Abstract
The objective of this work is to determine the extent to which various structural factors influence the yield of trapped free radicals, G(tfr), in DNA irradiated at 4 K. G(tfr) was measured in a series of 13 different oligodeoxynucleotides using electron paramagnetic resonance (EPR) spectroscopy. Each sample consisted of crystalline duplex DNA for which the crystal structure was verified to be that reported in the literature. We find that the G(tfr) of these samples is remarkably high, ranging from 0.55 to 0.75 μmol/J. The standard deviation in G(tfr) for a given crystal structure is generally small, typically less than ±10%. Furthermore, G(tfr) does not correlate with DNA base sequence, conformation, counterion or length of base stacking. Two observations point to the importance of DNA packing: (1) The radical yields in crystalline DNA are greater than those determined previously for DNA films (0.2 to 0.5 μmol/J); and (2) the variability in G(tfr) is less in DNA crystals than in DNA films. We conclude that closely packed DNA maximizes radical trapping by minimizing the interhelical solvent space. Furthermore, the high efficiency of electron and hole trapping at 4 K is not consistent with DNA possessing properties of a metallic conductor. Indeed, it behaves as an insulator, whether it is in A-, B-, or Z-form and whether base stacking is short- (8 bp) or long-range (> 1000 bp).
INTRODUCTION
The interaction of ionizing radiation with DNA and its hydration layer results in the production of free radical species trapped on either the bases or the sugar–phosphate backbone of the DNA (1, 2). The water surrounding DNA is separable into two regions, a hydration layer and a bulk water layer (3–5). Ionizing events in the bulk water layer create indirect damage to the DNA, primarily from reactions with the aqueous electron and hydroxyl radical (6–10). Electrons and holes created in the hydration layer have a high probability of transferring to the DNA molecule (5, 11, 12). This transferred damage cannot be differentiated from radical species produced by direct ionization of the DNA bases and sugar–phosphate backbone (5, 12). The collective damage produced by transfer from the hydration layer and that produced directly is referred to as direct-type damage. This paper presents a study of factors influencing the yield of direct-type damage.
The total free radical yield of DNA has been studied in several forms of DNA, including dilute aqueous solutions (8), lyophilized powders (4, 13), and films (14). All these experiments have demonstrated that DNA acts as an efficient trap of both electrons and holes. Several factors have been cited as influencing the ability of DNA to trap radiation-induced free radicals: level of hydration (4–6, 8, 9, 11, 13, 15, 16), conformation (A-, B-, Z- or other form of DNA) (9, 17, 18), sequence (19–24), and packing (14, 25, 26). Work by our group has suggested that of these factors, variability in DNA packing has the greatest effect on free radical yield.
The yield of trapped free radicals, G(tfr), depends on the method of sample preparation, even when the degree of DNA hydration is the same. All the methods bring DNA to a high concentration to promote aggregation. Upon aggregation, DNA can pack in various ways, and we have suggested that this variability leads to the large scatter observed in the data on radical yield (4, 11, 14). Because of the difficulties in characterizing the nature of DNA packing in films or powders, we undertook the investigation of crystalline DNA.
Crystals of oligodeoxynucleotides have been characterized by X-ray crystallography and provide definitive information on conformation, sequence and packing to nanometer-level (10−1 nm = 1 Å) resolutions. In the work reported here, over 50 samples of 13 different oligodeoxynucleotide crystals have been analyzed by electron paramagnetic resonance (EPR) spectroscopy. By employing Q-band EPR, picomoles of radicals are detected in microgram samples. G(tfr) was measured at 4 K after X irradiation at 4 K. A comparison of the results obtained from the different crystals provides information on the relative importance of sequence, conformation, base-stacking continuity, counterion, and packing on free radical trapping and electron migration in DNA.
MATERIALS AND METHODS
Oligonucleotides were purchased from TIBMolBiol, Midland Certified Reagent, and Ransom Hill Bioscience and were used for crystal growth without further purification. All of the sequences are, with one exception, self-complementary. For the one exception, distinct complementary strands were used for crystallization. Only duplex structures were studied, and each structure had been determined previously by X-ray crystallography. The sequences employed are: CTCTCGAGAG (27), CTCGAG (28), GCACGCGTGC (29), CCCTAGGG (30), GCGGGCCCGC (two forms) (31), CGCGCG (three forms) (32–34), GTGCGCAC (35), CGCACG:GCGTGC (36), CGCG (37), and CGCGAATTCGCG (38). The growth conditions for the various crystals may be found in the respective references; a brief summary of crystal characteristics and the content of the crystallization medium is given in the Table 1.
TABLE 1.
