Abstract
Radiation metabolomics employing mass spectral technologies represents a plausible means of high-throughput minimally invasive radiation biodosimetry. A simplified metabolomics protocol is described that employs ubiquitous gas chromatography-mass spectrometry and open source software including random forests machine learning algorithm to uncover latent biomarkers of 3 Gy γ radiation in rats. Urine was collected from six male Wistar rats and six sham-irradiated controls for 7 days, 4 prior to irradiation and 3 after irradiation. Water and food consumption, urine volume, body weight, and sodium, potassium, calcium, chloride, phosphate and urea excretion showed major effects from exposure to γ radiation. The metabolomics protocol uncovered several urinary metabolites that were significantly up-regulated (glyoxylate, threonate, thymine, uracil, p-cresol) and down-regulated (citrate, 2-oxoglutarate, adipate, pimelate, suberate, azelaate) as a result of radiation exposure. Thymine and uracil were shown to derive largely from thymidine and 2′-deoxyuridine, which are known radiation biomarkers in the mouse. The radiation metabolomic phenotype in rats appeared to derive from oxidative stress and effects on kidney function. Gas chromatography-mass spectrometry is a promising platform on which to develop the field of radiation metabolomics further and to assist in the design of instrumentation for use in detecting biological consequences of environmental radiation release.
INTRODUCTION
“Every senior leader, when you’re asked what keeps you awake at night, it’s the thought of a terrorist ending up with a weapon of mass destruction, especially nuclear.” U.S. Secretary of Defense Robert Gates (1).
The scenarios by which a nuclear or radiological device might be deployed in the United States have been discussed in some detail (1). The need is obvious for the development of high-throughput, noninvasive and rapid biomonitoring tools for the screening of mass populations in the event of such a catastrophe. As stated in an earlier report (2), the U.S. Homeland Security Council and the Office of Science and Technology Policy created the Weapons of Mass Destruction Medical Countermeasures Subcommittee to oversee the research and development of improved countermeasures (3). This Subcommittee listed the development of biomarkers and devices for biodosimetry among its highest priority areas of research (3).
Radiation metabolomics seeks to define patterns of metabolic changes that are associated with exposure to radiation. Although studies so far have been carried out in urine of irradiated and control animals (2, 4), it is anticipated that other metabolomes such as plasma, sweat, saliva or sebum may also be of value for detecting biomarkers of radiation exposure. This is important for radiation biomonitoring in the field where urine collection may be less practicable than e.g. saliva, sweat or sebum harvest in large populations. Nevertheless, proof of concept has been established in the mouse by employing urine analysis. Through harnessing the accurate mass determination of time-of-flight mass spectrometry (TOFMS) in combination with high-resolution ultra-performance liquid chromatography (UPLC), a number of biomarkers of γ-radiation exposure have been defined using doses ranging from 1 to 11 Gy (2, 4). For each sample analyzed, the UPLC-TOFMS methodology is capable of detecting in excess of 5,000 positive ions and 5,000 negative ions, each of them representing the protonated or deprotonated molecular ion of a distinct urinary metabolite that can be characterized by retention time, accurate mass and ion abundance. An unknown proportion of these ions arise from in-source fragmentation, adducts, dimers and isotopes. However, many of the highly polar metabolites present in urine elute in the first minute of the UPLC chromatogram, and this is not ideal for the analysis of these small polar analytes. An alternative approach is to use the high resolving power of capillary gas chromatography combined with mass spectrometry (GCMS). In this situation, the polar metabolites must first be rendered volatile by the process of chemical derivatization. Such an analysis is more time consuming than the rapid UPLC-TOFMS method, but nevertheless, the GCMS platform offers a useful adjunct to the UPLC-TOFMS approach. Each has its own advantages. Additionally, employing a low-mass resolution instrument, such as one with a single quadrupole mass analyzer, a GCMS has approximately 10% of the purchase price of a high-mass resolution UPLC-TOFMS instrument, putting GCMS-based metabolomics within the reach of more research groups.
In this report, a complete protocol for radiation metabolomics using low-mass resolution GCMS combined with random forests machine learning algorithm (5) is described. In the case of TOFMS systems, mass to charge ratios (m/z) for ions are produced with high mass resolution, typically to the fourth decimal place and with a typical mass error of <5 ppm. This alone may permit identification of the chemical nature of the ion or, at worst, narrow the solution to a number of empirical formulae. This process is discussed in detail elsewhere (6). In the case of a single quadrupole GCMS, which yields low-resolution mass spectra, either with nominal masses or to one decimal place, such a process is not possible. However, a GCMS instrument equipped with an electron impact ion source fragments the molecular ion to give a mass spectrum that is usually unique to a particular substance. Databases exist that contain in excess of 500,000 mass spectra and biological molecules, and their common chemical derivatives are contained within these databases, thus permitting preliminary identification of urinary analytes. Confirmation using authentic standards then follows.
We have applied this approach to radiation metabolomics in the rat to examine whether a similar pattern of radiation biomarkers emerged as described for the mouse (2, 4). Such insights are useful for the translation of laboratory investigations to human subjects and then into the field.
MATERIALS AND METHODS
Compounds
The following compounds were obtained from Sigma-Aldrich Chemie GmbH (Buchs, Switzerland): thymidine, thymine, uridine, 2′-deoxyuridine, uracil, cytidine, 2′-deoxycytidine, cytosine, citric acid, glyoxylic acid, p-cresol, arabitol, L-threonic acid, ferulic acid, azelaic acid, 2-oxoglutaric acid, pimelic acid, suberic acid, adipic acid, glycerol 3-phosphate, 3-chloro-4-hydroxyphenylacetic acid, O-methoxyamine hydrochloride, N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% trimethylchlorosilane (BSTFA/TMCS) and urease from jack beans (37.6 U/mg). Pyridine (purity ≥99.5%) was obtained from Merck KGaA (Darmstadt, Germany). Solvents and inorganic reagents were of the best available grade.
Animals
Male Wistar rats (240–260 g) were obtained from Charles River WIGA (Deutschland) GmbH (Sulzfeld, Germany) and were fed no. 3430 mouse and rat diet (Provimi Kliba AG, Kaiseraugst, Switzerland) ad libitum with free access to fresh drinking water and housed three to four per cage under a standard 12-h light, 12-h dark cycle. Animals were not selected for investigation until they had been housed at least 1 week in Bern. All animal handling and experimental protocols were designed for maximum possible well-being and were approved by the local ethics committee (Office for Agriculture and the Environment for Bern Canton).
Radiation Exposure
Rats were γ-irradiated in groups of three with a nominal dose of 3.0 Gy using a Gammacell® 40 Exactor (Best Theratronics, Ottawa, Canada). This research irradiator was fitted with two 137Cs sources (above and below) with an activity of 1800 Ci/67 TBq and delivering 1.0 Gy/min. Use of direct-reading dosimeters determined that the actual dose delivered to rats under the experimental conditions was 2.80–2.85 Gy. After irradiation, rats were immediately returned to their metabolic cages (see below). Sham irradiation was performed by placing rats similarly into the irradiator but without exposure to the γ-ray sources.
Urine Collection
During urine collection, rats were housed individually in plastic metabolic cages for 7 days. These cages permitted the collection of urine free from fecal contamination. Rats were monitored daily for outward signs of distress or adverse health effects, and their body weights were determined at the beginning and end of the investigation. Additionally, food and water consumption was monitored daily. Twenty-four-hour urines were collected at −4, −3, −2, and −1 days prior to irradiation and at +1, +2, and +3 days after irradiation. The purpose of this protocol was to permit acclimation of the rats to the metabolic cages (2, 4). Urine volumes were determined and the samples were diluted with purified water by rinsing the metabolic cages to a fixed volume of 20 ml and 14 ml for day 5 and all other days, respectively. Aliquots of urine were immediately frozen at −20°C until analyzed.
Sample Preparation for Gas Chromatography-Mass Spectrometry (GCMS)
For multivariate data analysis, urine samples were prepared for GCMS as follows: Urine (25 μl) was diluted fourfold with distilled water and 25 μl was added to an internal standard solution (25 μl of aqueous 200 μM 3-chloro-4-hydroxyphenylacetic acid) and the mixture blown to dryness under a gentle stream of nitrogen at room temperature in 0.3 ml Reacti-Vials (Thermo Fisher Scientific (Schweiz) AG, Basel, Switzerland). Pyridine (25 μl) was added to the dry residues, and the samples were reduced to dryness again under nitrogen to ensure complete removal of water. A two-step derivatization procedure was performed similarly to published methods (7, 8). O-Methoxyamine hydrochloride (0.5% w/v in pyridine; 25 μl) was added and the samples were heated in capped Reacti-Vials in a heating block at 75°C for 30 min. After cooling to room temperature, BSTFA/TMCS silylating reagent (25 μl) was added and the samples were capped and heated again at 75°C for 30 min, cooled to room temperature, and transferred to autoinjector vials, which were immediately capped and crimped.
