polyplexes were prepared at w/w 4. The first lane in each gel is the corresponding free nucleic acid.
Table 2.
Size and zeta potential of polyplexes.
| Nucleic acid | Hydrodynamic size (nm) | Zeta potential (mV) | ||
|---|---|---|---|---|
| PAA | rPAA | PAA | rPAA | |
| pDNA | 90.1 ± 2.6 | 68.8 ± 6.2 | 38.9 ± 1.2 | 39.9 ± 3.1 |
| mRNA | 350.2 ± 51.0 | 368.7 ± 33.9 | 46.9 ± 1.5 | 43.7 ± 2.6 |
| AON | 173.3 ± 9.1 | 176.2 ± 18.8 | 28.0 ± 0.9 | 32.0 ± 2.0 |
| siRNA | 175.1 ± 2.1 | 165.6 ± 6.7 | 39.2 ± 0.5 | 40.4 ± 0.6 |
3.3 Effect of GSH levels on transfection activity
All transfection studies were conducted with cells in log phase and results are reported as relative transfection to permit the use of non-reducible polyplexes as an internal control to account for any variations arising due to cell line-dependent factors, including differences in extent of cellular uptake, endosomal escape, and nuclear delivery. Polycation/nucleic acid ratio of 4 (w/w) was selected from initial optimization studies as a formulation that induced only minimal cell stress in all the cell lines. Cellular uptake of polyplexes was measured using quantitative real-time PCR [43] and no significant differences were observed between the two kinds of polyplexes (not shown), indicating that any observed differences would arise from differences in their subcellular processing.
3.3.1 Effect of GSH levels on transfection activity of pDNA polyplexes
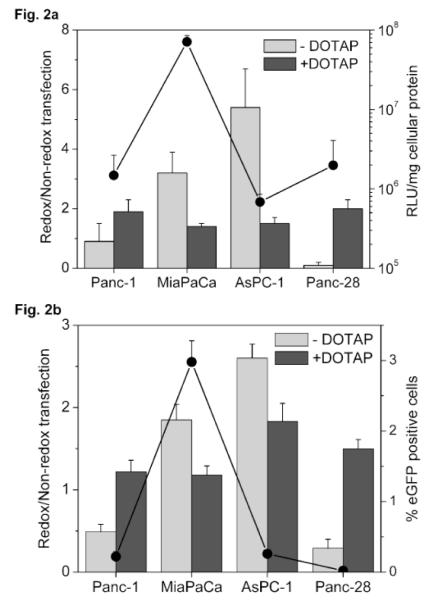
Transfections were conducted using pDNAs encoding for luciferase (Fig. 2a) and eGFP (Fig. 2b) to investigate if increased GSH levels would translate to an improvement in both expression levels and transfection frequency of reducible polyplexes. To study if any possible differences in the endosomal escape ability between the reducible and non-reducible polyplexes could mask the GSH effect, transfection was also conducted with ternary complexes of rPAA/pDNA and PAA/pDNA with DOTAP (dark grey bars in Fig. 2a). Cationic lipids mediate endosomal escape via destabilization of endosomal membrane followed by formation of charge neutral ion pairs with anionic lipids in the endosomal membrane, resulting in a release into cytoplasm [44]. Transfection activity of binary reducible polyplexes was similar, 3.2-fold higher and 5.4-fold higher than that of non-reducible polyplexes in Panc-1, MiaPaCa and AsPC-1, respectively. In contrast, transfection activity of reducible polyplexes was lower than non-reducible polyplexes in Panc-28, the cell line with the highest GSH concentration in the selected panel. The differences in relative transfection activity among Panc-1, MiaPaCa and AsPC-1 were statistically significant (P=0.003, ANOVA). Among the four cell lines, reducible polyplexes showed the best improvement in AsPC-1. Ternary DOTAP reducible complexes showed a 1.4- to 2.0-fold improvement in transfection activity but no significant differences in relative transfections were observed among the four cell lines. As shown on the secondary axis in Fig. 2a, the absolute levels of gene expression mediated by reducible polyplexes in the fastest growing cell line, MiaPaCa, were about 50-fold higher than in the other three cell lines. Although we observed no correlation between population doubling time and gene expression levels when results from all tested cell lines were analyzed, it was interesting to note that the fastest-growing cell line demonstrated superior transfection levels.
Figure 2. Transfection activity of pDNA polyplexes.