Free Radical Yields and Relevant Crystal Characteristics
| Crystal sequence | Conformation | Space group | Stacking continuitya | Growth mediumb | Number of crystal samples | Free radical yield (μm/J) | Referencec |
|---|---|---|---|---|---|---|---|
| CTCTCGAGAG | B | C2 | Y | CaAc | 4 | 0.70 ± 0.06 | 27 |
| CTCGAG | B | P6222 | Y | MgCI, Sp | 5 | 0.75 ± 0.14 | 28 |
| CGCGAATTCGCG | B | P212121 | N | Mg, Sp | 5 | 0.66 ± 0.06 | 38 |
| GCACGCGTGC | A | P6122 | N | Co, Sp | 5 | 0.57 ± 0.04 | 29 |
| CCCTAGGG | A | P43212 | N | MgCI, Sp | 6 | 0.70 ± 0.04 | 30 |
| GCGGGCCCGC (I) | A | P212121 | N | Sp | 5 | 0.55 ± 0.10 | 31 |
| GCGGGCCCGC (II) | A | P6122 | N | Co | 6 | 0.58 ± 0.08 | 31 |
| GTGCGCAC | A | P43212 | N | MgCI, Sp | 4 | 0.72 ± 0.07 | 35 |
| CACGCG:GTGCGC | Z | P212121 | Y | MgCI | 4 | 0.61 ± 0.03 | 36 |
| CGCGCG (I) | Z | P212121 | Y | MgCI, Sp | 4 | 0.73 ± 0.10 | 32 |
| CGCGCG (II) | Z | P212121 | Y | MgCI | 4 | 0.70 ± 0.06 | 34 |
| CGCGCG (III) | Z | P212121 | Y | Sp | 4 | 0.57 ± 0.05 | 33 |
| CGCG | Z | P212121 | Y | MgCI | 9 | 0.65 ± 0.15 | 37 |
Notes. CaAc, calcium acetate; MgCI, magnesium chloride; Sp, spermine tetrahydrochloride; Co, cobalt (III) hexaamine.
Y indicates the oligonucleotides exhibit continuous stacking, and N signifies they do not.
All crystal setups contained sodium cacodylate as a buffer.
Reference to crystallographic structure.
The crystals were collected from the mother liquor, and the excess liquor was removed by the application of fine paper wicks. Removal of the mother liquor, which contains 2-methyl-2,4-pentanediol as the precipitating agent, is critical to obtaining accurate sample weights. For several crystal types, a single crystal was too small for measurements, and polycrystalline samples had to be used. The uncertainty in determining the weight of polycrystalline samples is larger owing to the increased difficulty in removing the mother liquor and contaminating precipitants.
The space group and cell dimensions of representative crystals of each type were verified to conform to the dimensions and space group reported in the respective references cited above. This verification was made by collecting X-ray crystallographic precession data on an MSC R-Axis II X-ray detector with a cooling device that maintained the crystals at a temperature of 4°C or below. There is evidence that major structural changes do not occur at low temperatures. For example, a dodecamer d(CGCGAATTCGCG) crystal analyzed at room temperature and at 16 K maintained the same end-to-end winding and showed a slight reduction in groove widths, and spacing between neighboring helices (39). We believe, therefore, that the crystal structures measured at higher temperatures are applicable at 4 K with regard to the structural features under consideration in the work presented here.
The single crystal or polycrystalline samples were weighed on a Cahn C-30 balance to an accuracy of ±1 μg. The crystalline samples were then transferred to capillary tubes purchased from the Charles Supper Company. These tubes were selected for their thin quartz walls and subsequent low yields of free radical background. The capillaries were mounted with wax on an aluminum pin that attaches to the sample rod that is used for insertion into the Janis helium cryostat. The cryostat is designed for use at Q-band on a Varian E-12 EPR spectrometer (40).
Irradiations were carried out at 4 K with X rays generated by a Varian/Eimac OEG-76H tungsten-target tube operated at 70 keV and 20 mA. The dose rate was 26 kGy/h. Free radical yields were measured by comparing the intensity of the EPR signal at 4 K to the intensity of a ruby standard mounted on the side of the EPR cavity (see ref. 41 for a more complete description). The crystals were not visibly damaged by temperature cycling. Although the accuracy in relative yields is ± 10%, it is important to note that the absolute yields are less accurate, about ±25%.
RESULTS AND DISCUSSION
A selection of three EPR spectra, recorded at 4 K after X irradiation at 4 K, is shown in Fig. 1. The spectra are representative of A-, B- and Z-DNA in crystalline from. To our knowledge, this is the first reported EPR spectrum of Z-DNA.
FIG. 1.