GCMS Analysis
Samples were analyzed using an Agilent 6890N gas chromatograph fitted with an Agilent 7683B series automatic liquid sampler and an Agilent 5975B series mass selective detector (MSD). For the acquisition of data for multivariate data analysis, samples (1.0 μl) were injected in splitless mode after 3 min column equilibration onto an Agilent HP-5MS capillary column (0.25 μm film thickness; 60 m × 0.25 mm i.d.) subjected to a temperature program of 70°C for 3 min, 4°/min to 280°C, 3 min at 280°C, 10°/min to 300°C, 300°C for 10 min (total run time 70.5 min). The injector inlet was held at 300°C and the MSD transfer line at 280°C, and the carrier gas was helium (1.1 ml/min). Mass spectral data were collected in electron impact mode (69.92 eV) from 11.5 to 70.5 min between 35.0 and 800.0 m/z with the threshold for data acquisition set to 3000 for the elimination of ions with lowest abundance. The mass spectrometer source was set at 230°C and the quadrupole at 150°C. The MSD tune parameters were all within normal ranges. MSD ChemStation software version E.01.01.335 from Agilent Technologies Inc. (Santa Clara, CA) was employed for data analysis.
Bioinformatics
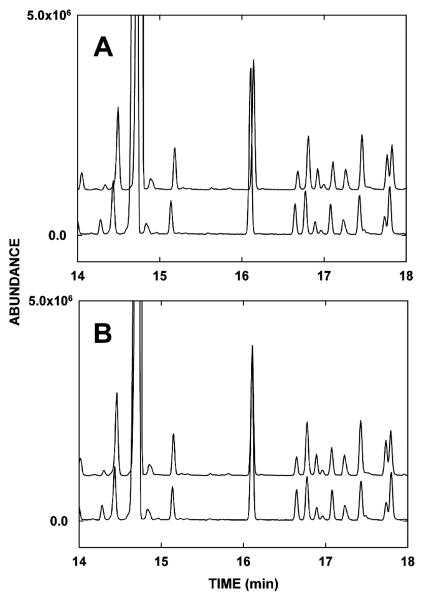
Raw GCMS data files obtained from rat urines collected at day +1 [first day after irradiation (n = 6) and sham irradiation (n = 6)] were exported from the ChemStation software to the AIA format, producing CDF files. MZmine, version 0.60, an open-source software (www.mzmine.sourceforge.net), was used for the pre-processing of the raw data in terms of peak alignment and peak detection. Three peak tables were created with MZmine for subjection to random forests analysis, differing in the time window that was covered. The first peak table encompassed the entire chromatogram, whereas the others were designed to eliminate the influence of the broad peak of urea (between 21.0 and 24.2 min) on the peak picking process. The cropping filter was used to select the appropriate time window and to restrict the mass range from 150 to 800 m/z. Peak detection was performed using the recursive threshold algorithm and the other parameters were set to extract a maximum of small chromatographic peaks and to retain a minimum of minor ions of each mass spectrum in the peak table. The fast aligner was run for the peak alignment. Chromatographic peaks present in less than four samples were eliminated from the peak table and empty gaps were filled estimating peak height and area for the empty slots in the table. For the first and third peak table created, the data were normalized using the peak height of the internal standard to compensate for variations in the derivatization efficiency during the sample preparation. The aligned chromatographic and spectral information that was obtained with this data processing were exported as text files and used for random forests analysis. Peak alignment is essential to avoid the introduction of false positives and false negatives among the highly important markers ranked by random forests. Figure 1A shows two raw nonaligned chromatograms and the same chromatograms after alignment with MZmine (Fig. 1B). It is clear that many peaks will be misclassified or appear as new peaks in the absence of peak alignment.
FIG. 1.
Schematic representation of the process of peak alignment. A section of the chromatograms of (panel A) urine of two control animals before peak alignment, exhibiting a shift in retention time of 0.034 min and (panel B) the same chromatograms after peak alignment with compensation for the shift of retention times between chromatograms.
Random forests analysis was performed in the R software environment. Samples were assigned according to their group membership, i.e. irradiated animals and control animals, and the calculations were based on 10,000 trees and variable importances averaged over 25 independent random forests. The average variable importance measure was used to rank the markers of decreasing importance. A fuller description of the random forests machine learning algorithm applied to metabolomic data appears elsewhere (6).
Verification, Identification and Confirmation
The variable importance score generated from the random forests algorithm was used to select promising signals, each providing the m/z value, the retention time, and the abundances of the respective ion in the two input groups. For the verification of the signals, the raw data generated during the GCMS analyses and the individual chromatograms were inspected. The ratio of the peak area (peak of interest area divided by the internal standard) was plotted for all animals over all 7 study days after correcting for the different dilution of the samples of day 5, using SigmaPlot for Windows Version 10.0 (Systat Software Inc., San Jose, CA).
The identification of peaks showing statistically significant differences between the two groups of animals on at least 1 day of the 3 days after the γ irradiation, and therefore fulfilling the criteria of potential biomarker candidates, was based on the spectral information of the peak of interest. The spectrum was compared with the almost 575,000 spectra stored in the NIST Mass Spectral Database (NIST/EPA/NIH Mass Spectral Library Version 2.0). Identified peaks were confirmed by analyzing aqueous solutions (typically 200 μM) of authentic standards under the same analytical conditions, and retention times and mass spectra were compared to demonstrate the biomarker identities unambiguously. To confirm the relationship between ion signal intensity generated in the mass analyzer and the amount of the analyte present in the sample, and thereby to exclude analytical artifacts stemming from variations in the derivatization efficiency during the sample preparation, selected biomarkers were spiked with known amounts of authentic standards.
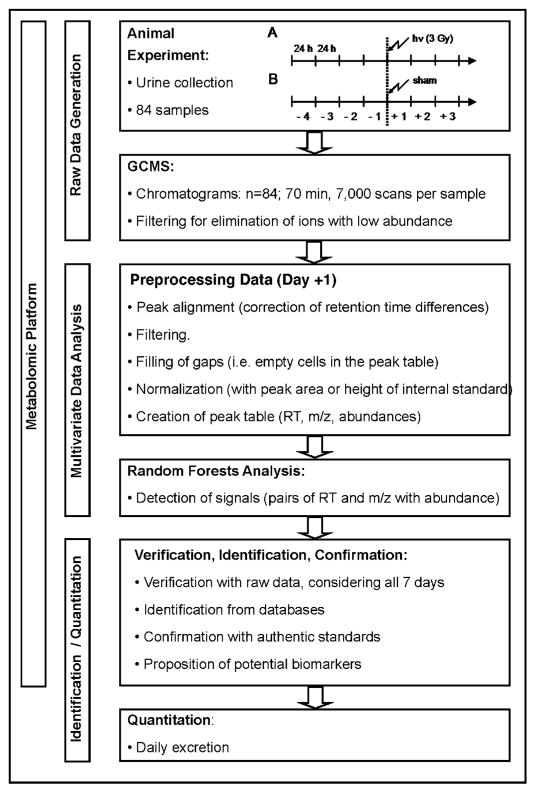
Finally, the relative concentrations (analyte peak area/internal standard peak area) of identified potential biomarkers were derived using 3-chloro-4-hydroxyphenylacetic acid as internal standard and expressed as daily excretion of the respective urinary constituent. This complete process is depicted in the flow diagram in Fig. 2. The GCMS apparatus is relatively inexpensive compared to the high-resolution UPLC-TOFMS platform that we have employed for metabolomic analysis hitherto (2, 4, 6, 9–14). Moreover, all bioinformatic software used was open-source.
FIG. 2.
Flow diagram showing the GCMS and random forests protocol for radiation metabolomics from the animal investigation through to biomarker quantification. The metabolomic platform comprises three main parts—raw data generation, multivariate data analysis, and biomarker identification/quantification.
Urea and Electrolyte Analysis
Samples of fresh urine from each rat on each of the 7 days of the study were sent for urea and electrolyte analysis at the Zentrallabor of the Inselspital, the local teaching hospital in Bern.
Statistical Analyses
For within-group analyses (e.g. sham compared to sham on different days), a paired Student’s t test was used. For comparisons between groups (e.g. sham day +1 compared to irradiated day +1), unpaired t tests were employed.