(a) Luciferase expression. Cells were transfected for 4 h with 0.5 μg pDNA complexed with PAA or rPAA at w/w 4 (binary polyplexes, −DOTAP) or with ternary complexes (+DOTAP). Luciferase expression was measured after 24 h as mean RLU/mg cellular protein (n=4). Redox/Non-redox is the ratio (± SD) of transfection activity of rPAA-to-PAA complexes. Secondary axis shows transfection in RLU/mg cellular protein mediated by binary rPAA polyplexes. (b) eGFP expression. Cells were transfected with eGFP pDNA as described above and the gene expression was measured 24 h after transfection as mean % GFP positive cells (n=4). Redox/Non-redox is the ratio (± SD) of transfection activity of rPAA-to-PAA complexes. Secondary axis shows transfection in % GFP positive cells mediated by binary rPAA polyplexes.
In transfections conducted with pDNA encoding for eGFP (Fig. 2b), binary reducible polyplexes showed a lower transfection activity than non-reducible polyplexes in Panc-1 and Panc-28. A significant 1.8- and 2.6-fold improvement in percentage of transfected cells by reducible polyplexes was observed in MiaPaCa and AsPC-1. Ternary reducible complexes (dark grey bars in Fig. 2b) mediated 1.2- to 1.8-fold improvement in transfection frequency compared to non-reducible complexes. The increase in relative transfection was statistically significant in AsPC-1 compared to Panc-1 (P=0.015) and MiaPaCa (P=0.007), and also in Panc-28 compared to Panc-1 (P=0.05) and MiaPaCa (P=0.01). As shown on secondary axis in Fig. 2b, the absolute transfection (% eGFP positive cells) mediated by binary reducible polyplexes were similar in Panc-1 and AsPC-1 and higher than in Panc-28. The observed trend in relative transfection activity was different in the case of binary and ternary complexes and we speculate that differences in kinetics and mode of endosomal escape between binary and ternary complexes and the subsequent rate-limiting processes that govern the nuclear delivery of pDNA might influence the observed trends. It has been demonstrated that incorporation of endosomal escape peptides in non-viral vectors resulted in increased endosomal escape rate constant by about two orders of magnitude [45].
3.3.2 Effect of GSH levels on transfection activity of mRNA polyplexes
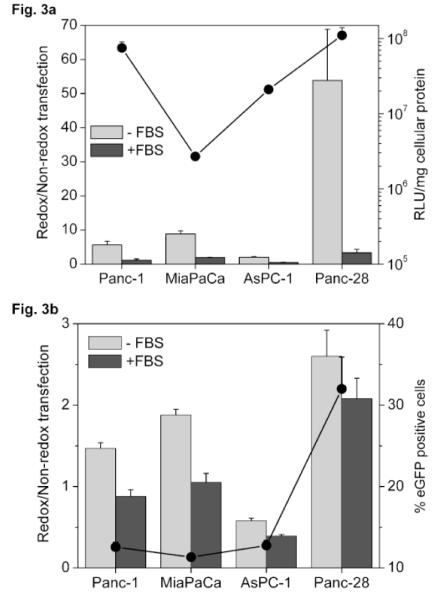
Differences among the studied cell lines were observed in both cytoplasmic and nuclear GSH levels. Polyplexes of mRNA were therefore investigated to find out if cytoplasmically-active nucleic acids will benefit more from the bioreducible nature of the polyplexes than pDNA polyplexes (Fig. 3). Reducible polyplexes showed substantial improvement in transfection in the absence of serum, with relative transfection of 6 in Panc-1, 9 in MiaPaCa, 2 in AsPC-1 and 54 in Panc-28 (light grey bars in Fig. 3a). Unlike pDNA polyplexes, the highest relative transfection (54) was seen in Panc-28, the cell line with the highest GSH concentration. Reducible and non-reducible polyplexes mediated similar levels of transfection in the presence of serum in Panc-1 and AsPC-1. Higher relative transfection was observed in MiaPaCa and Panc-28 (dark grey bars in Fig. 3a). The increase in relative transfection in MiaPaCa and Panc-28, compared to Panc-1, was statistically significant (P=0.03 and 0.02, respectively). The increase in relative transfection of mRNA complexes, irrespective of transfection conditions is in good agreement with our previous observations from the Bcl-2 overexpression model [21]. Transfections conducted with eGFP mRNA resulted in 1.5- to 2.6-fold improvement in activity of reducible complexes compared with non-reducible complexes in Panc-1, MiaPaCa and Panc-28 (Fig. 3b). No improvement was observed in AsPC-1. The trend in relative transfection was similar to that observed in luciferase mRNA transfection. The increase in relative transfection in MiaPaCa and Panc-28 was statistically significant, compared to Panc-1 (P=0.0004 and 0.002, respectively). The differences in relative transfection among all four cell lines were also statistically significant (P<0.0001). The % eGFP transfected cells is shown on the secondary axis in Fig. 3b. Eleven to thirteen % of cells were transfected in Panc-1, MiaPaCa and AsPC-1, whereas 32% of the cells were transfected in Panc-28, the cell line with the highest GSH concentration.