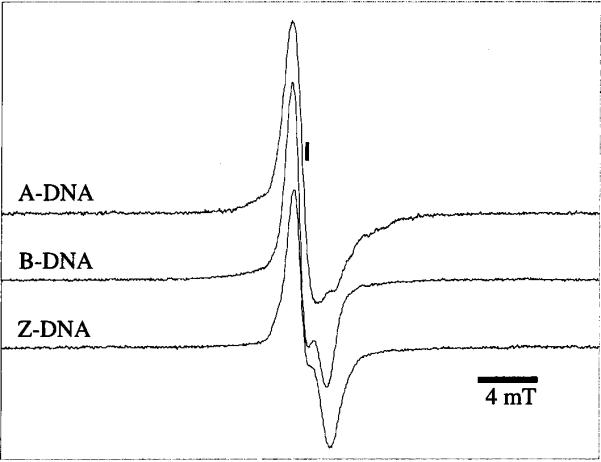
Single-scan 40 mT Q-band EPR spectra taken at 4 K of poly-crystalline A-DNA d(GCGGGCCCGC) (II) after a dose of 22 kGy (top spectrum), polycrystalline B-DNA d(CGCGAATTCGCG) after a dose of 17 kGy (middle spectrum), and polycrystalline Z-DNA d(CGCGCG) (I) after a dose of 22 kGy (bottom spectrum). The position of g = 2.0023 is indicated by the vertical line.
The EPR spectra of the crystalline oligodeoxynucleotides are similar to those of amorphous DNA; see for example ref. (42). We assume, therefore, that the free radical distribution in crystalline DNA is similar to that of amorphous DNA. Much work has been done to identify the free radicals trapped in amorphous DNA and their relative distribution. At the doses and LET employed here, previous work on DNA indicates that the yield of phosphate-centered radicals is very low (43, 44); also, the yield of deoxyribose-centered radicals is low (45) and difficult to measure (46). The majority of trapped damage in DNA is at the bases. The primary electron trap at low temperatures is cytosine (47–50), and the hole trap is guanine (2, 51, 52). Although EPR spectra of crystalline and amorphous DNA are qualitatively similar, there are relatively large quantitative differences.
Yields of Free Radicals Trapped at 4 K
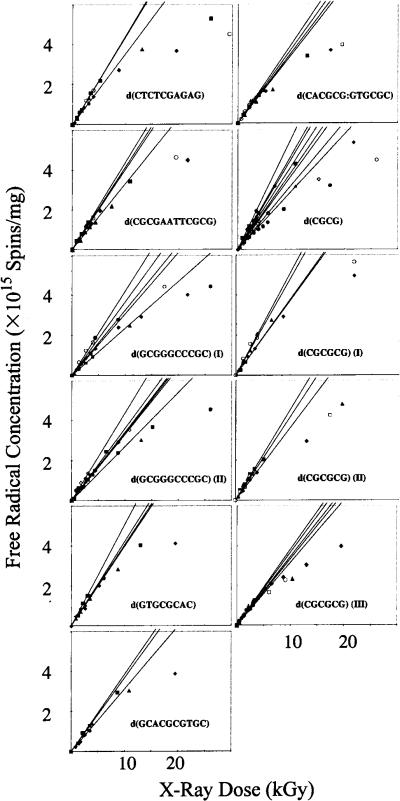
Dose–response curves for 11 different types of crystals are shown in Fig. 2. The curves of the two additional crystals included in Table 1 may be found in ref. (53). The data at and below 5.0 kGy were used to determine least-squares linear fits, the slopes of which are G(tfr) (the yield of trapped free radicals). The values of G(tfr), in micromoles of free radicals/joule, are reported in Table 1.
FIG. 2.
Free radical concentration as a function of X-ray dose for 11 different crystalline systems irradiated and measured at 4 K. The initial slopes of the individual data sets using the linear region of the data <5 kGy are plotted. From the slopes of these lines, we determine the free radical yields of the crystalline DNA samples.
The variation in G(tfr) is relatively small. Depending on the specific crystal type, the standard deviation varies from 5% to 23%. For the most part, this standard deviation conforms to the expected standard errors normally associated with the measurement of relative G values by EPR, an accuracy of ± 10%. The increased variation of certain samples is a function of the experimental difficulty of working with the crystal rather than a variation between samples. For example, the d(CGCG) tetramer forms small crystals that must be pooled in polycrystalline samples to determine yields. A review of the data in Fig. 2 shows that the larger standard deviations tend to contain one or two outliers. Regardless, the data are much more reproducible than those observed for amorphous samples of macromolecular DNA (with variations of 50–100%). It follows that the larger variations in the amorphous samples are a consequence of variables intrinsic to the sample and not an artifact of the measurement technique. In addition, it shows that the crystalline samples afford the opportunity to determine which variables are important in governing free radical trapping.
The variation in G(tfr) between the crystal types, given in Table 1, is only slightly greater than the standard deviations within each crystal type. The mean G(tfr) for all the crystalline samples collectively is 0.65 ± 10% μmol/J. The span of G(tfr) is from a minimum of 0.55 (μmol/J to a maximum of 0.75 μmol/J. Notably, the span is relatively small. Within that span, we look for a possible correlation between G(tfr) and intrinsic variables of crystalline DNA.
Effect of Tertiary Conformation
Samples listed in Table 1 are grouped according to conformation: A-, B- and Z-DNA. The mean value of G(tfr) is 0.62 ± 0.07 μmol/J for the five A-DNAs, 0.70 ± 0.04 μmol/J for the three B-DNAs, and 0.65 ± 0.06 μmol/J for the five Z-DNAs. Thus there is no significant difference between these three conformational groups. This result confirms our earlier conclusion, based on film DNA (14), that conformation plays at most a minor role in determining G(tfr) in DNA at 4 K.