RESULTS
Animal Performance
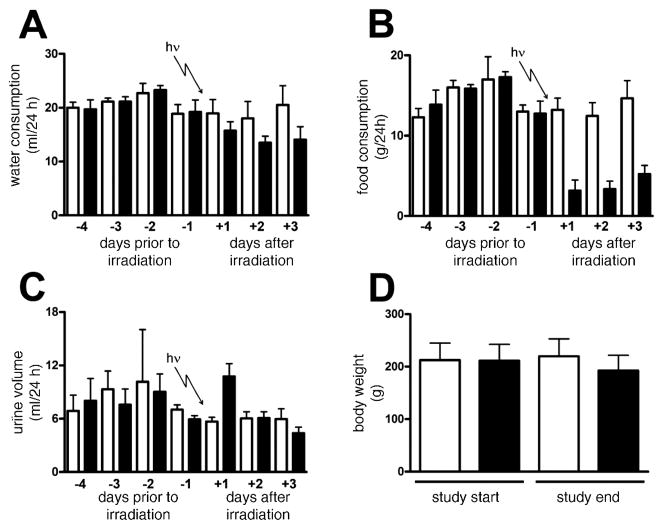
The sham treatment (control) animals did not show any outward signs of distress or adverse health effects during the entire study period. Food and water intake, urinary excretion and body weight for both the sham-irradiated and irradiated animals are depicted in Fig. 3. The animals randomly allocated to the treatment group did not differ in food and water intake, urinary excretion and body weight from the control animals prior to the γ irradiation. For the sham controls, water consumption (Fig. 3A), food intake (Fig. 3B), and urine volume (Fig. 3C) were comparable over the 7 days of the study. Control rats increased their body weight 3.2% from 244.8 ± 12.3 g (mean ± SD) to 252.7 ± 10.1 g during the study. In stark contrast, the irradiated animals experienced an 8.7% loss in body weight from 242.5 ± 6.0 g to 221.3 ± 9.1 g (t = 8.1, P = 0.0005). Despite this, the treated animals did not show obvious signs of sickness in terms of activity, mobility and appearance, nor did they appear to behave differently from the sham controls. Comparisons between irradiated and sham-irradiated rats on day +1 revealed no statistically significant difference in water consumption (Fig. 3A), but there was a marked reduction in food consumption in the irradiated rats on day +1 (Fig. 3B). Interestingly, urine output doubled in the irradiated rats compared both to sham-irradiated rats (Fig. 3C) and to their pre-irradiation urine output (day −1). Moreover, there was a trend toward lower water consumption in the irradiated animals across the 3 days postirradiation (Fig. 3A), and the food consumption remained depressed for these 3 days (Fig. 3B). However, the enhanced diuresis observed on day +1 in irradiated rats returned to normal levels on the subsequent 2 days (Fig. 3C).
FIG. 3.
Trends in water consumption (panel A), food consumption (panel B), and urine volume (panel C) for six sham-irradiated rats and six rats during 4 days of acclimation to metabolic cages and then after exposure to 3 Gy γ radiation. Body weight on days −4 and +3 is also shown (panel D). Open bars and filled bars are for sham-irradiated and irradiated rats, respectively. Error bars represent + 1 SD.
Changes in Urinary Urea and Electrolyte Excretion
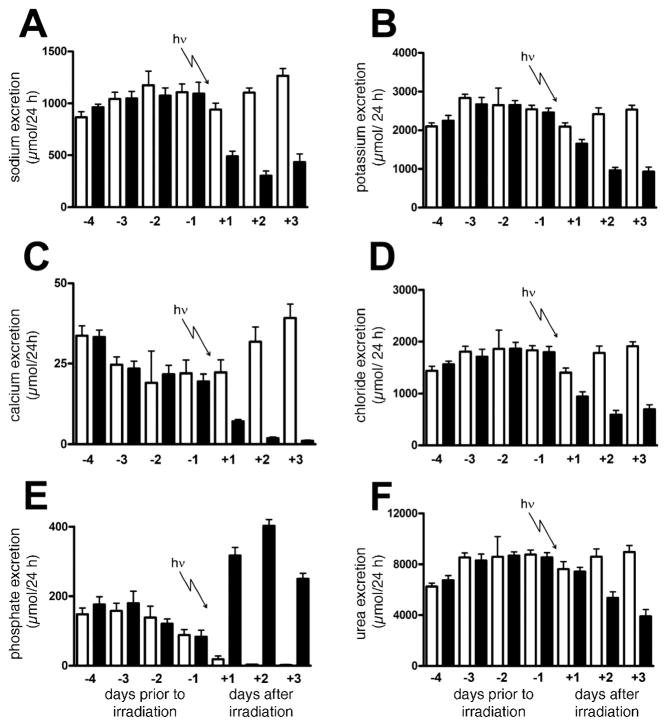
Figure 4 shows remarkable differences in the excretion of urea and selected electrolytes during the course of the study. Two factors must be considered. First, all 12 rats remained relatively inactive during their stay in the metabolic cages compared to their activity in their home cages in the animal facility, where they had been housed three or four per cage and with a larger living space, solid floor and bedding. Second is the effect of γ radiation. It would appear that some of the urinary electrolytes may be sensitive markers of both acclimation to the cage environment and the associated reduction in physical activity. There appeared to be an upward trend in Na+, Cl− and urea excretion in both groups of animal over the first 4 days (Fig. 4A, D and F, respectively). In contrast, there was an apparent fall in both Ca2+ and phosphate urinary excretion in the same period (Fig. 4C and E, respectively). In particular, compared to day −4, phosphate excretion was virtually extinguished in the sham animals by the 6th (Fig. 4E; day +2) and 7th (day +3) day of investigation. These changes in urea and other electrolyte excretion in the sham-irradiated rats were insignificant relative to those that occurred after irradiation.
FIG. 4.
Trends in sodium (panel A), potassium (panel B), calcium (panel C), chloride (panel D), phosphate (panel E) and urea (panel F) excretion for six sham-irradiated rats and six rats exposed to 3 Gy γ radiation after 4 days of acclimation to metabolic cages. Urine was collected daily on each of day −4, −3, −2, −1, +1, +2 and +3 relative to the radiation dose. Open bars and filled bars are for sham-irradiated and irradiated rats, respectively. Error bars represent + 1 SD.
Gamma-radiation exposure was associated with decreased urinary excretion of Na+, K+, Ca2+, Cl− and urea and with a massive increased excretion of phosphate, all of which persisted for the 3 days postirradiation. Considering, as a typical example, day +2 postirradiation, there were very significant differences between sham and irradiated rats in the excretion of Na+ (Fig. 4A), K+ (Fig. 4B), Ca2+ (Fig. 4C), Cl− (Fig. 4D), and urea (Fig. 4F). In contrast, there was a corresponding major increase in the urinary excretion of phosphate (Fig. 4E). The >130-fold difference in phosphate urinary excretion was due mainly to the virtual extinction of phosphate excretion in the sham controls (see above), and it rose to levels more than twice the values of those prior to irradiation (see Fig. 4E). Nevertheless, in the absence of γ-radiation exposure, one might have expected these rats to have similar miniscule urinary phosphate excretion values.
Effect of Starvation
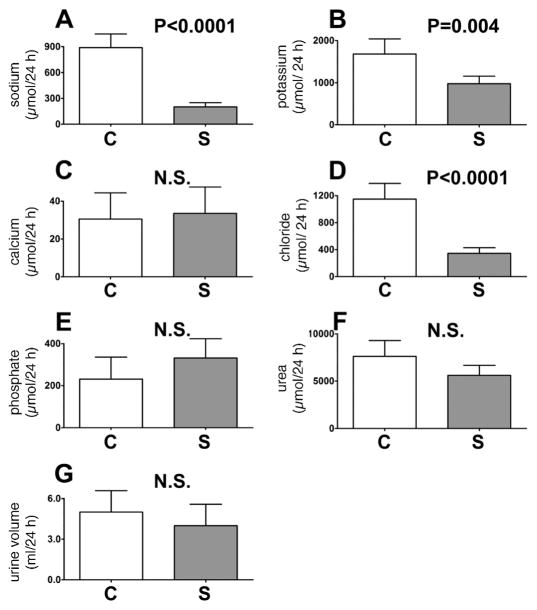
Urine (0–24 h) was collected from a further five rats that were permitted free access to drinking water but no food and an additional five control rats that had access to both food and water ad libitum. Urines were analyzed for their content of Na+, K+, Ca2+, Cl−, phosphate and urea; the 24-h output of each is shown in Fig. 5. These data provide clarification of the postirradiation observation in Fig. 4, in which the decreases in urinary excretion of Na+, K+ and Cl− may be due to the observed reduced food consumption postirradiation. However, the fall in Ca2+ excretion and rise in phosphate excretion postirradiation seems unlikely to be due to reduced food intake, based upon the results of this food withdrawal experiment (see Fig. 5C and E, cf. Fig. 4E and C).
FIG. 5.
Trends for sodium (panel A), potassium (panel B), calcium (panel C), chloride (panel D), phosphate (panel E), and urea (panel F) excretion, together with urine volume (panel G) over 0–24 h for five control rats permitted free access to food and water and five rats permitted free access to water but no food. Open bars and filled bars are for control (C) and starved (S) rats, respectively. Error bars represent +1 SD. P values from unpaired Student’s t tests are shown. NS means not significant (P > 0.05).