Figure 3. Transfection activity of mRNA complexes.
(a) Luciferase expression. Cells were transfected for 4 h with 1 μg Luc-mRNA complexed with PAA or rPAA (w/w 4) to which DOTAP was added to form ternary complexes. Luciferase expression was measured after 6 h as mean RLU/mg cellular protein (n=4). Redox/Non-redox is the ratio (± SD) of transfection activity of rPAA-to-PAA complexes. Secondary axis shows transfection in RLU/mg cellular protein mediated by rPAA complexes in the absence of serum. (b) eGFP expression. Cells were transfected with eGFP mRNA as described above and the gene expression was measured 24 h after transfection as mean % GFP positive cells (n=4). Redox/Non-redox is the ratio (± SD) of transfection activity of rPAA-to-PAA complexes. Secondary axis shows transfection in % GFP positive cells mediated by rPAA complexes in the absence of serum.
3.3.3 Effect of GSH levels on transfection activity of AON and siRNA
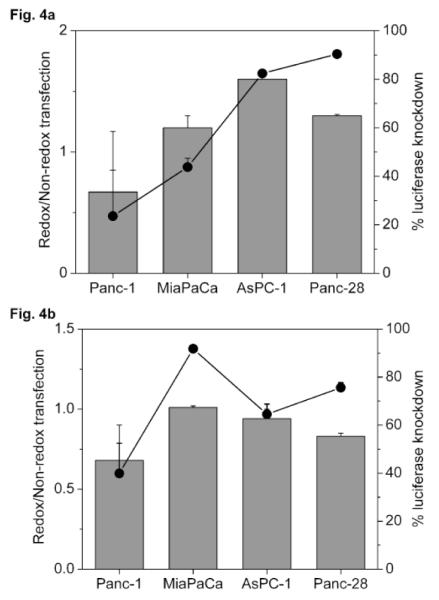
The above results suggest that intracellular site of action strongly influences the activity of reducible polyplexes. Disassembly of polyelectrolyte complexes is dependent on the molecular weight of polyelectrolytes (Fig. 1) and therefore polyplexes of short nucleic acids (AON, siRNA) were investigated next (Fig. 4). Fig. 4a shows that reducible AON complexes mediated lower luciferase knockdown than non-reducible complexes in Panc-1, whereas slightly increased knockdown was observed in MiaPaCa, AsPC-1 and Panc-28. The differences in relative transfection observed among the four cell lines was statistically significant (P=0.004, ANOVA). As shown on secondary axis in Fig. 4a, higher absolute levels of knockdown by reducible complexes were observed in cell lines with higher GSH levels. The effect of GSH levels on activity of cytoplasmically-active nucleic acids is shown in Fig. 4b on complexes of siRNA. Luciferase knockdown mediated by reducible complexes was equal to or slightly lower than non-reducible complexes in all four cell lines. An increase in absolute levels of luciferase knockdown was observed in MiaPaCa (2.3-fold), AsPC-1 (1.5-fold) and Panc-28 (1.9-fold) compared to Panc-1 and the increases were statistically significant (P = 0.002, 0.03 and 0.008, respectively).
Figure 4. Transfection activity of complexes of short nucleic acids (a) luciferase AON (b) luciferase siRNA.
Cells were co-transfected for 4 h in the absence of FBS with 0.5 μg luc-pDNA and (a) 0.075 μg AON (b) 0.2 μg siRNA targeted against luciferase, complexed with PAA/rPAA (w/w 4) to which DOTAP was added to form ternary complexes. Luciferase expression was measured after 24 h. Knockdown of luciferase expression was calculated by comparison with cells transfected with pDNA only. Redox/Non-redox is the ratio (± SD, n=4) of knockdown activity of rPAA-to-PAA complexes. Secondary axis shows % knockdown mediated by rPAA complexes.
