Abstract
The crystal structure of human endostatin reveals a zinc-binding site. Atomic absorption spectroscopy indicates that zinc is a constituent of both human and murine endostatin in solution. The human endostatin zinc site is formed by three histidines at the N terminus, residues 1, 3, and, 11, and an aspartic acid at residue 76. The N-terminal loop ordered around the zinc makes a dimeric contact in human endostatin crystals. The location of the zinc site at the amino terminus, immediately adjacent to the precursor cleavage site, suggests the possibility that the zinc may be involved in activation of the antiangiogenic activity following cleavage from the inactive collagen XVIII precursor or in the cleavage process itself.
Endostatin is a 20-kDa C-terminal globular domain of the collagen-like protein, collagen XVIII (1), first isolated from a hemangioendothelioma cell line for its ability to inhibit the proliferation of capillary endothelial cells (1). Purified recombinant murine endostatin generated in bacteria injected as an unfolded suspension, inhibited the growth of a variety of implanted, primary tumors in mice, but lacked antiproliferative activity on tumor cells themselves in vitro (1). Repeated cycles of endostatin-induced tumor regression and inhibitor-free regrowth did not generate endostatin resistant tumors consistent with an antiangiogenic mechanism targeting capillary epithelial cells rather than rapidly dividing tumor cells (2). Cycled treatment of several tumors in mice resulted in a persistent dormant state of the tumors (2). The x-ray structure of murine endostatin, to 1.5 Å resolution, revealed a structural similarity to the C-type lectin and E-selectin-like family of proteins, but endostatin lacks characteristic Ca2+-binding sites that are active in the carbohydrate-binding function of selectins (3). A surface rich in the basic residue arginine was identified as a potential heparin-binding site, which might antagonize the heparin-binding activity of the angiogenic protein, basic fibroblast growth factor (3, 4). Murine endostatin for x-ray studies was expressed in human tissue culture cells with four extra amino acids “APLA” at its N terminus. In the crystals, grown at pH 5.0, the first 10 residues and the last five residues were disordered (3) and no metal atoms were observed.
Several angiogenic and antiangiogenic proteins are stored as inactive precursors in the blood or basement membranes (reviewed in ref. 5). Endostatin is stored as the C-terminal portion of collagen XVIII (6), which is localized in the basement membrane around blood vessels (7). Fragments of collagen XVIII longer than endostatin do not inhibit endothelial cell proliferation (1). A balance of positive and negative signals appears to control angiogenesis (5, 8), but what factors regulate the activity of the various proteins, like endostatin, around tumors and normal cells is largely unknown.
We have determined the structure of human endostatin by x-ray crystallography, revealing a zinc site, and used atomic absorption spectroscopy to show that zinc is a constituent of both human and murine endostatin in solution. The location of the zinc site at the amino terminus, immediately adjacent to the precursor cleavage site, suggests the possibility that the zinc is important for the activation or antiangiogenic activity of endostatin.
METHODS
Expression, Purification, and Characterization of Human Endostatin.
Human endostatin was expressed in the NS/0 murine myeloma cell line as an Fc-endostatin fusion protein according to Lo et al. (9). Fc is a fragment of the human Ig γ-1. Polymerase chain reaction was used to adapt the endostatin cDNA for expression in the pdCs-Fc(D4K) vector, which contains the enterokinase recognition site (Asp)4-Lys (10) at the junction of the fusion protein. The forward primer was 5′-C AAG CTT CAC AGC CAC CGC GAC TTC C, where the AAGCTT (HindIII site) was followed by the sequence encoding the N terminus of endostatin. The reverse primer was 5′-C CTC GAG CTA CTT GGA GGC AGT CAT G, which was designed to put a translation stop codon (anticodon, CTA) immediately after the C terminus of endostatin, and this was followed by an XhoI site (CTCGAG). The PCR product was cloned and sequenced, and the HindIII–XhoI fragment encoding endostatin was ligated to the pdCs-Fc(D4K) vector. Stable clones expressing Fc(D4K)-endostatin were obtained by electroporation of NS/0 cells followed by selection in growth medium containing 100 nM methotrexate. Protein expression level was assayed by anti-human Fc ELISA as described (9) and confirmed by SDS/PAGE. The best producing clone was subcloned by limiting dilutions. 2. Fc-human endostatin was purified on Protein A Sepharose (Repligen, Needham, MA) by using 100 mM citric acid, pH 3.0 as elution buffer. Enterokinase and trypsin digestions were carried out, resulting in two forms of cleaved endostatin. Trypsin digestion was carried out at room temperature for 18 hr with an enzyme to protein (wt/wt) ratio of 1:200. The resulting endostatin was four amino acids (His-Ser-His-Arg) cleaved from the N terminus. Enterokinase digestion led to a product with one additional amino acid leucine at N terminus of endostatin. Both cleaved products were purified further by two additional steps; heparin Sepharose and SP Sepharose (Pharmacia). Soluble recombinant Fc-endostatin completely suppresses tumors in mice induced with Lewis Lung tumor (K.J. and J.F., manuscript in preparation).