GCMS
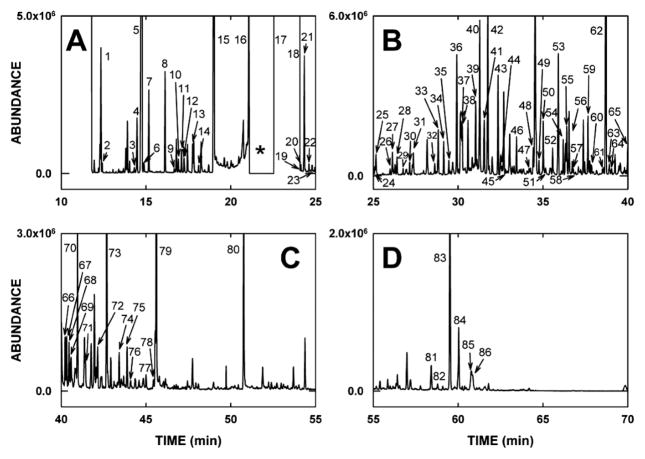
The goal of metabolomic studies is to investigate simultaneously as many metabolites present in the matrix of interest as possible. Therefore, sample preparation is crucial and must ensure that the minimum number of metabolites is lost during sample workup and that the risk of analytical artifacts is reduced. In this study diluted urines were used without extraction, which we believe to be the most suitable approach for this purpose. A two-step derivatization procedure similar to procedures described in the literature (7, 8) was adopted for the subsequent GCMS analyses. In the first step, aldehydes and ketones were stabilized as their methoxime derivatives, therefore enabling the identification of sugar compounds, whereas the second step improved the physical properties (volatility) of polar metabolites for GC by introducing trimethylsilyl groups onto polar functional groups. Possible variations in the efficiency of derivatization were compensated for adding an exogenous internal standard to the urine samples that was then used for the calculations of relative concentrations. This sample preparation in combination with a chromatographic method designed for high peak resolution enabled the separation of about 250 chromatographic peaks within 70 min with a total of more than 300 urinary metabolites per sample. A typical chromatogram is depicted in Fig. 6, divided into four panels for clarity. Peaks 1–86 were identified on the basis of comparison of their spectra to spectra of authentic compounds in mass spectral databases. The identity of these 86 urinary analytes, the number of methoxyamine (MOX) and trimethylsilyl (TMS) groups added, the molecular ion (M+•), the defining ions present in both unknown and authentic spectra, and the matches to the library spectrum for each analyte are given in Tables 1–3. It should be pointed out that an apparent poor match, for example for 2-hydroxybutanoic acid (25%) in Table 1, is due to the nature of the ion matching algorithm that the library searching software employs, whereby a spectrum that has a large number of low-abundance masses, typical of aliphatic compounds and sugars, is difficult to match to any authentic spectrum. This process is further compounded by high chromatographic background and/or incompletely resolved peaks. Thus human judgment must be employed.
FIG. 6.
A typical gas chromatogram of derivatized urine (see Materials and Methods) divided into four time quadrants (panels AD) and showing peaks 1 to 86 that were identified on the basis of their resultant mass spectra and comparison to mass spectral databases containing some 575,000 spectra. The identities of peaks 1 to 86 are given in Tables 1–3. * in panel A represents a time window when the electron multiplier detector was switched off to avoid collecting ions from the huge peak due to urea di-TMS derivative that eluted at 22.80–23.85 min (see Table 3).
TABLE 1.
GCMS Detection of Some Inorganic Acids, Aliphatic Acids and Dicarboxylic Acids
| Peak no. | Retention time (min) | Identity | No. MOX | No. TMS | M+• | Defining ions | Match |
|---|---|---|---|---|---|---|---|
| Inorganic acids | |||||||
| 2 | 12.39 | Boric acid | 0 | 3 | 278 | 263, 221, 205, 191, 175 | 93.9 |
| 17 | 22.55–22.75 | Phosphoric acid | 0 | 3 | 314 | 299, 189, 147 | 89.9 |
| Aliphatic acids and dicarboxylic acids | |||||||
| 1 | 12.34 | Glyoxylic acid | 1 | 1 | 175 | 160 | 92.1 |
| 3 | 14.30 | Pyruvic acid | 1 | 1 | 189 | 174 | 72.8 |
| 5 | 14.74 | Lactic acid | 0 | 2 | 234 | 219, 191, 147, 117 | 87.3 |
| 6 | 14.84 | 2-Hydroxybutanoic acid | 0 | 2 | 248 | 205, 147, 131 | 25.0 |
| 7 | 15.16 | Glycolic acid | 0 | 2 | 220 | 205, 177, 161, 147 | 73.6 |
| 10 | 16.90 | Fumaric acid | 0 | 2 | 260 | 260, 245 | 67.4 |
| 11 | 17.09 | Oxalic acid | 0 | 2 | 234 | 219, 190, 147 | 55.7 |
| 14 | 18.09 | Malonic acid | 0 | 2 | 248 | 233, 147 | 46.3 |
| 19 | 23.95 | Succinic acid | 0 | 2 | 262 | 247, 147 | 58.0 |
| 20 | 24.12 | Glyceric acid | 0 | 3 | 322 | 292, 205, 189 | 91.9 |
| 22 | 24.60 | 2,3-Dihydroxybutanoic acid | 0 | 3 | 336 | 292, 220, 203, 189, 147 | 50.6 |
| 23 | 24.79 | 5-Hydroxyhexanoic acid | 0 | 2 | 276 | 261, 204, 171, 147 | 76.7 |
| 29 | 26.75 | 2,4-Dihydroxybutanoic acid | 0 | 3 | 336 | 321, 219, 203, 189, 147, 103 | 83.2 |
| 31 | 27.36 | 3,4-Dihydroxybutanoic acid | 0 | 3 | 336 | 321, 233, 189, 147 | 94.5 |
| 34 | 29.15 | Malic acid | 0 | 3 | 350 | 335, 233, 189, 147 | 67.7 |
| 35 | 29.45 | Adipic acid | 0 | 2 | 290 | 275, 217, 172, 159, 147, 141 | 94.4 |
| 39 | 31.03 | Erythronic acid | 0 | 4 | 424 | 409, 319, 292, 205 | 89.3 |
| 41 | 31.53 | Threonic acid | 0 | 4 | 424 | 409, 319, 292, 205 | 90.0 |
| 42 | 31.73 | 2-Oxoglutaric acid | 1 | 2 | 364 | 349, 288, 247, 203 | 97.0 |
| 43 | 32.34 | Pimelic acid | 0 | 2 | 304 | 289, 217, 173, 147 | 87.5 |
| 49 | 34.76 | 3-Hydroxyadipic acid | 0 | 3 | 378 | 363, 247, 203, 147, 129 | 91.6 |
| 50 | 35.02 | Suberic acid | 0 | 2 | 318 | 303, 217, 187, 169, 147, 139,129 | 49.3 |
| 56 | 36.54 | cis-Aconitic acid | 0 | 3 | 390 | 375, 346, 285, 229 | 65.8 |
| 59 | 37.65 | Azelaic acid | 0 | 2 | 332 | 317, 273, 217, 201 | 96.3 |
| 60 | 37.86 | Ribonic acid | 0 | 5 | 526 | 333, 292, 217, 147 | 70.9 |
| 62 | 38.81 | Citric acid | 0 | 4 | 480 | 465, 375, 273, 245 | 86.2 |
| 66 | 40.23 | 2-O-Methylascorbic acid | 0 | 3 | 406 | 391, 361, 274, 205 | 88.9 |
| 67 | 40.32 | Hexose acid Ia | 0 | 4 | 466 | 361, 319, 217, 147, 129 | 76.8 |
| 73 | 42.69 | Hexose acid IIa | 0 | 6 | 628 | 433, 359, 333, 319, 305, 292 | 55.7 |
| 74 | 43.42 | Hexose acid IIIa | 0 | 6 | 628 | 433, 359, 333, 319 | 69.0 |
| 75 | 43.88 | Hexose acid IVa | 0 | 6 | 628 | 423, 393, 359, 333, 292, 217, 205 | 72.0 |
In the absence of authentic standards, it was not possible to distinguish between mannonic, gluconic, galactonic and glucaric acids on the basis of their mass spectra alone.
TABLE 3.