3.3.4 Increase in relative transfection is mediated by increased GSH levels
Reducible complexes of mRNA showed the largest improvement in transfection in Panc-28. To verify that the increase in activity was related to the changes in GSH, GSH levels were measured in cells treated with BSO (Fig. 5a), and transfection of mRNA complexes was studied in BSO-treated cells (Fig. 5b). GSH levels in cells incubated with BSO decreased to 26% of control untreated cells and a significant 50% decrease in transfection activity was observed as a result of the BSO treatment in the case of reducible complexes (P=0.007). BSO treatment had no significant effect on transfection of non-reducible complexes.
Figure 5. Effect of GSH inhibition on transfection activity of reducible mRNA complexes.
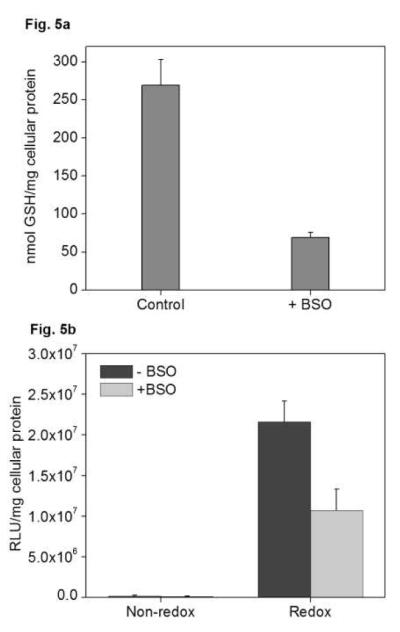
(a) GSH levels in BSO-treated cells. Panc-28 cells were incubated with 0.5 mM BSO for 24 h, following which cells were harvested and GSH levels were measured using an enzymatic recycling method. Results are expressed as mean ± SD (n=6). (b) Effect of BSO on mRNA transfection. Panc-28 cells were incubated with 5 mM BSO following which BSO-containing medium was removed and cells were washed with PBS prior to the addition of complexes. Cells were transfected for 3 h with 1 μg luc-mRNA complexed with PAA or rPAA (w/w 12) to which DOTAP was added to form ternary complexes (DOTAP/mRNA w/w 4). Luciferase expression was measured after 6 h as mean RLU/mg cellular protein (n=4).
4. DISCUSSION
Bioreducible polyplexes show advantages due to lower cytotoxicity [5, 7, 17, 46] and often increased activity. rPAA showed considerably lower cytotoxicity than PAA in all tested cell lines (manuscript in preparation). Because the properties of reducible polyplexes are related to intracellular disulfide reduction, one may anticipate that changes in redox state of cells would affect the activity of bioreducible polyplexes. The innate differences in GSH levels in the selected model allowed us to investigate its effect on activity of the polyplexes. We hypothesized that differences in overall GSH levels and its subcellular distribution will influence transfection activity of nucleic acids that are active in different subcellular compartments (nuclearly- and cytoplasmically-active nucleic acids). Because both size [47] and molecular weight [48, 49] of polyplexes have been reported to influence their stability, we conducted transfection studies using nucleic acids that differed in size, which would form polyplexes of different molecular weights (pDNA vs. AON and mRNA vs. siRNA).
Intracellular reduction of disulfides within the polyplexes has been clearly demonstrated in numerous studies. The reduction increases the rates of polyplex disassembly and thus promotes better availability of the delivered nucleic acids for transcriptional/translational machinery. A possible relationship between the increased extent and rates of polyplex disassembly following intracellular polycation degradation and activity of the nucleic acids with increasing GSH concentration offers a real opportunity to modulate polyplex activity in disease states like cancer that are associated with altered cellular redox state.
There were significant differences in both cytoplasmic and nuclear concentration of GSH among the studied cell lines. In general, increased GSH concentration will result in increased rates of polyplex disassembly regardless of whether the disassembly takes place in the cytoplasm or nucleus. For the increase in disassembly to result in increased activity of the delivered nucleic acid, however, it is important to consider other factors as well. For example, rapid disassembly of pDNA polyplexes in the cytoplasm may result in greater susceptibility of pDNA to cytoplasmic nuclease degradation and subsequently in lower transfection activity than if the disassembly occurred in the nucleus. As our results show, the lowest relative transfection with pDNA polyplexes was observed in the cell line with the highest GSH concentration (Panc-28). This finding suggests that there is an optimum rate of pDNA polyplex disassembly in the cytoplasm that leads to maximum transfection activity [50]. The observation of a gradual increase in relative transfection with increased GSH in Panc-1, MiaPaCa and AsPC-1 further supports the notion of optimal GSH concentration. The conclusion is also consistent with previous reports showing that increasing intracellular GSH by incubation with a membrane-permeable derivative of GSH may, depending on the cell line, lead to both enhancement and reduction of transfection [3, 7, 46, 51]. Our results also indicate that nuclear GSH levels are not a major factor that determines activity of reducible vs. non-reducible pDNA polyplexes. It is likely that pDNA polyplexes undergo partial disassembly in the cytoplasm, following which they might traffic to the nucleus, where the disassembly is completing. Alternatively, pDNA polyplexes may fully disassemble in the cytoplasm, following which pDNA may utilize the microtubule network to traffic to the nucleus [52]. It is important to re-emphasize that the reported and discussed transfection activities are normalized to transfection activity of non-reducible polyplexes in the respective cell line in order to minimize other effects, such as proliferation and transcription rate differences. The results therefore represent very closely the effect of the presence of disulfide bonds in the reducible polyplexes. Transfection activity of redox-responsive complexes typically is more sensitive to the presence of serum than that of non-reducible complexes. The relative transfection activity was lower in the presence of serum when compared in the absence of serum, in agreement with previous observations [7, 21, 46].