Structure Determination.
The structure was determined from crystals obtained at 4°C by hanging drop vapor diffusion, with equal volumes of 10 mg/ml protein in 20 mM Tris·HCl (pH 8.5) and 150 mM NaCl and 50 mM Tris⋅HCl (pH 8.5), 6% polyethylene glycol 8000, 1.5 M NaCl, and 2 mM MgCl2 mixed at room temperature in the drops and equilibrated against 0.5 ml of the latter buffer. Crystals (C2 a = 92.76 Å, b = 74.27 Å, c = 137.80 Å, β = 102.56) have four monomers in the asymmetric unit. Crystals were transferred to 15% polyethylene glycol 6,000, 20% glycerol, 50 mM Tris⋅HCl, pH 8.5, and 150 mM NaCl for ≈20 sec before flash cooling in a stream of cold nitrogen. Data were collected at the Cornell High Energy Synchronton Source (CHESS) A1 station (λ = 0.92 Å) with the 80 mm 2K CCD (bined mode) (0.5 degree oscillations). Data were integrated and scaled with denzo and scalepack (17). Most of the subsequent processing used the ccp4 programs (11, 12).
Murine endostatin (3) was used in molecular replacement calculations by using amore (13) (residues Q138–S308 corresponding to human P8–S177) with the 17 sequence differences made alanine. Four monomers were located with final correlation coefficient of 45.9% and R factor of 41.4% by using data from 12.0–3.5 Å. Initial electron density maps (2Fo-Fc and Fo-Fc) were calculated from the rigid body-refined search model. The Fo-Fc map showed four strong positive peaks (>7σ), suggesting metals, located in pairs, near the monomer-monomer interfaces of two noncrystallographic dimers. Some electron density for metal ligands was evident in both 2Fo-Fc and Fo-Fc electron density maps, and some electron density could be seen at the dimer interface, but it was discontinuous at that stage. Two residues from the N terminus and one from the C terminus were deleted from the initial search model to reduce model bias. A monomer envelope was made based on the model by using mama (11, 12) and checked with program o (14). An electron density map calculated with sfal (15) was fourfold averaged inside the envelope by using ave (11, 12) and extended to the whole unit cell. New phases were calculated from the averaged electron density to initiate iterative noncrystallographic real space averaging and solvent flattening with dm (11, 12). After 12 cycles, the correlation converged (from 0.5 to 0.8). This improved electron density map showed that the N-terminal histidines were metal ligands. Cycles of model building and refinement included a bulk-solvent correction coupled with conventional positional refinement, simulated annealing, and individual B factor refinement. Because of the low resolution of the data and a low observation to parameter ratio, strict NCS constraints were used at all stages of refinement. After three rounds of rebuilding (Rfree = 0.315%; Rwork = 0.295%; see Table 1), the N-terminal nine residues (not observed in the murine structure; ref. 3) and two from C terminus were built. This permitted construction of a nonoverlapping monomer envelope in the dimer interface near the metal sites. Iterative fourfold averaging starting from a map with phases calculated by omitting the nine N-terminal residues and using this new envelope generated clear electron density for the entire 9-aa loop around the zinc (Fig. 5). For refinement, data with |Fobs| (observed structure factor amplitude) > 2 σ were used; for map calculation, data with |Fobs|>0 were used. The final Rfree is 27.5% and Rwork 24.0%. The final model contains residues from 1 to 178 and one zinc. An N-terminal residue preceding the endostatin sequence and the five C-terminal endostatin residues are not observed (Table 1). All non-glycine residues are in the allowed region of the Ramachandran plot, with 83% in the most favorable regions, 16% in the allowed region, and with Asp-76, a zinc ligand, in the generally allowed region.