GCMS Detection of Some Phenols, Purines, Pyrimidines, Sugars and Miscellaneous Compounds
| Peak no. | Retention time (min) | Identity | No. MOX | No. TMS | M+• | Defining ions | Match |
|---|---|---|---|---|---|---|---|
| Phenols | |||||||
| 4 | 14.45 | Phenol | 0 | 1 | 166 | 166, 151 | 79.0 |
| 12 | 17.24 | 3-Hydroxypyridine | 0 | 1 | 167 | 167, 152 | 98.0 |
| 13 | 17.75 | p-Cresol | 0 | 1 | 180 | 180, 165 | 42.7 |
| 52 | 35.57 | 5-Hydroxyindole | 0 | 2 | 277 | 277, 205, 147 | 71.4 |
| Purines and pyrimidines | |||||||
| 21 | 24.33 | Uracil | 0 | 2 | 256 | 256, 255, 241, 147 | 77.1 |
| 28 | 26.39 | Thymine | 0 | 2 | 270 | 270, 255, 147, 113 | 91.1 |
| 38 | 30.24 | Cytosine | 0 | 2 | 255 | 254, 240, 170 | 97.0 |
| 79 | 45.60 | Uric acid | 0 | 4 | 456 | 456, 441 | 89.5 |
| 80 | 50.76 | Pseudouridine | 0 | 5 | 604 | 589, 424, 357, 217 | 85.5 |
| Sugars and derivatives | |||||||
| 47 | 34.18 | Galactosone | 2 | 4 | 524 | 307, 217, 205, 189, 147, 103 | 60.9 |
| 48 | 34.42 | Ribose | 1 | 4 | 467 | 307, 277, 233, 277 | 31.0 |
| 55 | 36.40 | Fucose | 1 | 4 | 481 | 321, 160, 147 | 64.8 |
| 68 | 40.46 | Gluconolactone | 0 | 4 | 466 | 466, 361, 305, 217 | 35.8 |
| 71 | 41.37 | Gulonolactone | 0 | 6 | 466 | 466, 361, 333, 230, 217, 204 | 54.9 |
| 81 | 58.41 | Lactose (D-galactose-β-D-glucose) | 0 | 8 | 918 | 451, 361, 331, 319, 305, 291, 271, 243, 217, 204, 191, 169 | 18.7 |
| 82 | 58.79 | Maltose (D-glucose-α-D-glucose) | 0 | 8 | 918 | 361, 271, 243, 217, 204, 191, 169, 147, 129 | 21.0 |
| 83 | 59.52 | Turanose (D-glucose-α (1→3)-D-fructose | 0 | 7 | 846 | 451, 361, 345, 331, 319, 291, 271, 257, 243, 231, 217, 204 | 47.7 |
| 84 | 60.04 | Matches also to turanose | 0 | 7 | 846 | 361, 331, 319, 291, 271, 243, 231, 217, 204 | 28.0 |
| 85 | 60.80 | Sucrose (β-D-fructofuranosyl-α-D-glucopyranoside) | 0 | 8 | 918 | 451, 437, 361, 271, 217, 217, 204, 191, 147 | 63.7 |
| 86 | 60.86 | Matches also to sucrose | 0 | 8 | 918 | 361, 217, 204, 191, 169, 147, 129, 103 | 32.9 |
| Miscellaneous | |||||||
| 15 | 19.03 | Ureaa | 0 | 3 | 276 | 276, 261, 245, 173, 147 | 97.9 |
| 18 | 22.80–23.85 | Ureaa | 0 | 2 | 204 | 189, 147 | 96.5 |
| 27 | 26.11 | Isethionic acid | 0 | 2 | 270 | 255, 147 | 83.6 |
| 33 | 28.81 | Parabanic acid | 0 | 2 | 258 | 258, 243, 215, 188, 158, 100 | 97.1 |
| 40 | 31.27 | Creatinine | 0 | 3 | 329 | 329, 314, 171, 143 | 93.5 |
| 69 | 40.58 | 2,4-Dihydroxyestrone | 0 | 3 | 518 | 518, 428 | 96.0 |
Urea forms a major di-TMS derivative and an earlier running minor tri-TMS derivative.
Table 1 lists two inorganic acids and 31 aliphatic and dicarboxylic acids that appear in Fig. 6A–C. Table 2 lists five polyols, 11 amino acid peaks, and 11 aromatic acids and derivatives that appear in Fig. 6A–C. Table 3 lists four phenols, five purines and pyrimidines, and 11 sugars and derivatives, together with six miscellaneous peaks that occur in Fig. 6A–D. It was these chromatograms and the mass spectral data they contain from 84 rat urine samples (12 rats × 7 days) that were subjected to random forests analysis.
TABLE 2.
GCMS Detection of Some Polyols, Amino Acids, Aromatic Acids and Their Derivatives
| Peak no. | Retention time (min) | Identity | No. MOX | No. TMS | M+• | Defining ions | Match |
|---|---|---|---|---|---|---|---|
| Polyols | |||||||
| 36 | 29.92 | Erythritol | 0 | 4 | 410 | 307, 217, 147 | 42.4 |
| 54 | 36.19 | Arabitol | 0 | 5 | 512 | 332, 319, 307, 217, 205, 147 | 37.9 |
| 70 | 40.96 | Mannitol | 0 | 6 | 614 | 319, 217, 205, 147 | 21.1 |
| 76 | 44.09 | Scyllo-inositol | 0 | 6 | 612 | 318, 305, 217, 204, 191, | 66.5 |
| 78 | 45.54 | Myo-inositol | 0 | 6 | 612 | 432, 367, 305, 265 | 73.9 |
| Amino acids and derivatives | |||||||
| 8 | 16.14 | Alanine | 0 | 2 | 233 | 218, 190, 147, 116, | 79.0 |
| 9 | 16.66 | Glycine | 0 | 2 | 219 | 204, 176, 147, 102 | 87.3 |
| 24 | 25.08 | Serine | 0 | 3 | 321 | 278, 218, 204 | 73.2 |
| 25 | 25.14 | Pipecolic acid | 0 | 2 | 273 | 230, 156 | 48.0 |
| 26 | 25.98 | Threonine | 0 | 3 | 335 | 291, 218, 203, 147, 117, 101 | 68.2 |
| 30 | 27.14 | β-Alanine | 0 | 3 | 305 | 290, 248, 174, 147 | 96.6 |
| 32 | 28.61 | Aminomalonic acid | 0 | 3 | 335 | 320, 292, 248, 218 | 96.0 |
| 37 | 30.17 | 5-Oxoproline | 0 | 2 | 273 | 258, 230, 156 | 73.2 |
| 45 | 32.80 | Anthranilic acida | 0 | 2 | 281 | 266, 147, 134 | 93.2 |
| 51 | 35.15 | Anthranilic acida | 0 | 1 | 209 | 209, 194, 176, 150, 119, 92 | 46.5 |
| 61 | 38.52 | 5-Hydroxytryptophan | 0 | 4 | 508 | 290, 202 | 68.2 |
| Aromatic acids and derivatives | |||||||
| 16 | 21.06 | Benzoic acid | 0 | 1 | 194 | 179, 135, 105, 77 | 81.5 |
| 44 | 32.61 | 3-Hydroxyphenylacetic acid | 0 | 2 | 296 | 296, 281, 252, 191 | 86.1 |
| 46 | 33.45 | 4-Hydroxyphenylacetic acid | 0 | 2 | 296 | 281, 252, 225, 179 | 78.9 |
| 53 | 35.90 | 3-(3-Hydroxyphenyl)-propanoic acid | 0 | 2 | 310 | 310, 295, 205, 192, 177 | 98.0 |
| 57 | 36.80 | 3-(4-Hydroxyphenyl)-propanoic acid | 0 | 2 | 310 | 310, 192, 179 | 88.5 |
| 58 | 36.93 | Vanillic acid | 0 | 2 | 312 | 312, 297, 282, 267, 253, 223, 193 | 94.8 |
| 63 | 38.96 | Hippuric acid | 0 | 1 | 251 | 206, 190, 117, 105 | 82.7 |
| 64 | 39.06 | 1H-Indole-3-carboxylic acid | 0 | 2 | 305 | 305, 290 | 89.0 |
| 65 | 40.02 | Phenacetylglycine | 0 | 1 | 265 | 265, 250, 221, 174 | 97.8 |
| 72 | 42.15 | 1H-Indole-3-acetic acid | 0 | 2 | 319 | 319, 304, 202, 186, 170, 156 | 60.8 |
| 77 | 45.00 | Ferulic acid | 0 | 2 | 338 | 338, 323, 308, 293, 279, 249 | 68.9 |
Anthranilic acid formed both mono- and di-TMS derivatives.
Bioinformatics
A crucial step in the multivariate data analysis is the pre-processing of the raw GCMS data that creates a peak table containing pairs of retention times and m/z, together with the corresponding abundance of the respective ion. During this data transformation, shifts in retention times between different chromatograms are compensated for and corresponding peaks are aligned (see Fig. 1). Furthermore, the complexity of the mass spectra obtained using a single quadrupole mass analyzer with electron impact ionization has to be reduced during this step, because large chromatographic peaks in excess of 100 fragment ions are typically recorded. This will result in multiple entries in the peak table stemming from one component. The pre-processing was designed to include small chromatographic peaks in the peak table while reducing the influence of large peaks and their fragments on the multivariate data analysis. For this purpose three peak tables were created to be subjected to random forests analysis that differed in the time window covered. The first peak table (Table 4) encompassed the entire chromatogram. The random forests analysis of the entire chromatogram was dominated by multiple ions arising mainly from three urinary compounds. One was found to be urea that coeluted with inorganic phosphate (22.7 min), which was excreted in much higher amounts in the treated group of animals at day 5 and was found to be one of the peaks with highest abundance in this group. The other was identified as citrate (38.8 min) and was confirmed by the analysis of the authentic standard. Citrate was one of the most abundant peaks in the urine samples obtained from the control rats at day 5 and was down-regulated in the treated group. The other peak tables covered only a part of the chromatogram to eliminate the influence of the broad peak of urea, phosphate and/or citrate on the peak picking process (Tables 5 and 6). The focus in this study was the investigation of potential biomarkers that are excreted in higher amounts in the treated group compared to the control animals.
TABLE 4.