Studies using mRNA complexes revealed a large increase in relative transfection activity, which is attributed to increased disassembly of the complexes mediated by cytoplasmic GSH. The increase in relative transfection of mRNA was of a significantly higher magnitude than that observed for pDNA complexes. The differences in the magnitude of improvement in relative transfection may be primarily attributed to differences in specific rate-limiting steps involved in the delivery of mRNA compared with pDNA. Our data clearly document that disassembly is a more critical barrier for mRNA polyplexs than for pDNA polyplexes and confirm a previously reported hypothesis [14]. Following entry of pDNA polyplexes into the nucleus, exchange reaction with chromosomal DNA might result in disassembly, leading to release of free pDNA. Alternatively, polyplexes might undergo exchange reactions with cytoplasmic proteins leading to release of free pDNA, following which cytoplasmic transcription factors may bind and translocate it to the nucleus. A study identified 15 cytoplasmic proteins that bind to PEI/DNA polyplexes resulting in their disassembly and leading to release of pDNA [53]. In addition, cytoplasmic transcription factors may bind to sequence-specific motifs on the DNA, forming protein-DNA complexes, and ultimately traffic the DNA into the nucleus [54]. In contrast, lack of any such specific mechanism and the high disassembly stability in case of mRNA polyplexes [14] may require additional built-in or a stimulus-controlled mechanism of disassembly.
In addition to differences in intracellular GSH levels, the cell lines used in this study also exhibited differences in the content of redox-active thiols on the surface of plasma membrane. Based on the transfection results for mRNA and pDNA complexes, we conclude that cell surface thiols do not adversely affect activity of reducible polyplexes prior or during cellular internalization.
Disassembly of polyelectrolyte complexes is strongly dependent on the molecular weight of polyelectrolytes and thus we predicted that the benefits of intracellular disulfide reduction would be most pronounced in the case of high-molecular-weight nucleic acids like pDNA and mRNA. The reduction of disulfides in bioreducible complexes is likely to have less significant effect on the activity of short nucleic acids such as AON and siRNA due to their inherently lower affinity to the polycations [48, 49]. Thus as expected, the knockdown mediated by reducible complexes was not significantly higher compared to non-reducible complexes. However, increased levels of knockdown observed in cell lines with higher GSH concentrations encourage the use of redox-sensitive vectors for delivery of antisense and siRNA therapeutics.
If increasing GSH concentration in the tested cell lines resulted in enhanced activity of mRNA complexes, depleting intracellular GSH by inhibiting its de novo synthesis by BSO should have the opposite effect. Our observations on BSO-mediated inhibition of cytoplasmic GSH indicated that the GSH-mediated polyplex disassembly is responsible for increased activity of reducible mRNA complexes. Several published observations have reported similar extents of influence of GSH synthesis inhibition on activity on redox-sensitive polyplexes [3, 4, 7, 10, 20]. Typically, inhibition of GSH biosynthesis has resulted in milder effects on activity of reducible polyplexes. Although no clear explanations are currently available regarding why no dramatic changes are observed, it has been shown that both GSH-dependent and GSH-independent pathways are involved in intracellular disulfide reduction and that the relative contribution of these pathways are cell-line dependent [13]. A recent report quantified intracellular thiols in human cervical carcinoma cells and their results indicate that protein thiols represent a larger redox pool than glutathione [55].