Table 1.
Statistics for data collection and refinement
| Data collection | |
|---|---|
| Resolution, Å | 25.0–2.9 (3.0–2.9) |
| Mosaicity, ° | 0.75 |
| Unique reflections | 19089 (1336) |
| Completeness, % | 89.8 (59.0) |
| Average I/s, I | 14.4 (6.4) |
| Redundancy | 2.9 (1.9) |
| Rsym, % | 7.1 (13.8) |
| Refinement (10.0–3.0 Å) | |
| Reflections (free) | 18175 (1272) |
| Rwork (Rfree), % | 24.0 (27.5) |
| Non-hydrogen protein atoms | 1382 × 4 = 5528 |
| rms deviations | |
| Bonds, Å | 0.013 |
| Angles, ° | 2.03 |
| B-factor, Å2 | 8.43 |
I/s(I) is the mean reflection intensity/estimated error. Rsym = Σ|Ii − 〈I〉/ΣIi, where Ii is the intensity of an individual reflection and 〈I〉 is the average intensity of that reflection. Rcryst = Σ| |Fp|−|Fc| |/Σ|Fp|, where Fc is the calculated structure factor. Rfree is equivalent to Rcryst but calculated for a randomly chosen 7% of reflections that were omitted from the refinement process.
Figure 5.
Electron density of the zinc site. This electron density map was calculated with the terminal residues 1–9 and 178–183 omitted, using the murine molecular replacement model. The map was iteratively averaged (fourfold) and solvent flattened with a molecular envelope determined from the final structure.
RESULTS
Expression and Structure Determination.
Human endostatin has been expressed as a secreted fusion protein, with the Fc domain of IgG-1, in a murine myeloma cell and released from the Fc portion by enterokinase digestion. N-terminal sequencing and mass spectrometry indicate that one leucine precedes the natural N-terminal histidine (residue 132 [ref. (3)] here called residue 1) of the purified, full length (1–183) endostatin. The three-dimensional structure was determined and refined to 3.0 Å resolution by using the mouse endostatin coordinates to initiate phase determination by molecular replacement and the presence of four endostatin molecules in the asymmetric unit of the crystal for iterative fourfold noncrystallographic real-space averaging and solvent flattening. The final model (statistics in Table 1) is missing the nonendostatin leucine at the N-terminal and the five C-terminal residues 179MTASK183.
Human Endostatin Contains Zinc.
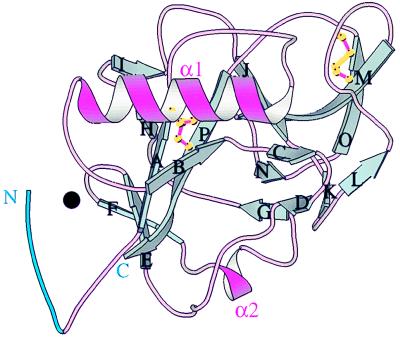
The human endostatin monomer structure is centered on a seven-stranded β-sheet (E, F, A, P, J, M, O in Fig. 1), like the murine molecule (3), the last five strands (A-O) being analogous to the five central strands in the carbohydrate recognition domain of mammalian C-type lectins (reviewed in ref. 16). One side of the β-sheet contains the α1 α helix (Fig. 1) and the other side is covered by short β sheets, loops, and a second α helix, α2. The loops following strand J and preceding strand M follow very different paths from those in carbohydrate recognition domains, which contain Ca2+ sites and are involved in carbohydrate binding (16). The N-terminal six residues of human endostatin (blue in Fig. 1) are ordered around a zinc atom (black sphere, Fig. 1) (see below). The indication that zinc might be a constituent of endostatin came from studies by one of us (T.B.) indicating that mouse endostatin expressed in yeast was differentially susceptible to proteolytic removal of residues 1–3 (His, Thr, and His) by a contaminating protease, only when treated with EDTA at 37°C (T.B. and J.F., unpublished data). This prompted us to grow human endostatin crystals at higher pH in which the N-terminal histidines would not be charged. Data from a new crystal form of mouse endostatin grown at pH 8.5 also shows a zinc site (E. Hohenester, unpublished data).