Random Forests Results for the Peak Table Spanning the Complete Chromatogram (11.9–70.0 min)
| Ranka | Retention time (min) | m/z | Changeb | Identityc |
|---|---|---|---|---|
| 1, 2, 4, 10, 11, 12, 20 | 38.82 | 465, 211, 150, 171, 333, 347, 393 | down | Citric acid |
| 3 | 20.93 | 248 | up | n.a.d |
| 5, 6, 7, 18, 19 | 22.70 | 283, 150, 225, 253, 211 | up | Phosphoric acid |
| 8 | 23.30 | 299 | up | n.a. |
| 9, 15, 17 | 21.05 | 189, 157, 173 | down | n.a. |
| 14 | 40.32 | 319 | up | Hexose acid Ie |
| 16 | 12.34 | 160 | up | Glyoxylic acid |
Importance of the random forests signal (i.e. position in the table of the top components).
Up- or down-regulation of the component in the irradiated animals compared to the sham control animals.
Confirmation with authentic standard.
Not assigned.
In the absence of authentic standards it was not possible to distinguish between mannonic, gluconic, galactonic and glucaric acids on the basis of their mass spectra alone.
TABLE 5.
Random Forests Results for a Peak Table Spanning 11.9–21.0 Minutes of the Chromatogram
| Ranka | Retention time (min) | m/z | Changeb | Identityc |
|---|---|---|---|---|
| 1 | 12.34 | 160 | up | Glyoxylic acid |
| 2, 3, 15 | 21.00 | 189, 157, 171 | down | n.a.d |
| 4 | 20.94 | 248 | up | n.a. |
| 5 | 17.05 | 156 | down | n.a. |
| 6 | 15.14 | 177 | up | Glycolic acid |
| 7, 10, 11, 14 | 14.72 | 150, 219, 191, 175 | up | Lactic acid |
| 8 | 20.79 | 261 | down | n.a. |
| 9 | 20.47 | 247 | up | n.a. |
| 12 | 20.77 | 189 | down | n.a. |
| 13 | 17.74 | 165 | up | p-Cresol |
Importance of the random forests signal (i.e. position in the table of the top components).
Up- or down-regulation of the component in the irradiated animals compared to the sham control animals.
Confirmation with authentic standard.
Not assigned.
TABLE 6.
Random Forests Results for a Peak Table Spanning 24.2–70.0 Minutes of the Chromatogram
| Ranka | Retention time (min) | m/z | Changeb | Identityc |
|---|---|---|---|---|
| 1, 3, 6–12, 15, 16, 18, 19, 32, 37, 60, 85 | 38.86 | 333, 421, 465, 257, 305, 363, 150, 231, 171, 375, 347, 211, 393, 319, 285, 273, 183 | down | Citric acid |
| 2, 47 | 40.32 | 319, 217 | up | Hexose acid Id |
| 5, 57 | 40.83 | 174, 245 | down | n.a.e |
| 13, 84, 86 | 36.20 | 277, 217, 307 | down | Arabitol |
| 14 | 31.54 | 205 | up | Threonic acid |
| 17, 20 | 26.97 | 174, 172 | up | n.a. |
| 21 | 61.24 | 204 | down | n.a. |
| 22 | 50.08 | 204 | down | n.a. |
| 23 | 64.19 | 204 | down | n.a. |
| 24, 25 | 47.45 | 171, 261 | down | n.a. |
| 26, 44, 53, 64, 68 | 45.00 | 219, 308, 175, 338, 293 | down | Ferulic acid |
| 27 | 53.70 | 259 | up | n.a. |
| 28, 30, 34, 48, 54 | 37.68 | 171, 317, 217, 152, 201 | down | Azelaic acid |
| 29, 39, 40, 45, 51, 67 | 31.78 | 156, 229, 304, 186, 198, 171 | down | 2-Oxoglutaric acid |
| 31 | 62.06 | 398 | down | n.a. |
| 33, 71 | 55.85 | 287, 185 | down | n.a. |
| 35, 42, 72 | 56.42 | 171, 185, 275 | down | n.a. |
| 36 | 53.12 | 273 | down | n.a. |
| 38, 41, 50, 56 | 32.38 | 173, 155, 217, 289 | down | Pimelic acid |
| 43, 46, 95 | 28.17 | 243, 154, 174 | up | n.a. |
| 49 | 53.70 | 273 | down | n.a. |
| 52 | 52.72 | 273 | down | n.a. |
| 55 | 42.76 | 245 | down | n.a. |
| 58 | 50.45 | 204 | down | n.a. |
| 59 | 24.31 | 241 | up | Uracil |
| 61, 90 | 42.37 | 318, 217 | down | n.a. |
| 62 | 56.16 | 287 | down | n.a. |
| 63, 69, 77, 78, 97 | 58.42 | 243, 217, 361, 319, 204 | down | Lactose |
| 65 | 58.78 | 204 | down | Maltose |
| 66, 74, 75 | 35.02 | 169, 303, 187 | down | Suberic acid |
| 70 | 58.42 | 160 | down | Lactose |
| 73 | 41.37 | 293 | down | Gulonolactone |
| 76 | 59.84 | 204 | down | n.a. |
| 79 | 38.73 | 150 | down | Citric acidf |
| 80 | 36.40 | 219 | up | Fucose |
| 81 | 26.40 | 255 | up | Thymine |
| 82, 83, 94, 100 | 34.52 | 174, 238, 326, 188 | up | n.a. |
| 87 | 56.35 | 261 | down | n.a. |
| 88 | 35.85 | 173 | down | n.a. |
| 89 | 35.65 | 306 | down | n.a. |
| 91 | 29.45 | 172 | down | Adipic acid |
| 92 | 43.12 | 261 | down | n.a. |
| 93 | 47.75 | 185 | down | n.a. |
| 96 | 39.28 | 318 | down | n.a. |
| 98 | 37.24 | 357 | up | Glycerol 3-phosphate |
| 99 | 39.28 | 159 | down | In peak of hit 96 |
Importance of the random forests signal (i.e. position in the table of the top components).
Up- or down-regulation of the component in the irradiated animals compared to the sham control animals.
Confirmation with authentic standard.
In the absence of authentic standards it was not possible to distinguish between mannonic, gluconic, galactonic and glucaric acids on the basis of their mass spectra alone.
Not assigned.
This is a second entry of m/z = 150 for citric acid found by random forests (see first row of table) with a slightly different retention time (38.73 compared to 38.86 min). This is due to the large size of the citric acid peak.
Up-regulated Urinary Biomarkers
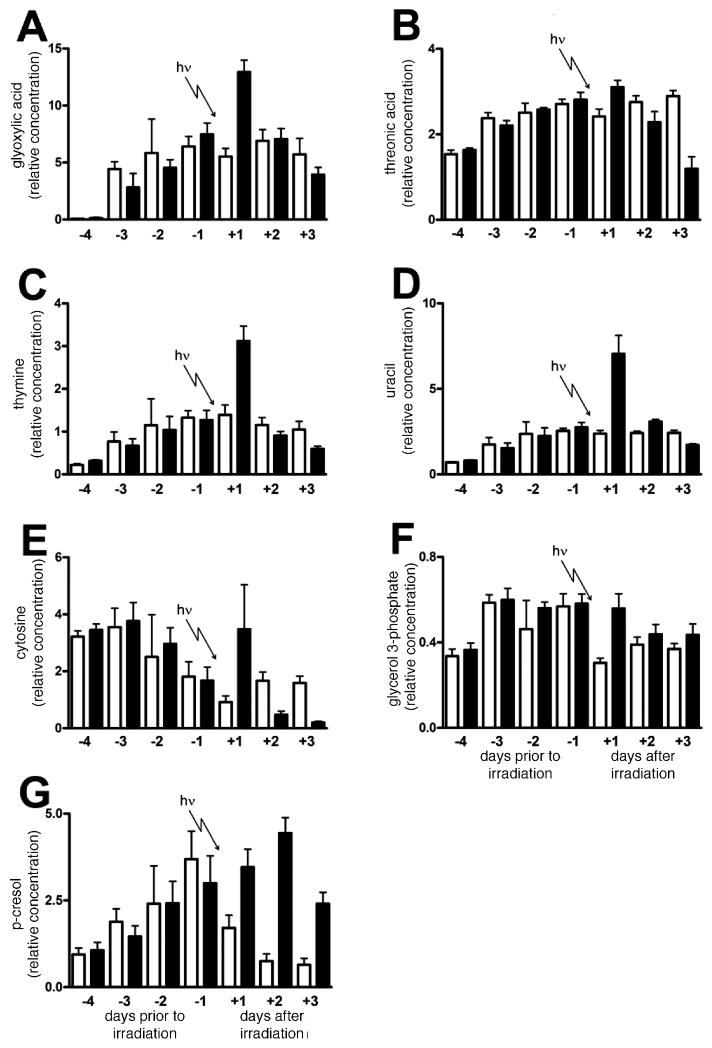
Perusal of the top 100 hits in the random forests findings in Tables 4–6 shows the urinary ions that were either up- or down-regulated as a result of γ irradiation of rats with 3 Gy. The following up-regulated biomarkers of γ irradiation were confirmed with authentic standards: phosphoric acid (confirmed also by independent electrolyte analysis, Fig. 4E), glyoxylic acid, p-cresol (4-methylphenol), threonic acid [(2R,3S)-2,3,4-trihydroxybutanoic acid], uracil, thymine and glycerol 3-phosphate. Of these, glyoxylic acid, threonic acid, thymine, uracil, p-cresol and glycerol 3-phosphate were established as robust positive biomarkers of γ irradiation of rats with 3 Gy on the basis that they showed statistically significant increases on the first day postirradiation. The urinary excretion of these six biomarkers in irradiated and sham-irradiated rats across the 7 days of the investigation is shown in Fig. 7A–D, F and G. Glyoxylic acid was elevated from 5.5 ± 1.7 units in sham-irradiated animals to 12.9 ± 2.5 units in irradiated animals (t = 5.93; P = 0.0001). Threonic acid was elevated from 2.4 ± 0.4 units to 3.1 ± 0.4 (t = 2.91; P = 0.016).