A final consideration for interpreting the results of this study is the effect of cell growth phase on intracellular GSH levels. All the results reported here were obtained with cells in log phase. Considering that the GSH concentrations were consistently higher during log phase compared with stationary growth phase, it remains to be determined how the cell growth phase affects the activity of reducible polyplexes, especially because of its relevance for in vivo activity.
5. CONCLUSIONS
We hypothesized that increased GSH levels present in cancer cells will selectively improve the activity of redox-sensitive gene delivery systems and tested our hypothesis in a panel of four different human pancreatic cancer cell lines with innate differences in GSH levels. Overall, the results presented here demonstrate that redox-sensitive polyplexes improve delivery of all major types of nucleic acids and that increased innate GSH levels can be advantageously utilized to improve delivery of mRNA in particular. These findings encourage further exploitation of reducible delivery systems to confirm the enhanced selectivity in vivo and demonstrate the promise of these systems to deliver nucleic acid therapeutics to tumors with altered redox state.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (5R01 CA109711). We thank Jun Chen for help with synthesis and molecular weight analysis of polycations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15(16):1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- [2].Neu M, Germershaus O, Behe M, Kissel T. Bioreversibly crosslinked polyplexes of PEI and high molecular weight PEG show extended circulation times in vivo. J Control Release. 2007;124(12):69–80. doi: 10.1016/j.jconrel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- [3].Neu M, Germershaus O, Mao S, Voigt KH, Behe M, Kissel T. Crosslinked nanocarriers based upon poly(ethylene imine) for systemic plasmid delivery: In vitro characterization and in vivo studies in mice. J Control Release. 2007;118(3):370–380. doi: 10.1016/j.jconrel.2007.01.007. [DOI] [PubMed] [Google Scholar]
- [4].Christensen LV, Chang CW, Yockman JW, Conners R, Jackson H, Zhong Z, Feijen J, Bull DA, Kim SW. Reducible poly(amido ethylenediamine) for hypoxia-inducible VEGF delivery. J Control Release. 2007;118(2):254–261. doi: 10.1016/j.jconrel.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Manickam DS, Oupicky D. Multiblock reducible copolypeptides containing histidine-rich and nuclear localization sequences for gene delivery. Bioconjug Chem. 2006;17(6):1395–1403. doi: 10.1021/bc060104k. [DOI] [PubMed] [Google Scholar]
- [6].McKenzie DL, Smiley E, Kwok KY, Rice KG. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconjug Chem. 2000;11(6):901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- [7].Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33(9):e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].You YZ, Manickam DS, Zhou QH, Oupicky D. Reducible poly(2-dimethylaminoethyl methacrylate): synthesis, cytotoxicity, and gene delivery activity. J Control Release. 2007;122(3):217–225. doi: 10.1016/j.jconrel.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McKenzie DL, Kwok KY, Rice KG. A potent new class of reductively activated peptide gene delivery agents. J Biol Chem. 2000;275(14):9970–9977. doi: 10.1074/jbc.275.14.9970. [DOI] [PubMed] [Google Scholar]
- [10].Jeong J. Hoon, Christensen LV, Yockman JW, Zhong Z, Engbersen JF, Kim W. Jong, Feijen J, Kim S. Wan. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28(10):1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]
- [11].Zhong Z, Song Y, Engbersen JF, Lok MC, Hennink WE, Feijen J. A versatile family of degradable non-viral gene carriers based on hyperbranched poly(ester amine)s. J Control Release. 2005;109(13):317–329. doi: 10.1016/j.jconrel.2005.06.022. [DOI] [PubMed] [Google Scholar]
- [12].Kakizawa Y, Harada A, Kataoka K. Glutathione-Sensitive Stabilization of Block Copolymer Micelles Composed of Antisense DNA and Thiolated Poly(ethylene glycol)-block-poly(L-lysine): A Potential Carrier for Systemic Delivery of Antisense DNA. Biomacromolecules. 2001;2(2):491–497. doi: 10.1021/bm000142l. [DOI] [PubMed] [Google Scholar]
- [13].Biaglow JE, Donahue J, Tuttle S, Held K, Chrestensen C, Mieyal J. A Method for Measuring Disulfide Reduction by Cultured Mammalian Cells: Relative Contributions of Glutathione-Dependent and Glutathione-Independent Mechanisms. Analytical Biochemistry. 2000;281(1):77–86. doi: 10.1006/abio.2000.4533. [DOI] [PubMed] [Google Scholar]
