Abstract
The cell adhesion molecule Close Homologue of L1 (CHL1) is important for apical dendritic projection and laminar positioning of pyramidal neurons in caudal regions of the cerebral cortex. The p21-activated kinase (PAK 1-3) subfamily of serine/threonine kinases has also been implicated in regulating cell adhesion, migration, and morphology. Immunofluorescence staining in mouse embryonic brain showed that PAK1-3 was expressed in embryonic cortex and colocalized with CHL1 during neuronal migration and differentiation. To investigate a cooperative function for CHL1 and PAK in pyramidal cell differentiation or migration, a dominant-negative PAK mutant (PAK1 AID) that inhibits PAK1-3 kinase activity while coexpressing a green fluorescent protein (GFP) reporter was electroporated into the lateral ventricles of wild type and CHL1 null mutant mouse embryos (E14.5), then brain slices were cultured and neurons analyzed for laminar positioning and morphology by confocal microscopy after 3 days in vitro. Expression of PAK1 AID in CHL1 mutant cortex inactivated PAK and caused embryonic cortical neurons to branch profusely in the intermediate zone and cortical plate. The number of nodes, terminals and length of leading processes/apical dendrites of CHL1 mutant embryos expressing PAK1 AID increased dramatically, compared to CHL1 mutants without PAK1 AID, or wild type embryos with or without PAK1 AID. These findings suggest that CHL1 and PAK1-3 kinase cooperate, most likely in independent pathways, in regulating morphological development of the leading process/apical dendrite of embryonic cortical neurons.
Keywords: CHL1, PAK, electroporation, cerebral cortex development, neuronal branching
Introduction
The neural cell recognition molecule Close Homolog of L1 (CHL1) is a member of the L1 family of neural cell adhesion molecules, which includes L1, CHL1, NrCAM, and Neurofascin {Maness, 2007 #3564}. CHL1 is required for laminar positioning and dendritic projection of pyramidal neurons in the posterior region of the developing mouse neocortex, where CHL1 is expressed in a high-caudal to low-rostral gradient (Demyanenko et al., 2004). Deep layer pyramidal neurons of CHL1-minus mice are malpositioned to lower laminar positions in the visual and somatosensory cortex, and develop misoriented, often inverted, apical dendrites (Demyanenko et al., 2004). Although CHL1 potentiates cell migration through β1 integrins, Src and ERK MAP kinase in culture (Buhusi et al., 2003, Demyanenko et al., 2004), the molecular mechanism by which CHL1 regulates neuronal process outgrowth and positioning in the developing neocortex is unclear.
The p-21-activated kinases (PAK) are potential signaling effectors in cortical neurons, as they interact with many of the molecular components involved in migration, process outgrowth, and spine morphogenesis (Nikolic, 2008). PAKs comprise a family of serine-threonine kinases consisting of Group I PAK1-3, which share extensive homology and phosphorylation sites, and Group II PAK 4-6, which vary from the canonical structure (Jaffer and Chernoff, 2002). Phosphorylation of PAK1 at Thr423, and equivalent sites in PAK2,3, is necessary for stabilization of the active kinase conformation (Zenke et al., 1999, Lei et al., 2000), however kinase-dependent and kinase-independent functions of PAK have been identified (Bokoch, 2003, Zhang et al., 2005, Higuchi et al., 2008). PAK1-3 are downstream effectors of the Rho GTPases Rac1 and Cdc42, which play a major role in cell motility, process outgrowth, and dendrite and axon guidance (Nikolic et al., 1998, Edwards et al., 1999, Rashid et al., 2001, Bokoch, 2003, Nikolic, 2008). Loss of PAK1 function disrupts the morphology of migrating neurons, which accumulate in the IZ and deep cortical layers (Causeret et al., 2009).
We speculated that PAK1-3 may function coordinately with CHL1 as an important determinant of cortical pyramidal neuron differentiation. PAK kinase activity has been implicated in controlling premature neuronal differentiation and promoting migration (Hayashi et al., 2002, Sakakibara and Horwitz, 2006, Cobos et al., 2007, Hayashi et al., 2007). Moreover, PAK1 is involved in L1- and integrin-dependent haptotactic cell migration in vitro (Schmid et al., 2004, Schmid and Maness, 2008). To investigate a coordinate function for PAK and CHL1 in morphological differentiation and positioning of embryonic cortical pyramidal neurons, we examined PAK 1-3 and CHL1 expression in the developing mouse neocortex, and analyzed the effects of electroporation-mediated gene transfer of a dominant negative PAK1 autoinhibitory domain (AID) mutant on neurons in CHL1 null mutant and wild type mice. We show here that combined loss of CHL1 and inhibition of PAK1-3 kinase induced abnormal branching of developing cortical neurons, thus providing evidence that CHL1 cooperates with PAK 1-3, most likely in independent pathways, to regulate morphology of embryonic cortical neurons important for normal neuronal development.