Figure 1.
The structure of human endostatin. β-strands (cyan) are labeled in sequential order A–P, α helices are violet, and connecting loops are pink. Residues 1–6 are blue; zinc is a black circle. Human and murine endostatin are very similar (rms deviation = 0.46 Å for 196 Cα pairs; cf. Fig. 3B in ref. 3).
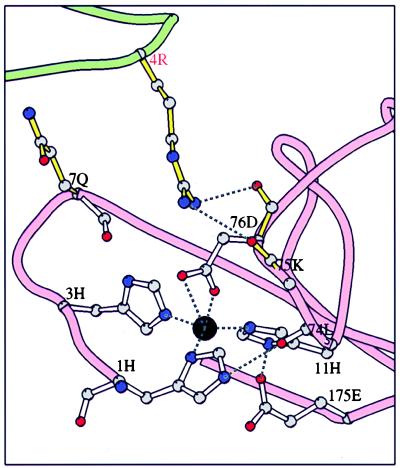
The zinc-binding site of human endostatin is tetrahedral with three zinc ligands from the N-terminal loop, histidine-1, -3, and -11, and a fourth ligand, aspartic acid-76 from the loop between the E and F β-strands (Fig. 1 and Fig. 2). The zinc ligands are conserved in the three known endostatin sequences: human, murine, and dog. No zinc atoms have been found in other members of the C-lectin family of proteins and none of the Ca2+ ions of those molecules are found near the endostatin zinc position, despite the clear structural similarity among these molecules. The endostatin zinc-binding site most closely resembles the structural zinc site of the matrix metalloproteinases such as human fibroblast collagenase (18–20), which have three histidine and one aspartic acid ligand, and the active site zinc of proteases, such as thermolysin and carboxypeptidase A, which have two histidines and one glutamic acid as zinc ligands, with the fourth position occupied by a bound, activated water molecule that is the attacking nucleophile in the proteolytic mechanism (reviewed in refs. 21 and 22). The possibility that histidine-1 at the N terminus of endostatin might be replaced by a water molecule generating a protease at some stage, such as in the precursor form of the molecule, is discussed below.
Figure 2.
The N-terminal loop and zinc binding site of human endostatin. The zinc (black circle) ligands histidine-1, -3, and -11 and aspartic acid-76, as well as the second shell of interactions that position the zinc ligands, glutamic acid-175, residue 11 carbonyl oxygen, and arginine-4 from the N-terminal loop of an adjacent molecule in a dimer are shown as ball-and-stick models. The zinc is coordinated approximately tetrahedrally between the three histidines and one aspartic acid. Both oxygens of the aspartic acid may bond to the zinc, as shown, but the electron density at this resolution is not unambiguous.
Atomic absorption spectroscopy confirmed that zinc is a constituent of the Fc-endostatin of both human and murine endostatin in solution (Table 2). Enterokinase treated Fc-human endostatin, which yields full length endostatin, also showed approximately one atom of zinc per endostatin monomer. Trypsin cleaved Fc-human endostatin, which results in loss of the first four residues of endostatin contains no detectable zinc (Table 2). These results are consistent with the human endostatin crystal structure that shows histidines at residues 1 and 3 to be two of the zinc ligands.
Table 2.