FIG. 7.
Trends in glyoxylic acid (panel A), threonic acid (panel B), thymine (panel C), uracil (panel D), cytosine (panel E), glycerol 3-phosphate (panel F), and p-cresol (panel G) excretion for six sham-irradiated rats and six rats during 4 days of acclimation to metabolic cages and then after exposure to 3 Gy γ radiation. For the determination of uracil (panel D), urine was treated with urease to remove the large interfering peak of urea that prevented measurement of uracil. Open bars and filled bars are for sham-irradiated and irradiated rats, respectively. Error bars represent + 1 SD.
Because we had observed in random forests that thymine and uracil had elevated excretion, we chose to examine the data for the third pyrimidine base cytosine (retention time 30.24 min, Table 3) that had not appeared in the top 100 hits in the random forests analysis (Tables 4–6). Its excretion profile is depicted in Fig. 7E. It can be seen that there is a wide variance in cytosine elevated excretion on day +1 after irradiation, but on days +2 and +3 the excretion clearly falls below control levels. This was a similar finding to that for 2′-deoxycytidine excretion in mice, where the excretion fell below control values after γ irradiation with both 2 and 3 Gy (2).
Origins of the Pyrimidine Bases in Urine
Thymine, uracil and cytosine may have been excreted as the free base or, alternatively, may have been formed during sample workup from their corresponding nucleosides or deoxynucleosides. We postulated this because it has been reported that thymidine and 2′-deoxyuridine are biomarkers for γ irradiation in the mouse (2, 4). Using authentic standards for the nucleosides uridine and cytidine, together with the 2′ deoxynucleosides thymidine, 2′-deoxycytidine and 2′-deoxyuridine, the conversion of these compounds to the pyrimidine bases uracil, thymine and cytosine was examined under the conditions of urine derivatization with methoxyamine followed by BSTFA/TMCS (see above). Cytidine (<1% deribosylation) and uridine (8%) were stable during derivatization. However, 2′-deoxycytidine (70%), 2′-deoxyuridine (52%), and thymidine (54%) were all converted in our assay to a significant degree to their corresponding pyrimidine bases. This makes it likely that the pyrimidines observed in urine after γ irradiation of rats with 3 Gy originated from the corresponding 2′-deoxynucleosides. While thymidine has been readily detected in human urine by high-performance liquid chromatography (without derivatization), levels of thymine were undetectable (15).
Down-regulated Urinary Biomarkers
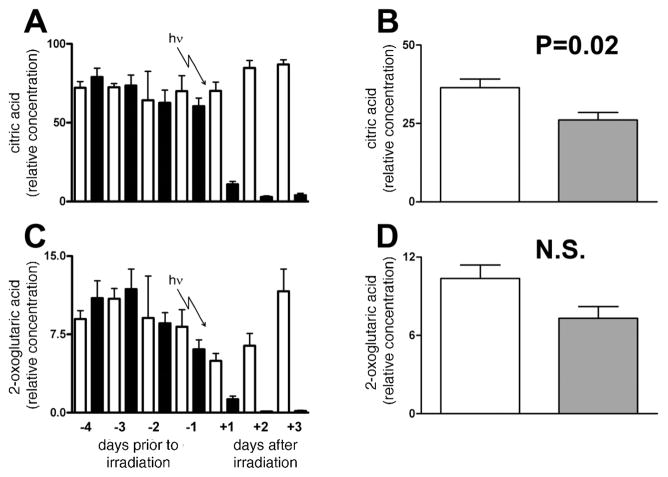
Table 6 clearly shows that citric acid was the most important analyte identified by random forests analysis as differing between irradiated and sham-irradiated rats on day +1, with 17 ions from citric acid tetra-TMS derivative (Table 1) identified independently. Citric acid was highly significantly down-regulated after γ irradiation of rats with 3 Gy (Fig. 8A). The potential role of reduced food consumption in this phenomenon was examined in the urine of the five starved and five control rats described above. As shown in Fig. 8B, complete food withdrawal for 24 h did not mimic the reduced excretion of citric acid observed after irradiation. However, there was a small and statistically significant decline in citric acid excretion after starvation for 24 h. To determine whether this was due to a lowered turnover of the citric acid cycle, we examined the data for 2-oxoglutarate, another citric acid cycle intermediate found in urine. Table 6 shows that 2-oxoglutarate is also a down-regulated biomarker of radiation with six ions identified by random forests, from 5.0 ± 1.7 units to 1.3 ± 0.8 units after irradiation (t = 4.74; P = 0.0008). The urinary excretion data for 2-oxoglutarate are illustrated in Fig. 8C. The changes mimic the changes seen in citric acid excretion in Fig. 8A. Starvation of rats for 24 h did not produce a significant fall in 2-oxoglutarate excretion (Fig. 8D). It would appear likely, therefore, that the decline of citric and 2-oxoglutaric acid urinary excretion is related to γ radiation. Other notable down-regulated biomarkers included the C6–C9 dicarboxylic acids, adipic, pimelic, suberic and azelaic acids (Table 6).
FIG. 8.
Trends in citric acid (panel A) and 2-oxoglutaric acid (panel C) excretion for six sham-irradiated rats and six rats during 4 days of acclimation to metabolic cages and then after exposure to 3 Gy γ radiation. Open bars (□) and filled bars (■) are for sham irradiated and irradiated rats, respectively. Also shown is the effect of 0–24 h starvation for five control rats permitted free access to food and water and five rats permitted free access to water but no food. Open bars and filled bars are for control and starved rats, respectively. Error bars represent + 1 SD. A P value from an unpaired Student’s t test is shown. NS means not significant (P > 0.05).
DISCUSSION
By employing a simplified metabolomics protocol that uses low-mass resolution GCMS instrumentation and open-source software for bioinformatics, we demonstrate here that biomarkers of γ irradiation of rats can be detected and validated. Up-regulated biomarkers included glyoxylic acid, threonic acid, thymine, uracil and glycerol 3-phosphate. Down-regulated biomarkers included citric acid and 2-oxoglutaric acid, together with the dicarboxylic acids adipic, pimelic, suberic and azelaic acids. In addition, urinary urea and electrolyte analysis revealed that Na+, K+, Ca2+, Cl− and urea excretion all declined after irradiation, while phosphate excretion was enhanced. It is of interest to evaluate potential sources of this postirradiation metabolomic phenotype to understand better the biological processes that contribute to it. Regarding elevated urinary excretion of the pyrimidine bases thymine and uracil, we have already reported that their corresponding 2′-deoxynucleosides thymidine and 2′-deoxyuridine show increased excretion in the mouse after γ irradiation at 1–3 Gy (2), and we have postulated that they are elevated due to nitrosative deamination of the aminopyrimidine cytidine (2). We report here that these corresponding 2′-deoxynucleosides undergo deribosylation under the conditions of our sample preparation and derivatization and yield the pyrimidine bases. In all probability, the observed pyrimidines arose from the DNA 2′-deoxynucleosides but not from the RNA nucleosides, e.g. uridine. This would be consistent with our previous reports in the mouse (2, 4) and with the known composition of human urine (15).
The enhanced excretion of p-cresol is of interest in that it is a known product of protein degradation whereby tyrosine is converted to p-cresol by the gut microbiota and then excreted in the urine (16). In a previous study (4), the elevated urinary excretion of a related phenolic metabolite, 3-hydroxy-2-methylbenzoic acid 3-O-sulfate, was reported in mice exposed to 3 Gy. This novel urinary metabolite was almost certainly contributed by gut floral metabolism. Gamma irradiation of mice has been reported to alter both the composition and size of the gut microbiota (17, 18) and presumably, therefore, its metabolic capacity.
The increased urinary excretion of glyoxylic acid may represent evidence of lipid peroxidation of polyunsaturated fatty acids (19). This would fit with the nitrosative deamination of cytidine to 2′-deoxyuridine hypothesis as proposed previously (2) in that it also involves the generation of reactive oxygen species (ROS) by ionizing radiation. The other up-regulated biomarker, threonic acid, is an oxidation product of ascorbic acid via dehydroascorbic acid (20), the latter being produced when ascorbic acid reacts with ROS. This is yet further evidence that ROS is involved in the generation of radiation metabolomic urinary biomarkers. The final biomarker identified, glycerol 3-phosphate (Fig. 7F), should not be considered a biomarker of radiation exposure since it has been identified on the basis of a reduced urine excretion in the sham-irradiated animals with an unremarkable excretion in the irradiated animals. The reason for the decline in excretion in the sham-irradiated animals is not known.