- [14].Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Peptide-mediated RNA delivery: a novel approach for enhanced transfection of primary and post-mitotic cells. Nucleic Acids Res. 2001;29(18):3882–3891. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- [16].Manickam D. Soundara, Oupicky D. Polyplex gene delivery modulated by redox potential gradients. J Drug Target. 2006;14(8):519–526. doi: 10.1080/10611860600834409. [DOI] [PubMed] [Google Scholar]
- [17].Manickam D. Soundara, Bisht HS, Wan L, Mao G, Oupicky D. Influence of TAT-peptide polymerization on properties and transfection activity of TAT/DNA polyplexes. J Control Release. 2005;102(1):293–306. doi: 10.1016/j.jconrel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- [18].Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- [19].Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22(5):471–480. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- [20].Balakirev M, Schoehn G, Chroboczek J. Lipoic acid-derived amphiphiles for redox-controlled DNA delivery. Chem Biol. 2000;7(10):813–819. doi: 10.1016/s1074-5521(00)00030-2. [DOI] [PubMed] [Google Scholar]
- [21].Manickam DS, Hirata A, Putt DA, Lash LH, Hirata F, Oupicky D. Overexpression of Bcl-2 as a proxy redox stimulus to enhance activity of non-viral redox-responsive delivery vectors. Biomaterials. 2008;29(17):2680–2688. doi: 10.1016/j.biomaterials.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JF. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: synthesis and in vitro gene transfer properties. J Control Release. 2006;116(2):130–137. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- [23].Hong CY, You YZ, Wu DC, Liu Y, Pan CY. Thermal control over the topology of cleavable polymers: From linear to hyperbranched structures. J Am Chem Soc. 2007;129(17):5354–5355. doi: 10.1021/ja070871+. [DOI] [PubMed] [Google Scholar]
- [24].Fariss MW, Reed DJ. High-performance liquid chromatography of thiols and disulfides: dinitrophenol derivatives. Methods Enzymol. 1987;143:101–109. doi: 10.1016/0076-6879(87)43018-8. [DOI] [PubMed] [Google Scholar]
- [25].Jiang XM, Fitzgerald M, Grant CM, Hogg PJ. Redox control of exofacial protein thiols/disulfides by protein disulfide isomerase. J Biol Chem. 1999;274(4):2416–2423. doi: 10.1074/jbc.274.4.2416. [DOI] [PubMed] [Google Scholar]
- [26].Grande S, Luciani AM, Rosi A, Palma A, Giovannini C, Sapora O, Guidoni L, Viti V. Metabolism of glutathione in tumour cells as evidenced by 1H MRS. FEBS Lett. 2007;581(4):637–643. doi: 10.1016/j.febslet.2007.01.025. [DOI] [PubMed] [Google Scholar]
- [27].Ortega A, Ferrer P, Carretero J, Obrador E, Asensi M, Pellicer JA, Estrela JM. Down-regulation of glutathione and Bcl-2 synthesis in mouse B16 melanoma cells avoids their survival during interaction with the vascular endothelium. J Biol Chem. 2003;278(41):39591–39599. doi: 10.1074/jbc.M303753200. [DOI] [PubMed] [Google Scholar]
- [28].Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- [29].Hwang C, Lodish HF, Sinskey AJ. Measurement of glutathione redox state in cytosol and secretory pathway of cultured cells. Methods Enzymol. 1995;251:212–221. doi: 10.1016/0076-6879(95)51123-7. [DOI] [PubMed] [Google Scholar]
- [30].Kosower NS, Kosower EM. The glutathione status of cells. Int Rev Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- [31].Bellomo G, Vairetti M, Stivala L, Mirabelli F, Richelmi P, Orrenius S. Demonstration of nuclear compartmentalization of glutathione in hepatocytes. Proc Natl Acad Sci U S A. 1992;89(10):4412–4416. doi: 10.1073/pnas.89.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Smith CV, Jones DP, Guenthner TM, Lash LH, Lauterburg BH. Compartmentation of glutathione: implications for the study of toxicity and disease. Toxicol Appl Pharmacol. 1996;140(1):1–12. doi: 10.1006/taap.1996.0191. [DOI] [PubMed] [Google Scholar]
- [33].Soboll S, Grundel S, Harris J, Kolb-Bachofen V, Ketterer B, Sies H. The content of glutathione and glutathione S-transferases and the glutathione peroxidase activity in rat liver nuclei determined by a non-aqueous technique of cell fractionation. Biochem J. 1995;311(Pt 3):889–894. doi: 10.1042/bj3110889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Markovic J, Borras C, Ortega A, Sastre J, Vina J, Pallardo FV. Glutathione is recruited into the cell nucleus in early phases of cell proliferation. J Biol Chem. 2007;282(28):20416–20424. doi: 10.1074/jbc.M609582200. [DOI] [PubMed] [Google Scholar]