Measurements of zinc in endostatin
| Protein | N terminus sequence | Endostatin concentration, μM | Zinc concentration, μM | Zinc/endostatin monomer |
|---|---|---|---|---|
| hE-EK | LHSHRD–– | 190 | 176 | 0.9 |
| hE-Tryp | D–– | 180 | 14 | 0.07 |
| hFc-hE | 19 | 18 | 0.9 | |
| mFc-mE | 90 | 79 | 0.9 |
Zinc determinations were performed by flame atomic absorption spectrometry with a Perkin–Elmer 2280 instrument. hE-EK, human endostatin derived from Fc construct by enterokinase digestion. hE-Tryp, human endostatin obtained from Fc construct by trypsin digestion. Fc-hE and Fc-mE denote human and murine Fc constructs, respectively. For more details, see Methods.
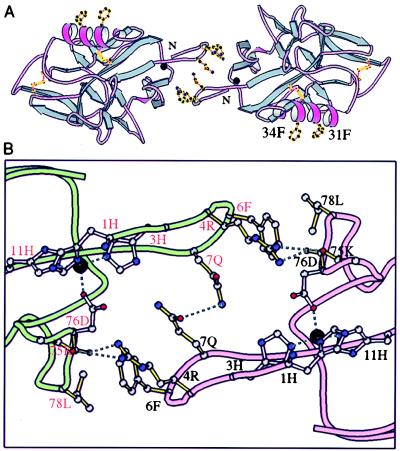
Endostatin Dimers in the Crystal.
A number of twofold symmetric dimers is evident in the packing of the four endostatin monomers in the asymmetric unit of the human endostatin crystals. One dimer interface is formed by the contact of two solvent exposed phenylalanines on the α1 α helix from each monomer, a prominent nonpolar patch suggested as a potential interdomain interaction site in murine endostatin (3) (residues F31 and F34 in Fig. 3). Another dimeric contact, that would be zinc dependent, is formed between the projecting N-terminal loops of two monomers; each loop ordered around a zinc ion (Fig. 3A, center). This contact is formed primarily by three residues that project from the rim of the N-terminal loop: arginine-4, phenylalanine-6, and glutamine-7 (Fig. 3B). The glutamines at residue 7 of each monomer contact each other forming a hydrogen bond at the center of the interface. Each ring of the phenylalanines at residue 6 contacts a nonpolar patch containing leucine-78 of the adjacent monomer. Arginine-4 from each monomer forms hydrogen bonds with two main chain carbonyl oxygens on the loop containing the aspartic acid zinc ligand at residue 76 (carbonyls at residue 75 and 76) (Fig. 3B). This dimeric interaction could only occur if the N-terminal loops were ordered as the result of binding zinc. No dimers of human endostatin released from Fc-endostatin by enterokinase were observed by gel filtration chromatography. Whether these dimeric contacts observed in crystal packing occur in vivo, e.g., when bound to a receptor, is unknown.
Figure 3.
A zinc dependent dimer in human endostatin crystals. (A) The zinc (black circle) site N-terminal loops of two monomers contact across a central dyad axis (colors as in Fig. 1). Glutamine-7, phenylalanine-6, and arginine-4 of the loop project from one monomer to the next. Also shown are two phenylalanine rings, residues 31 and 34, that project from an endostatin α helix and form another dimeric contact in the crystal (see text). (B) Contacts at the interface of the dimer seen in crystals of human endostatin. Zinc (black sphere) ligands have open bonds, interface residues have solid colored bonds. The path of the polypeptide chains of the two monomers are shown as pink and green tubes. The solvent accessible surface buried in this dimer interface is 403 Å2 per monomer (probe size = 1.4 Å).
Tentative Heparin-Binding Site.
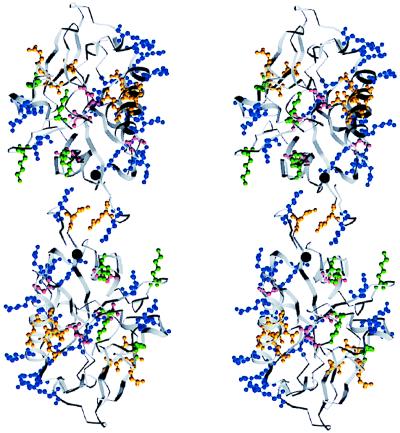
One surface of endostatin contains 11 of the 15 positively charged arginine residues (Fig. 5 in ref. 3) and has been proposed to contain the heparin-binding site. The arrangement of this patch of residues on the zinc dependent dimer of human endostatin is shown in Fig. 4.