Effect of Base Stack Length
The length of base stacking for each crystal type depends on both the size and the lattice packing of the oligodeoxynucleotide. Packing in 7 of the 13 crystals in Table 1 is such that base stacking is continuous. That is, the terminal base pair of one helix abuts the terminal base pair of the adjacent helix. At this intermolecular contact, the π-bonding distance is similar to that within the oligomer. For the oligomers packed end to end, base stacking runs continuously through the crystal lattice and the length of base stacking corresponds to crystal size (10–1000 μm). There are 6 crystal structures in Table 1 in which the packing is not end to end. In those crystals, the length of base stacking corresponds to the number of base pairs in the oligomer: 8, 10 or 12 bp.
There is no difference in values of G(tfr) observed for those systems that are continuously stacked compared to those that are not. The mean values for the entries in Table 1 with continuous stacking and discontinuous stacking are 0.67 ± 0.06 and 0.63 ± 0.07 μmol/J, respectively. As discussed further below, the fact that the free radical yields are large, and remain so under conditions of continuous stacking, provides insight into the range and mechanism of electron and hole migration.
Effect of DNA Counterion
Seven of the crystals under study in this paper were grown with spermine tetrachloride present in the crystallization mother liquor (see Table 1). There has been considerable discussion in the crystallography community as to the presence of additives such as sperrnine in the disordered regions of certain crystals. Shui et al. (38) recently detected the presence of ordered spermine in the major groove of a dodecamer structure of B-DNA, and they suggest that the spermine would be present in essentially all crystals grown using spermine. The possibility that spermine might affect G(tfr) in the DNA crystalline system was studied using the CGCGCG hexamer.
Three different crystal types of CGCGCG were grown in the presence of magnesium andor spermine: type (I), Mg++ + spermine; type (II), Mg++; and type (III), spermine. There was no discernible correlation in the free radical yield that could be ascribed to the presence or absence of spermine, as may be seen from Table 1.
Another counterion used is cobalt (III) hexaamine. The influence of cobalt as an impurity in the crystal was studied in the d(GCACGCGTGC) crystal. Crystals were grown from mother liquor containing from 0.5 to 5.0 mM cobalt hexaamine. Cobalt hexaamine is bright orange in color. The crystals grown from solutions containing 5.0 mM cobalt hexaamine were bright orange, whereas crystals grown from 0.5 mM cobalt hexaamine were almost clear. Although we did not quantify the degree of cobalt doping in these crystals, there is little question that the highly transparent yellow crystals contain considerably less cobalt than the dark orange crystals. The radical yields for the dark orange and pale yellow crystals were the same.
A further check of the effects of cobalt was done using the d(GCGGGCCCGC) crystals. Two crystals of this sequence were grown. Those with cobalt hexaamine in the growth medium [type (II)] are orange; the others without this additive [type (I)] are colorless. The crystals form different structures; the cobalt-containing crystal are in a hexagonal space group, while the cobalt-free crystals are in an orthorhombic space group. The yields of these two crystals were the same (Table 1).
Finally, the EPR spectra were examined for the presence of the broad EPR spectrum of , which is paramagnetic; formation of would occur if the diamagnetic were competitive with DNA as an electron trap. There are extremely weak features in the wings of the spectra that are likely due to reduced cobalt. Given that we do not know the crystalline environment(s) of reduced cobalt, and therefore the symmetry and strength of the crystal fields, no analysis of these spectral features was attempted. With a nuclear spin of 7/2 and principal g values that range from 2 to 5, the powder spectrum would normally be very broad. We presume that comprises a small fraction of the total unpaired spin population. Because G(tfr) is insensitive to the presence of , we conclude that the influence of Co+++ on radical trapping by DNA is negligible.
Effect of Base Sequence
The oligodeoxynucleotides used in these experiments were guanine- and cytosine-rich, because these crystals grow most easily. For those crystals containing exclusively CG base pairs, the yields were G(tfr) = 0.63 ± 0.07 μmol/J (the average of six different crystal types). Note that there is little difference between the sequences with alternating CG and those containing GGG. This is of interest because the oxidizing potential of G within DNA has been observed to decrease in the order GGG < GGX < XGX (22). Thus GGG should be a preferred site of hole trapping.
When one or more AT base pairs are introduced into the sequence, the mean yield is G(tfr) = 0.67 ± 0.06 μmol/J (a total of seven samples). In one oligomer, there is stretch of four AT base pairs, d (CGCGAATTCGCG), and G(tfr) remains in this range. This is of interest because Giese and colleagues (21, 54, 55) have presented evidence that AT-rich regions act as possible barriers to hole transfer by tunneling. By their measure, the rate of transfer by tunneling drops about an order of magnitude per AT base pair. Thus 4 bp effectively shut down this type of hole transfer.