Experimental Procedures
Mice
Generation and propagation of mice in which the CHL1 gene was knocked out by homologous recombination was described previously (Montag-Sallaz et al., 2002, Demyanenko et al., 2004). CHL1 homozygous null and wild type controls were on a C57Bl/6 genetic background. All experiments were approved by the animal care committee of The University of North Carolina at Chapel Hill, USA (in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80-23 revised 1996).
Immunofluorescence Staining
Affinity-purified rabbit polyclonal antibodies raised against a 19-residue peptide mapping at the carboxyl terminus of PAK1, which cross reacts with PAK2 and PAK3 (Santa Cruz Biotechnology, Inc.: sc-881), was used for detection of PAK1-3. This antibody was previously characterized in neuronal cultures (Rashid et al., 2001). Antibody 2601 to active PAK1-3 isoforms phosphorylated at Thr423, Thr402, or Thr421, respectively, was from Cell Signaling Technology, Inc. (Danvers, MA). CHL1/L1CAM2/CALL antibody (R&D systems: AF2147) was used for detection of CHL1. Specificity of this antibody against CHL1 was confirmed previously by us and others (Demyanenko et al., 2004, Nikonenko et al., 2006, Wright et al., 2007). These antibodies do not cross-react with other L1 family members in embryonic mouse brain or transfected HEK293 cells, and show no nonspecific immunostaining of homozygous CHL1-minus brain (Nikonenko et al., 2006). Rabbit polyclonal anti-green fluorescent protein (GFP) antibody (Invitrogen: A11122) was used to enhance GFP detection in neurons electroporated with pCIG2 plasmids. RC2 antibody was obtained from the Developmental Hybridoma Bank (University of Iowa).
Timed pregnant mice were deeply anesthetized with avertin (2.5% 0.02 mL/g in saline, intraperitoneally), and embryos were removed and immersion fixed in 4% paraformaldehyde overnight. Sections were cut on a vibratome at 60 µm, fixed in ice-cold methanol (10 min), washed with PBS, and permeabilized with blocking solution: 0.2% Triton X-100, 0.5% fish skin gelatin, and 10% donkey serum for 1 hr at room temperature. Sections were stained overnight with anti-PAK1-3 (1:250) and/or anti-CHL1 (1:100) antibodies in blocking solution/PBS (1:1). Sections were washed and incubated with FITC or TRITC- conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc) for 1 hr in a light protected chamber. Sections were washed and embedded in Vectashield (Vector Labs) and visualized by confocal microscopy. Immunofluorescent staining was performed on 4 embryos of each genotype.
Plasmids
Amino acids 83–149 of hPAK1 cDNA, comprising the auto-inhibitory domain (AID) of PAK 1 (Zhao et al., 1998) were amplified by PCR from plasmid pCMV6-myc-PAK1(83–149)(Sells et al., 1997) (gift of Tina Leisner, UNC Chapel Hill) and inserted into pCIG2 (Hand et al., 2005) (gift of Dante Bortone, UNC Chapel Hill) at XhoI and XmaI to make the pCIG2 PAK1 AID plasmid, which expresses the AID domain under the chicken β-actin promoter with a CMV early-immediate enhancer. EGFP is expressed from an internal IRES sequence in the same plasmid containing PAK1 AID, ensuring coexpression. Inserts were verified by DNA sequencing.