Figure 4.
A positively charged surface formed by arginines on the human endostatin dimer. Stereo diagram, surface glutamines (yellow), asparagines (cyan), lysines (green), and arginines (blue) are shown.
Structural Similarities to Other Proteins.
A search of the structural database using the program dali (23, 24) found that endostatin has structural similarity to the carbohydrate recognition domains of the C-type lectin family of proteins (16). Murine endostatin was found (3) to be most similar to E-selectin (dali score Z = 3.1), an endothelial cell-specific membrane adhesion glycoprotein (25) and lithostathine, an inhibitor of stone formation in the pancreas (26). Although endostatin shares only a statistically insignificant 9% sequence identity with these proteins, the structural similarity indicates an evolutionary relationship.
A similar search with the human endostatin structure found slightly greater structural similarity to two proteins of the same C-type lectin family, human tetranectin, a trimeric plasminogen-binding protein (Z = 3.8) (27) and coagulation factors IX/X-binding protein from snake venom (Z = 3.7) (28, 29). The α-carbon positions of these two proteins can be superimposed on human endostatin with rms deviation values of 3.8 Å and 3.1 Å, respectively (best 87 and 79 Cα pairs). Tetranectin has structural similarity to the collectins, like mannose-binding proteins (16), that contain trimeric stems of collagen repeats and an α helical coiled coil. Although no α helical coiled coil is evident in its sequence, the precursor of endostatin, collagen XVIII, is also a trimeric molecule with a long stem of collagen repeats (6, 30). Tetranectin binds both plasminogen and carbohydrates (31–33) and has been colocalized with plasminogen at the invasive front of cutaneous melanoma lesions (34), suggesting that it may target plasminogen to particular cells or extracellular matrices (27), but its biological role remains unknown. Coagulation factors IX/X-binding protein is an anticoagulant that binds in a Ca2+-dependent fashion to block the coagulation cascade (35). Tetranectin and the anticoagulant are both Ca2+ proteins and do not have zinc sites, but like many proteins that control angiogenesis (1, 5, 8, 36–38), they are associated with blood clotting enzymes suggesting an evolutionary relationship between the regulation of angiogenesis and of coagulation (1).
DISCUSSION
The finding that zinc is a constituent of endostatin suggests that it may be a requirement either for the activation of endostatin from its precursor or for its antiangiogenic activity. Zinc-binding sites in proteins can be simply structural or more directly functional. Even if the endostatin zinc were primarily structural it still could have consequences for activity either indirectly by stabilizing the tertiary structure or more directly if the N-terminal loop structured around the zinc were involved in activity, such as through endostatin dimer formation or through interaction of this loop with a currently unknown target protein. This idea can be tested by mutation of the arginine, phenylalanine, and glutamine, which project from the N-terminal loop to form the dimer (Fig. 3).
A more direct enzymatic function of the zinc site currently appears unlikely for two reasons: first, endostatin has four zinc ligands (Fig. 2), one of which (such as the N-terminal histidine) would need to be replaced by water to create a zinc-protease-like site; second, when we superimpose the zinc site of carboxypeptidase A on endostatin, we do not find candidates for either (i) the general base equivalent to glutamic acid 270 of carboxypeptidase A, required in the proteolytic mechanisms to abstract a proton from the zinc bound water forming the nucleophilic hydroxide ion that attacks the substrate carbonyl carbon or (ii) a side chain or backbone amide equivalent to arginine-127 of carboxypeptidase A, to stabilize the negative charge developed on the carbonyl oxygen of a substrate in the tetrahedral intermediate in the proteolysis. It is not impossible that these elements required for catalysis could be supplied by parts of precursor collagen XVIII, whose structure is unknown, and that the zinc site could be involved either in the cleavage of the precursor or in some activity following cleavage. Activation of some matrix prometalloproteinases occurs by the conversion of a structural, tetradentate, protein-zinc site to a tridentate-zinc site. Water then becomes the additional ligand after proteolytic removal of a segment containing the fourth protein-zinc ligand (20, 21, 39). In the collagenase-like astacins and serralysins, one finds substrate displacement of a fifth tyrosine-zinc ligand during catalysis.