The lack of an effect due to deeper trapping sites (GGG) and electron transfer barriers (AATT) is consistent with our earlier observations on single crystals of d(CGATCG):anthracycline (53). In these crystals of continuously stacked B-DNA, anthracycline is intercalated between each of the adjacent CG pairs, giving two per hexamer. The anthracycline redox potentials are such that all of the electrons and holes are trapped by the intercalated anthracycline (41). The G(tfr) of this crystal is 0.4 μmol/J. That this value is actually less than the values reported in Table 1 has interesting ramifications that will not be discussed here. The relevant point here is that un-doped DNA is as good a trap as or even a better trap than doped DNA. Thus, at least at 4 K, the naturally occurring base sequences are sufficient to trap electrons and holes with extraordinary efficiency.
Effect of Packing
Within the 13 crystals studied so far, there are no obvious packing effects that account for the variations in G(tfr). As a whole, however, crystalline DNA has proven to be a more efficient radical trapping agent than other forms of DNA studied previously. In addition, the uniform packing of the crystalline DNA has eliminated the extreme scatter seen in previous studies of the free radical yields in DNA (11, 14, 16).
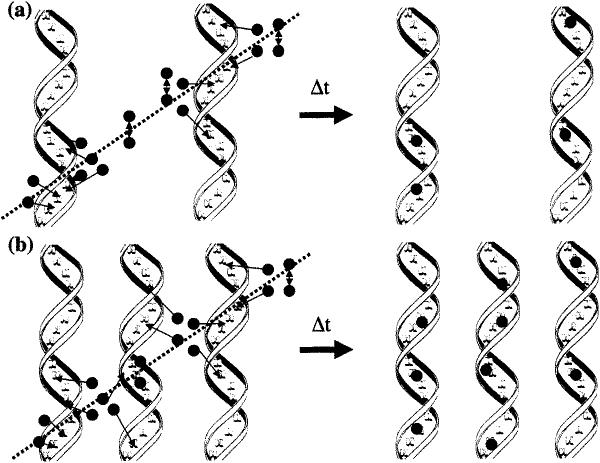
DNA in the form of film has values of G(tfr) ranging from 0.2 μmol/J to 0.5 μmol/J (at hydration levels comparable to the crystals) (14). In lyophilized DNA, G(tfr) reaches 0.6 μmol/J. It was proposed that lyophilized DNA is a more efficient radical trap than film DNA because of an increase in DNA packing density (56). A direct test of that proposal is lacking, in part because there are no methods by which the local density of DNA packing can be measured in amorphous samples. Our finding here, i.e. the relatively large values of G(tfr) found for all 13 crystalline DNAs, supports this proposal. The crystalline lattices allow for a high density of packing, close to if not at the maximum density achievable. Commensurate with high packing density is the reduction in the interstitial space occupied by solvent. As the interstitial space is reduced, there is an increased probability that free radicals formed in a given spur will be isolated on DNA (see Fig. 3). Absent trapping by DNA, the probability of free radicals combining within the solvent-occupied space is very high; this is known because the yield of solvent-centered radicals is very small. Thus a decrease in radical combination reactions is accompanied by an increase in DNA trapped radicals and relatively high values of G(tfr).
FIG. 3.
Model of free radical trapping in (panel a) film and (panel b) crystalline DNA. As ionizing radiation passes through the DNA matrix, it produces holes and electrons, represented by dark circles. In the film DNA, DNA spacing is expected to be variable, with the occurrence of large interstitial water spaces where increased combination reactions occur, and a lower number of free radicals are trapped on the DNA after time t. In the crystalline lattice, DNA spacing is homogeneous and large islands of solvent are absent; thus holes and electrons created in the hydration regions have increased probability of being trapped.
Range of Electron Transfer
A recent report by Kelly and Barton (57) concludes that small variations in stacking and energetics result in “profound changes” in the distance dependence of electron transfer. Our results lead to another conclusion. DNA remains an effective trap of holes and electrons at 4 K over a wide range of conformations and stacking interactions. If DNA were a good conductor, the yield of trapped radicals should be effectively zero. One might argue that the conductive properties of DNA change dramatically in going from single crystals at 4 K to dilute aqueous solution at 300 K. But if DNA is a molecular wire, i.e. behaves as a metallic conductor, the conductivity should increase with decreasing temperature. That is not observed in our experiments. The results reported here do not support the proposal that DNA can act as a conductor or that small variations in stacking have large effects on the rate of electron transfer in DNA.
That DNA is not a molecular wire (electrons and holes do not travel long distances at low resistance) does not preclude long-range migration of electrons or holes. Indeed, there is little question that raising the temperature above 4 K extends electron and hole migration distances and that this migration is activated by heating. The thermal annealing properties of oligodeoxynucleotide crystals will be the subject of a future report.