The reduced excretion of citric acid, 2-oxoglutaric acid and the four C6–C9 dicarboxylic acids is of interest for a number of reasons. At first sight, it would appear that these compounds reflect the reduced caloric intake by the irradiated rats, but this was not substantiated by the starvation experiment (Fig. 8). These data may reflect changes in renal tubular cells. In rats fasted for 18–24 h, Na+, K+ and urea excretion has been reported to be reduced (21), although we did not confirm the urea difference in our own experiments (Fig. 5F). Our observations (Fig. 4) clearly go beyond the effects of starvation. It is well known that 2-oxoglutaric acid (α-ketoglutarate) is an important driver of renal organic anion inward transport and dicarboxylic acid exchange in tubular cells in a Na+-dependent fashion (22, 23). The changes in Na+ excretion observed here after irradiation, plus a decline in renal tubular energy production and thus 2-oxoglutaric acid availability, may be the reasons underlying a proportion of the urinary radiation metabolomic phenotype that includes reduced excretion of dicarboxylic acids.
The reduced excretion of Ca2+ and massive increase in phosphate excretion, against a background of reduced excretion in the sham-irradiated rats (Fig. 4C and E), is worthy of special comment. The high urinary ratio of Ca2+/phosphate in the sham-irradiated rats is completely reversed in the irradiated animals, suggesting that these animals have developed a high plasma Ca2+/phosphate ratio by preserving Ca2+ but excreting phosphate. This is likely to be mediated by parathyroid hormone (PTH). The mechanism of release of PTH by γ radiation is unknown.
Overall, the effect of 3 Gy γ radiation on male rats was to produce a number of verifiable up-regulated and down-regulated urinary metabolomic biomarkers. The radiation metabolomic signature included phosphate (↑), glyoxylate (↑), threonate (↑), p-cresol (↑), thymine (↑), uracil (↑), citrate (↓), 2-oxoglutarate (↓), adipate (↓), pimelate (↓), suberate (↓), and azelaate (↓). We propose that the elevated urinary concentrations of thymine and uracil detected in this study are surrogates for elevated urinary excretion of thymidine and 2′-deoxyuridine, due to chemical deribosylation under the conditions of sample preparation employed. This means that mice and rats behave similarly with respect to the postirradiation production of deaminated 2′-deoxynucleosides, by a mechanism that has been discussed elsewhere (2). This encourages the belief that human subjects may also react similarly to these doses of γ radiation and that metabolomics may provide a practical means of rapid noninvasive biomonitoring of exposure to ionizing radiation.
Finally, it should be pointed out that in this report we describe methods for the conduct of metabolomic studies that can yield abundant urinary biomarkers and that would, in all probability, generate biomarkers using other metabolomes, such as sweat, sebum, plasma or other biofluids. The protocol described here uses low-mass-resolution gas chromatography-mass spectrometry and open-source software to uncover a metabolomic fingerprint of radiation exposure. This same protocol would also work with simple capillary gas chromatography without a mass-selective detector, providing that the investigator had a rich supply of authentic endogenous compounds, as many laboratories do. Our initial studies in this field have employed high mass resolution orthogonal quadrupole time-of-flight mass spectrometry coupled to ultra-high-resolution chromatography (2, 4). This capital expense is not within the reach of all laboratories. The methods described here not only offer a cost-effective alternative but also can provide a substantial yield of biomarkers. It is hoped that this will encourage the application of metabolomic protocols in the field of radiation research.
Acknowledgments
The authors wish to thank Professor Bernhard Lauterburg in Bern for his support of this work and helpful discussions. The expert technical assistance of Jürg Müller is also acknowledged. We thank Prof. Dr. Bendicht, Wermuth, Director of the Zentrallabor, Inselspital, Bern, for providing analyses of urinary urea and electrolytes. This work was performed as part of the Columbia University Center for Medical Countermeasures against Radiation (P.I. David Brenner) and funded by NIH (NIAID) grant U19 AI067773-02 and also supported in part by the National Cancer Institute Intramural Research Program. ADP is supported by the Pharmacology Research Associate in Training program, National Institute of General Medical Sciences. JRI is grateful to U.S. Smokeless Tobacco Company for a grant for collaborative research.
References
- 1.Graham B, Talent J, Allison G, Cleveland R, Rademaker S, Roemer T, Sherman W, Sokolski H, Verma R. The Report of the Commission on the Prevention of WMD Proliferation and Terrorism. Vintage Books; New York: 2008. World at Risk. [Google Scholar]
- 2.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Gonzalez FJ, Idle JR. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pellmar TC, Rockwell S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat Res. 2005;163:115–123. doi: 10.1667/rr3283. [DOI] [PubMed] [Google Scholar]
- 4.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 6.Patterson AD, Idle JR. A metabolomic perspective of small molecule toxicity. In: Marrs TC, Ballantyne B, Syversen T, editors. General and Applied Toxicology. Wiley; Chichester: 2009. [Google Scholar]
- 7.Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Wang G, Du Y, Zhu L, JA GC/MS analysis of the rat urine for metabonomic research. J Chromatogr B Anal Technol Biomed Life Sci. 2007;854:20–25. doi: 10.1016/j.jchromb.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39:581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Krausz KW, Idle JR, Gonzalez FJ. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J Biol Chem. 2008;283:4543–4559. doi: 10.1074/jbc.M706299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Krausz KW, Shah YM, Idle JR, Gonzalez FJ. Serum metabolomics reveals irreversible inhibition of fatty acid beta-oxidation through the suppression of PPARalpha activation as a contributing mechanism of acetaminophen-induced hepatotoxicity. Chem Res Toxicol. 2009;22:699–707. doi: 10.1021/tx800464q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Shah YM, Morimura K, Krausz KW, Miyazaki M, Richardson TA, Morgan ET, Ntambi JM, Idle JR, Gonzalez FJ. Metabolomics reveals that hepatic stearoyl-CoA desaturase 1 downregulation exacerbates inflammation and acute colitis. Cell Metab. 2008;7:135–147. doi: 10.1016/j.cmet.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Jr, Gonzalez FJ, Idle JR. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80:665–674. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhen Y, Krausz KW, Chen C, Idle JR, Gonzalez FJ. Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Mol Endocrinol. 2007;21:2136–2151. doi: 10.1210/me.2007-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tavazzi B, Lazzarino G, Leone P, Amorini AM, Bellia F, Janson CG, Di Pietro V, Ceccarelli L, Donzelli S, Giardina B. Simultaneous high performance liquid chromatographic separation of purines, pyrimidines, N-acetylated amino acids, and dicarboxylic acids for the chemical diagnosis of inborn errors of metabolism. Clin Biochem. 2005;38:997–1008. doi: 10.1016/j.clinbiochem.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Bone E, Tamm A, Hill M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr. 1976;29:1448–1454. doi: 10.1093/ajcn/29.12.1448. [DOI] [PubMed] [Google Scholar]
- 17.Brook I, Elliott TB, Ledney GD, Shoemaker MO, Knudson GB. Management of postirradiation infection: lessons learned from animal models. Mil Med. 2004;169:194–197. doi: 10.7205/milmed.169.3.194. [DOI] [PubMed] [Google Scholar]
- 18.Brook I, Walker RI, MacVittie TJ. Effect of antimicrobial therapy on bowel flora and bacterial infection in irradiated mice. Int J Radiat Biol Relat Stud Phys Chem Med. 1988;53:709–716. doi: 10.1080/09553008814551081. [DOI] [PubMed] [Google Scholar]
- 19.Maekawa M, Kawai K, Takahashi Y, Nakamura H, Watanabe T, Sawa R, Hachisuka K, Kasai H. Identification of 4-oxo-2-hexenal and other direct mutagens formed in model lipid peroxidation reactions as dGuo adducts. Chem Res Toxicol. 2006;19:130–138. doi: 10.1021/tx050236m. [DOI] [PubMed] [Google Scholar]
- 20.Deutsch JC. Spontaneous hydrolysis and dehydration of dehydroascorbic acid in aqueous solution. Anal Biochem. 1998;260:223–229. doi: 10.1006/abio.1998.2700. [DOI] [PubMed] [Google Scholar]
- 21.Murayama Y, Sakai F. Effects of fasting on the Henele’s loop function of rat kidney. Jpn J Pharmacol. 1975;25:475–477. doi: 10.1254/jjp.25.475. [DOI] [PubMed] [Google Scholar]
- 22.Pritchard JB. Intracellular alpha-ketoglutarate controls the efficacy of renal organic anion transport. J Pharmacol Exp Ther. 1995;274:1278–1284. [PubMed] [Google Scholar]
- 23.Shuprisha A, Lynch RM, Wright SH, Dantzler WH. Real-time assessment of alpha-ketoglutarate effect on organic anion secretion in perfused rabbit proximal tubules. Am J Physiol. 1999;277:F513–F523. doi: 10.1152/ajprenal.1999.277.4.F513. [DOI] [PubMed] [Google Scholar]