Understanding the role of zinc in the activity or regulation of the activity of endostatin may inform studies of the regulation of angiogenesis and may have practical application in the design of experimental antiangiogenic therapies. Strategies for stabilizing the zinc-binding loop with other metals may be useful to stabilize the protein, especially if the loop contacts a receptor. Protein engineering to create a covalent dimer by a disulfide link between residues at position 7 (Fig. 3b) or by forming a single chain dimer based on the proximity of the C and N termini of respective monomers, might produce a more potent protein.
Acknowledgments
We thank Richard Crouse and Steven Sluter for excellent technical support and other members of the Harrison/Wiley group and Wei Wen for useful discussions. Y.-H.D. has been supported by the Howard Hughes Medical Institute and the National Institutes of Health. E.H. acknowledges receipt of a long-term fellowship from the Human Frontier Science Program Organization. The research is supported by the National Institutes of Health, the Howard Hughes Medical Institute (D.C.W.), a National Cancer Institute grant (J.F.), a grant from EntreMed to the Children’s Hospital, and the European Community (EC). D.C.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Data deposition: The atomic coordinates of human endostatin have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1bnl).
References
- 1.O’Reilly M S, Boehm T, Shing Y, Fukai N, Vasios G, Lane W S, Flynn E, Birkhead J R, Olsen B R, Folkman J. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 2.Boehm T, Folkman J, Browder T, O’Reilly M S. Nature (London) 1997;390:404–407. doi: 10.1038/37126. [DOI] [PubMed] [Google Scholar]
- 3.Hohenester E, Sasaki T, Olsen B R, Timpl R. EMBO J. 1998;17:1656–1664. doi: 10.1093/emboj/17.6.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown K J, Parish C P. Biochemistry. 1994;33:13918–13927. doi: 10.1021/bi00250a047. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Folkman J. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 6.Oh S P, Kamagata Y, Muragaki Y, Timmons S, Ooshima A, Olsen B R. Proc Natl Acad Sci USA. 1994;91:4229–4233. doi: 10.1073/pnas.91.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muragaki Y, Timmons S, Griffith C M, Oh S P, Fadel B, Quertermous T, Olsen B R. Proc Natl Acad Sci USA. 1995;92:8763–8767. doi: 10.1073/pnas.92.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouck N. Biochim Biophys Acta. 1996;1287:63–66. doi: 10.1016/0304-419x(96)00005-4. [DOI] [PubMed] [Google Scholar]
- 9.Lo K-M, Sudo Y, Chen J, Li Y, Lan Y, Kong S-M, Chen L, An Q, Gillies S G. Protein Eng. 1998;11:495–500. doi: 10.1093/protein/11.6.495. [DOI] [PubMed] [Google Scholar]
- 10.LaVallie E R, Rehemtulla A, Racie L A, DiBlasio E A, Ferenz C, Grant K L, Light A, McCoy J M. J Biol Chem. 1993;268:23311–23317. [PubMed] [Google Scholar]
- 11.Jones T A. In: Molecular Replacement: Proceedings of the CCP4 Study Weekend. Dodson E J, Gover S, Wolf W, editors. Darsbury, UK: SERC; 1992. pp. 91–105. [Google Scholar]
- 12.Kleywegt G J, Jones T A. In: Halloween … Masks and Bones in From First Map to Final Model. Bailey S, Hubbard R, Walker D, editors. SERC Darnesbury Laboratory; 1994. pp. 59–66. [Google Scholar]