CONCLUSIONS
This study has demonstrated that crystalline DNA is very efficient at trapping electrons and holes at 4 K. The yields of trapped radicals (0.55–0.75 μmol/J) are relatively independent of the base sequence, conformation, counterion, and length of base stacking. These results support the proposal that closely packed DNA maximizes radical trapping by minimizing the interhelical solvent space.
Furthermore, the high efficiency of electron and hole trapping at 4 K is not consistent with the DNA possessing properties of a metallic conductor. Indeed, it behaves as an insulator, whether it is in A-, B- or Z-form and whether base stacking is short- (8 bp) or long-range (>1000 bp).
ACKNOWLEDGMENTS
We thank Kermit Mercer for his invaluable technical assistance, Tom Colby and Michael Strickler for assistance in verifying crystal structures, and Michael T. Milano for his thoughtful discussions. This investigation was supported by PHS Grant 2-R01-CA32546, awarded by the National Cancer Institute, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
REFERENCES
- 1.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; New York: 1987. [Google Scholar]
- 2.Becker D, Sevilla MD. The chemical consequences of radiation damage to DNA. Adv. Radiat. Biol. 1993;17:121–180. [Google Scholar]
- 3.Schneider B, Patel K, Berman H. Hydration of the phosphate group in double-helical DNA. Biophys. J. 1998;75:2422–2434. doi: 10.1016/S0006-3495(98)77686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Becker D, Sevilla MD. The influence of hydration on the absolute yields of primary ionic free radicals in γ-irradiated DNA at 77 K. I. Total radical yields. Radiat. Res. 1993;135:146–154. [PubMed] [Google Scholar]
- 5.La Vere T, Becker D, Sevilla MD. Yield of OH in gamma-irradiated DNA as a function of DNA hydration: Hole transfer in competition with OH formation. Radiat. Res. 1996;145:673–680. [PubMed] [Google Scholar]
- 6.Ito T, Baker SC, Stickley CD, Peak JG, Peak MJ. Dependence of the yield of strand breaks induced by γ-rays in DNA on the physical conditions of exposure: Water content and temperature. Int. J. Radiat. Biol. 1993;63:289–296. doi: 10.1080/09553009314550391. [DOI] [PubMed] [Google Scholar]
- 7.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 8.Swarts SG, Becker D, Sevilla MD, Wheeler KT. Radiation-induced DNA damage as a function of hydration. II. Base damage from electron loss centers. Radiat. Res. 1996;145:304–314. [PubMed] [Google Scholar]
- 9.Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT. Radiation-induced DNA damage as a function of hydration. I. Release of unaltered bases. Radiat. Res. 1992;129:333–344. [PubMed] [Google Scholar]
- 10.Milligan JR, Wu CC, Ng J-Y, Aguilera J, Ward JF. Characterization of the reaction rate coefficient of DNA with the hydroxyl radical. Radiat. Res. 1996;146:510–513. [PubMed] [Google Scholar]
- 11.Mroczka N, Bernhard WA. Hydration effects on free radical yields in DNA X-irradiated at 4 K. Radiat. Res. 1993;135:155–159. [PubMed] [Google Scholar]
- 12.Becker D, La Vere T, Sevilla MD. ESR detection at 77 K of the hydroxyl radical in the hydration layer of gamma-irradiated DNA. Radiat. Res. 1994;140:123–129. [PubMed] [Google Scholar]
- 13.Wang W, Yan M, Becker D, Sevilla MD. The influence of hydration on the absolute yields of primary free radicals in gamma-irradiated DNA at 77 K. II. Individual radical yields. Radiat. Res. 1994;137:2–10. [PubMed] [Google Scholar]
- 14.Milano MT, Bernhard WA. The effect of packing and conformation on free radical yields in films of variably hydrated DNA. Radiat. Res. 1999;151:39–49. [PMC free article] [PubMed] [Google Scholar]
- 15.Hüttermann J, Roehrig M, Köhnlein W. Free radicals from irradiated lyophilized DNA: Influence of water of hydration. Int. J. Radiat. Biol. 1992;61:299–313. doi: 10.1080/09553009214550981. [DOI] [PubMed] [Google Scholar]
- 16.Mroczka NE, Bernhard WA. Electron paramagnetic resonance investigation of X-irradiated poly(U), poly(A) and poly(A):poly(U): Influence of hydration, packing and conformation on radical yield at 4 K. Radiat. Res. 1995;144:251–257. [PubMed] [Google Scholar]
- 17.Michalik V, Tartier L, Spotheim-Maurizot M, Charlier M. Reaction of hydroxyl radical with B- and Z-DNA. In: Fuciarelli AF, Zimbrick JD, editors. Radiation Damage in DNA: Structure/Function Relationships at Early Times. Battelle Press; Columbus, OH: 1995. pp. 19–27. [Google Scholar]