- [35].Sipos B, Moser S, Kalthoff H, Torok V, Lohr M, Kloppel G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: towards the establishment of an in vitro research platform. Virchows Arch. 2003;442(5):444–452. doi: 10.1007/s00428-003-0784-4. [DOI] [PubMed] [Google Scholar]
- [36].Frazier ML, Fernandez E, de Llorens R, Brown NM, Pathak S, Cleary KR, Abbruzzese JL, Berry K, Olive M, Le Maistre A, Evans DB. Pancreatic adenocarcinoma cell line, MDAPanc-28, with features of both acinar and ductal cells. Int J Pancreatol. 1996;19(1):31–38. doi: 10.1007/BF02788373. [DOI] [PubMed] [Google Scholar]
- [37].Donoghue N, Yam PT, Jiang XM, Hogg PJ. Presence of closely spaced protein thiols on the surface of mammalian cells. Protein Sci. 2000;9(12):2436–2445. doi: 10.1110/ps.9.12.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Feener EP, Shen WC, Ryser HJ. Cleavage of disulfide bonds in endocytosed macromolecules. A processing not associated with lysosomes or endosomes. J Biol Chem. 1990;265(31):18780–18785. [PubMed] [Google Scholar]
- [39].Ryser HJ, Mandel R, Ghani F. Cell surface sulfhydryls are required for the cytotoxicity of diphtheria toxin but not of ricin in Chinese hamster ovary cells. J Biol Chem. 1991;266(28):18439–18442. [PubMed] [Google Scholar]
- [40].Sahaf B, Heydari K, Herzenberg LA. The extracellular microenvironment plays a key role in regulating the redox status of cell surface proteins in HIV-infected subjects. Arch Biochem Biophys. 2005;434(1):26–32. doi: 10.1016/j.abb.2004.11.015. [DOI] [PubMed] [Google Scholar]
- [41].Laragione T, Bonetto V, Casoni F, Massignan T, Bianchi G, Gianazza E, Ghezzi P. Redox regulation of surface protein thiols: identification of integrin alpha-4 as a molecular target by using redox proteomics. Proc Natl Acad Sci U S A. 2003;100(25):14737–14741. doi: 10.1073/pnas.2434516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Weitz DA, Huang JS, Lin MY, Sung J. Dynamics of Diffusion-Limited Kinetic Aggregation. Phys Rev Lett. 1984;53:1657–1660. [Google Scholar]
- [43].Zhou QH, You YZ, Wu C, Huang Y, Oupicky D. Cyclic RGD-targeting of reversibly stabilized DNA nanoparticles enhances cell uptake and transfection in vitro. J Drug Target. 2009:1–10. doi: 10.1080/10611860902807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu Y, Szoka FC., Jr. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35(18):5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- [45].Moore NM, Sheppard CL, Barbour TR, Sakiyama-Elbert SE. The effect of endosomal escape peptides on in vitro gene delivery of polyethylene glycol-based vehicles. J Gene Med. 2008;10(10):1134–1149. doi: 10.1002/jgm.1234. [DOI] [PubMed] [Google Scholar]
- [46].Read ML, Bremner KH, Oupicky D, Green NK, Searle PF, Seymour LW. Vectors based on reducible polycations facilitate intracellular release of nucleic acids. J Gene Med. 2003;5(3):232–245. doi: 10.1002/jgm.331. [DOI] [PubMed] [Google Scholar]
- [47].Wan L, Manickam DS, Oupicky D, Mao G. DNA Release Dynamics from Reducible Polyplexes by Atomic Force Microscopy. Langmuir. 2008;24(21):12474–12482. doi: 10.1021/la802088y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci U S A. 2007;104(41):16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16(1):163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4(5):438–446. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- [51].Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, Engbersen JF, Feijen J. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjug Chem. 2006;17(5):1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- [52].Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2003;100(7):3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Iida T, Mori T, Katayama Y, Niidome T. Overall interaction of cytosolic proteins with the PEI/DNA complex. J Control Release. 2007;118(3):364–369. doi: 10.1016/j.jconrel.2006.12.027. [DOI] [PubMed] [Google Scholar]
- [54].Lam AP, Dean DA. Cyclic stretch-induced nuclear localization of transcription factors results in increased nuclear targeting of plasmids in alveolar epithelial cells. J Gene Med. 2008;10(6):668–678. doi: 10.1002/jgm.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hansen RE, Roth D, Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.