- 13.Navaza J, Saludjian P. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 14.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Collaborative Computational Project. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 16.Weis W I, Taylor M E, Drickamer K. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter C W Jr, Sweet R M, editors. Vol. 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 18.Lovejoy B, Cleasby A, Hassell A M, Longley K, Luther M A, Weigl D, McGeehan G, McElroy A B, Drewry D, Lambert M H, et al. Science. 1994;263:375–377. doi: 10.1126/science.8278810. [DOI] [PubMed] [Google Scholar]
- 19.Lovejoy B, Hassell A M, Luther M A, Weigl D, Jordan S R. Biochemistry. 1994;33:8207–8217. doi: 10.1021/bi00193a006. [DOI] [PubMed] [Google Scholar]
- 20.Stocker W, Grams F, Baumann U, Reinemer P, Gomis-Ruth F X, McKay D B, Bode W. Protein Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallee B L, Auld D S. Proc Natl Acad Sci USA. 1990;87:220–224. doi: 10.1073/pnas.87.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallee B L, Auld D S. Biochemistry. 1993;32:6493–6500. doi: 10.1021/bi00077a001. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 24.Holm L, Sander C. Nucleic Acids Res. 1994;22:3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 25.Graves B J, Crowther R L, Chandran C, Rumberger J M, Li S, Huang K-S, Presky D H, Familletti P C, Wolitzky B A, Burns D K. Nature (London) 1994;367:532–538. doi: 10.1038/367532a0. [DOI] [PubMed] [Google Scholar]
- 26.Bertrand J A, Pignol D, Bernard J-P, Verdier J-M, Dagorn J-C, Fontecilla-Camps J C. EMBO J. 1996;15:2678–2684. [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen B B, Kastrup J S, Rasmussen H, Holtet T L, Graversen J H, Etzerodt M, Thøgersen H C, Larsen I K. FEBS Lett. 1997;412:388–396. doi: 10.1016/s0014-5793(97)00664-9. [DOI] [PubMed] [Google Scholar]
- 28.Mizuno H, Fujimoto Z, Koizumi M, Kano H, Atoda H, Morita T. Nat Struct Biol. 1997;4:438–441. doi: 10.1038/nsb0697-438. [DOI] [PubMed] [Google Scholar]
- 29.Morita T, Atoda H, Sekiya F. Natural Toxins. New York: Plenum; 1996. [Google Scholar]
- 30.Rehn M, Pihlajaniemi T. Proc Natl Acad Sci USA. 1994;91:4234–4238. doi: 10.1073/pnas.91.10.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemmensen I, Petersen L C, Kluft C. Eur J Biochem. 1986;156:327–333. doi: 10.1111/j.1432-1033.1986.tb09586.x. [DOI] [PubMed] [Google Scholar]
- 32.Kluft C, Jie A F, Los P, de-Wit E, Havekes L. Biochem Biophys Res Commun. 1989;161:427–433. doi: 10.1016/0006-291x(89)92616-8. [DOI] [PubMed] [Google Scholar]
- 33.Kluft C, Los P, Clemmensen I. Thromb Res. 1989;55:233–238. doi: 10.1016/0049-3848(89)90440-4. [DOI] [PubMed] [Google Scholar]
- 34.De Vries T J, De Wit P E J, Clemmensen I, Verspaget H W, Weidle U H, Bröcker E B, Ruiter D J, Van Muijen G N P. J Pathol. 1996;179:260–265. doi: 10.1002/(SICI)1096-9896(199607)179:3<260::AID-PATH586>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 35.Atoda H, Morita T. J Biochem (Tokyo) 1989;106:808–813. doi: 10.1093/oxfordjournals.jbchem.a122935. [DOI] [PubMed] [Google Scholar]
- 36.Good D J, Polversini P J, Rastinejad F, LeBeau M M, Lemons R S, Frazier W A, Bouck N P. Proc Natl Acad Sci USA. 1990;87:6624–6628. doi: 10.1073/pnas.87.17.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Reilly M S, Holmgren L, Shing Y, Chen C, Rosenthal R A, Cao Y, Moses M, Lane W S, Sage E H, Folkman J. Cold Spring Harbor Symp Quant Biol. 1994;59:471–482. doi: 10.1101/sqb.1994.059.01.052. [DOI] [PubMed] [Google Scholar]
- 38.O’Reilly M S, Holmgren L, Shing Y, Chen C, Rosenthal R A, Moses M, Lane W S, Cao Y, Sage E H, Folkman J. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 39.Springman Proc Natl Acad Sci USA. 1990;87:364–368. doi: 10.1073/pnas.87.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]