- 18.Sy D, Savoye C, Begusova M, Michalik V, Charlier M, Spotheim-Maurizot M. Sequence-dependent variations in DNA structure modulate radiation-induced strand breakage. Int. J. Radiat. Biol. 1997;72:147–155. doi: 10.1080/095530097143365. [DOI] [PubMed] [Google Scholar]
- 19.O'Neill P, Melvin T, Botchway SW. Migration of radiation damage in DNA. In: Fuciarelli AF, Zimbrick JD, editors. Radiation Damage in DNA: Structure/Function Relationships at Early Times. Battelle Press; Columbus, OH: 1995. pp. 65–72. [Google Scholar]
- 20.Fuciarelli AF, Sisk EC, Miller JH, Zimbrick JD. Radiation-induced electron migration in nucleic acids. Int. J. Radiat. Biol. 1994;66:505–509. doi: 10.1080/09553009414551531. [DOI] [PubMed] [Google Scholar]
- 21.Meggers E, Michel-Beyerle ME, Giese B. Sequence dependent long range hole transport in DNA. J. Am. Chem. Soc. 1998;120:12950–12955. [Google Scholar]
- 22.Saito I, Nakamura T, Nakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. Mapping of the hot spots for DNA damage by one-electron oxidation: Efficacy of GG doublets and GGG triplets as a trap for long-range hole migration. J. Am. Chem. Soc. 1998;120:12686–12687. [Google Scholar]
- 23.Melvin T, Cunniffe SMT, O'Neill P, Parker AW, Roldan-Arojna T. Guanine is the target for direct ionisation damage in DNA, as detected by using excision enzymes. Nucleic Acids Res. 1998;26:4935–4942. doi: 10.1093/nar/26.21.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalik V, Spotheim-Maurizot M, Charlier M. Calculation of hydroxyl radical attack on different forms of DNA. J. Biomol. Struct. Dyn. 1995;13:565–575. doi: 10.1080/07391102.1995.10508867. [DOI] [PubMed] [Google Scholar]
- 25.Bernhard WA, Barnes J, Mercer KR, Mroczka N. The influence of packing on free radical yields in crystalline nucleic acids: The pyrimidine bases. Radiat. Res. 1994;140:199–214. [PubMed] [Google Scholar]
- 26.Bernhard WA, Mroczka N, Barnes J. Combination is the dominant free radical process initiated in DNA by ionizing radiation: An overview based on solid-state EPR studies. Int. J. Radiat. Biol. 1994;66:491–497. doi: 10.1080/09553009414551511. [DOI] [PubMed] [Google Scholar]
- 27.Goodsell DG, Grzeskowiak K, Dickerson RE. Crystal structure of CTCTCGAGAG. Implications for the structure of the Halli-day junction. Biochemistry. 1995;34:1022–1029. doi: 10.1021/bi00003a037. [DOI] [PubMed] [Google Scholar]
- 28.Wahl MC, Rao ST, Sundaralingam M. Crystal structure of the B-DNA hexamer d(CTCGAG): Model for an A-to-B transition. Biophys. J. 1996;70:2857–2866. doi: 10.1016/S0006-3495(96)79855-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ban C, Sundaralingam M. Crystal structure of the self-complementary 5′-purine start decamer d(GCACGCGTGC) in the ADNA conformation. Biophys. J. 1996;71:1222–1227. doi: 10.1016/S0006-3495(96)79351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tippin DB, Sundaralingam M. Structure of d (CCCTAGGG): comparison with nine isomorphous octamer sequences reveals four distinct patterns of sequence-dependent intermolecular interactions. Ada Crystallogr. D. 1996;52:997–1003. doi: 10.1107/S0907444996005033. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan B, Sundaralingam M. Evidence for crystal environment dominating base sequence effects on DNA conformation: Crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures. Biochemistry. 1993;32:11458–11468. doi: 10.1021/bi00093a025. [DOI] [PubMed] [Google Scholar]
- 32.Wang A, Quigley G, Kolpak F, Crawford J, van Boom J, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 33.Egli M, Williams LD, Gao Q, Rich A. Structure of pure-spermine form of Z-DNA (magnesium free) at 1 Å resolution. Biochemistry. 1991;30:11388–11402. doi: 10.1021/bi00112a005. [DOI] [PubMed] [Google Scholar]
- 34.Gessner RV, Frederick CA, Quigley GJ, Rich A, Wang AH-J. The molecular structure of the left-handed Z-DNA double helix at 1.0-Å atomic resolution. J. Biol. Chem. 1989;264:7921–7935. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- 35.Bingman C, Li X, Zon G, Sundaralingam M. Crystal and molecular structure of d(GTGCGCAC): investigation of the effect of base sequence on the conformation of octamer duplexes. Biochemistry. 1992;31:12803–12812. doi: 10.1021/bi00166a014. [DOI] [PubMed] [Google Scholar]
- 36.Sadasivan C, Gautham N. Sequence-dependent microheterogeneity of Z-DNA: The crystal and molecular structures of d(CACGCG)d(CGCGTG) and d(CGCACG)d(CGTGCG) J. Mol. Biol. 1995;248:918–930. doi: 10.1006/jmbi.1995.9894. [DOI] [PubMed] [Google Scholar]
- 37.Crawford JL, Kolpak FJ, Wang A, Quigley G, van Boom J, van der Marel G, Rich A. The tetramer d(CpGpCpG) crystallizes as a left-handed double helix. Proc. Natl. Acad. Sci. USA. 1980;77:4016–4020. doi: 10.1073/pnas.77.7.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shui X, McFail-Isom L, Hu GG, Williams LD. The BDNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 39.Drew HR, Samson S, Dickerson RE. Structure of B-DNA dodecamer at 16 K. Proc. Natl. Acad. Sci. USA. 1982;79:4040–4044. doi: 10.1073/pnas.79.13.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer KR, Bernhard WA. Design and operation of a variable temperature accessory for Q-band ESR. J. Magn. Reson. 1987;74:66–71. [Google Scholar]
- 41.Milano MT, Hu GG, Williams LD, Bernhard WA. Migration of electrons and holes in crystalline d(CGATCG)-anthracycline complexes X-irradiated at 4 K. Radiat. Res. 1998;150:101–114. [PubMed] [Google Scholar]
- 42.Weiland B, Hüttermann J. Free radicals from X-irradiated `dry' and hydrated lyophilized DNA as studied by electron spin resonance spectroscopy: Analysis of spectral components between 77K and room temperature. Int. J. Radiat. Biol. 1998;74:341–358. doi: 10.1080/095530098141483. [DOI] [PubMed] [Google Scholar]
- 43.Becker D, Razskazovskii Y, Callaghan MU, Sevilla MD. Electron spin resonance of DNA irradiated with a heavy-ion beam (16O8+): Evidence for damage to the deoxyribose phosphate back-bone. Radiat. Res. 1996;146:361–368. [PubMed] [Google Scholar]
- 44.Steenken S, Goldbergerova L. Photoionization of organic phosphates by 193 nm laser light in aqueous solution: Rapid intramolecular H-transfer to the primarily formed phosphate radical. A model for ionization-induced chain-breakage in DNA? J. Am. Chem. Soc. 1998;120:3928–3934. [Google Scholar]
- 45.Weiland B, Hüttermann J, Malone ME, Cullis PM. Formation of Cl' located sugar radicals from X-irradiated cytosine nucleosides and -tides in BeF2 glasses and frozen aqueous solutions. Int. J. Radiat. Biol. 1996;70:327–336. doi: 10.1080/095530096145067. [DOI] [PubMed] [Google Scholar]
- 46.Close D. Where are the sugar radicals in irradiated DNA? Radiat. Res. 1997;147:663–673. [PubMed] [Google Scholar]
- 47.Bernhard WA. Initial sites of one-electron attachment in DNA. In: Fielden EM, O'Neill P, editors. The Early Effects of Radiation on DNA. Springer-Verlag; Berlin and Heidelberg: 1991. pp. 141–154. [Google Scholar]
- 48.Steenken S. Electron-transfer-induced acidity/basicity and reactivity changes of purine and pyrimidine bases. Consequences of redox processes for DNA base pairs. Free Radic. Res. Commun. 1992;16:349–379. doi: 10.3109/10715769209049187. [DOI] [PubMed] [Google Scholar]
- 49.Cullis PM, Evans P, Malone ME. Electron addition to DNA-thymine vs cytosine radical anion? Chem. Commun. 1996:985–986. [Google Scholar]
- 50.Wang W, Sevilla MD. Protonation of nucleobase anions in gamma-irradiated DNA and model systems. Which base is the ultimate sink for the electron? Radiat. Res. 1994;138:9–17. [PubMed] [Google Scholar]
- 51.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solutions. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 52.Jovanovic SV, Simic MG. One-electron redox potentials of purines and pyrimidines. J. Phys. Chem. 1986;90:974–978. [Google Scholar]
- 53.Debije MG, Milano MT, Bernhard WA. DNA responds to ionizing radiation as an insulator not as a `molecular wire'. Angew. Chem. Int. Ed. 1999;37:2752–2756. [PMC free article] [PubMed] [Google Scholar]
- 54.Meggers E, Kusch D, Spichty M, Wille U, Giese B. Electron transfer through DNA in the course of radical-induced strand cleavage. Angew. Chem. Int. Ed. 1998;37:460–462. doi: 10.1002/(SICI)1521-3773(19980302)37:4<460::AID-ANIE460>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 55.Giese B, Wesseley S, Spormann M, Lindemann U, Meggers E, Michel-Beyerle ME. On the mechanism of long-range electron transfer through DNA. Angew. Chem. Int. Ed. 1999;38:996–998. doi: 10.1002/(SICI)1521-3773(19990401)38:7<996::AID-ANIE996>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 56.Milano MT, Bernhard WA. The influence of packing on free radical yields in solid-state DNA: Film compared to lyophilized frozen solution. Radiat. Res. 1999;152:196–206. [PMC free article] [PubMed] [Google Scholar]
- 57.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]