Electroporation
Ex vivo electroporation (Swartz et al., 2001) was performed as described (Hand et al., 2005). Embryos at E14.5 were removed and placed in ice-cold HBSS from deeply anesthetized (Avertin 2.5%, 0.02 ml/gm) time-pregnant CHL1 mutant and wild type mice. A solution containing DNA, either pCIG2 or pCIG2 PAK AID at 2 µg/µL in PBS (pH 7.3) with Fast green (0.01%) as tracer (Sigma) was injected into the lateral ventricles of each hemisphere of embryos using glass micropipettes for 5 – 10 pulses at 3–4 msec/pulse 55V (100 msec duration with 500 msec intervals between each pulse) using a ECM 830 electroporator (BTX Harvard Apparatus) and 7 mm stainless steel tweezers electrodes (Harvard Apparatus, Holliston, MA). The positive electrode was situated over the dorsal lateral ventricle while the negative electrode was situated along the neck on the contralateral side, causing selective uptake of the plasmid DNA into the cells lining the lateral ventricles. Brains were dissected out and mounted in 3% low-melting point agarose in complete HBSS, then sectioned at 300 µm on a Vibratome. Slices were mounted on organotypic slice culture membranes previously coated with poly-L-lysine (0.1 mg/mL) and laminin (0.01 mg/mL), then incubated in 35 mm dishes containing 1.2 mL slice culture medium (Polleux and Ghosh, 2002) in 5% CO2 at 37°C. Sections were incubated for 2–4 days in culture. Following incubation, sections were fixed in 4% PFA in PBS (pH 7.4). Fixed sections were washed in PBS, blocked in 10% goat serum, 3% BSA, and 0.3% Triton X-100 for 1 hr at room temperature, then incubated with GFP antibody (1:1000) in blocking solution at 4°C overnight, washed in PBS, and incubated with secondary antibody (Invitrogen Corporation: A11008, 1:400) in blocking solution for 1 hr at room temperature. Than sections were washed in PBS, mounted in Vectashield, and analyzed using confocal Zeiss 510 microscope. Phospho-PAK1 activity was analyzed in IRES-EGFP positive cells after 3 days in culture. Sections (300 µm) were fixed in 4% PFA in PBS (pH=7.4) for 2 hours. Fixed sections were washed in PBS, blocked in 10% goat serum, 3% BSA, 3% Triton X-100 overnight at 4°C, then incubated with phospho-PAK1 (Thr423)/PAK2 (Thr402) antibody (1:250) in blocking solution at 4°C. After 48 hours sections were washed in PBS and incubated with secondary Alexa Fluor 546 antibody (Invitrogen, A11010, 1:400) in blocking solution overnight at 4°C. Sections were again washed in PBS, mounted in Vectashield, and analyzed by confocal microscopy.
Imaging and Neurolucida analysis
Confocal images were captured on a Zeiss LSM 510 (Confocal/Multiphoton Microscopy Core Facility, Neuroscience Center, UNC Chapel Hill) or on a LSM5 Pascal Confocal Laser Scanning Microscope (Microscopy Services Laboratory, Department of Pathology UNC Chapel Hill). Confocal Z-stacks (from 20 images) were captured using 20x objective. Projection images were calibrated for Neurolucida, fully integrated software for performing anatomical mapping and neuron tracing. To quantify the number of nodes, ends and total length of cell processes, the Auto Neuron tracing function of Neurolucida was used. Corresponding means from experimental and control plasmid electroporations were compared by Student's t-test for significant differences (p<0.05). Parametric one-way analysis of variance was used to confirm mean differences in measurements across mouse plasmid groups with Tukey adjustment of p-values for multiple comparisons. Statistical analysis was performed with SAS statistical software, version 9.2 (SAS Institute Inc., Cary, NC).
For analysis of neuronal positioning, EGFP-positive cells were scored in a region of the neocortex that extended 500 µm medially from the neocortex/archicortex boundary. Maximum-brightness projections were made of each confocal Z-stack image using ImageJ software (NIH). The boundaries of the ventricular and pial surfaces were marked manually and the centers of the soma of EGFP-expressing cells were marked automatically. Automatic marking was done by applying a band pass filter, thresholding, applying a watershed filter, and using the Analyze Particles feature in ImageJ. Bins corresponding to the cortical plate and marginal, intermediate, subventricular, and ventricular zones were identified from DAPI staining, and the position of EGFP-positive cells was determined as described (Hand et al., 2005). Corresponding bins from experimental and control plasmid electroporations were compared by Student's t-test (p<0.05). Statistical assistance was obtained from Dr. Robert Hamer (Department of Biostatistics School of Public Health, UNC at Chapel Hill and biostatistician Wing K. Chiu (UNC Lineberger Comprehensive Cancer Center).
Results
PAK 1-3 expression in the developing cerebral cortex
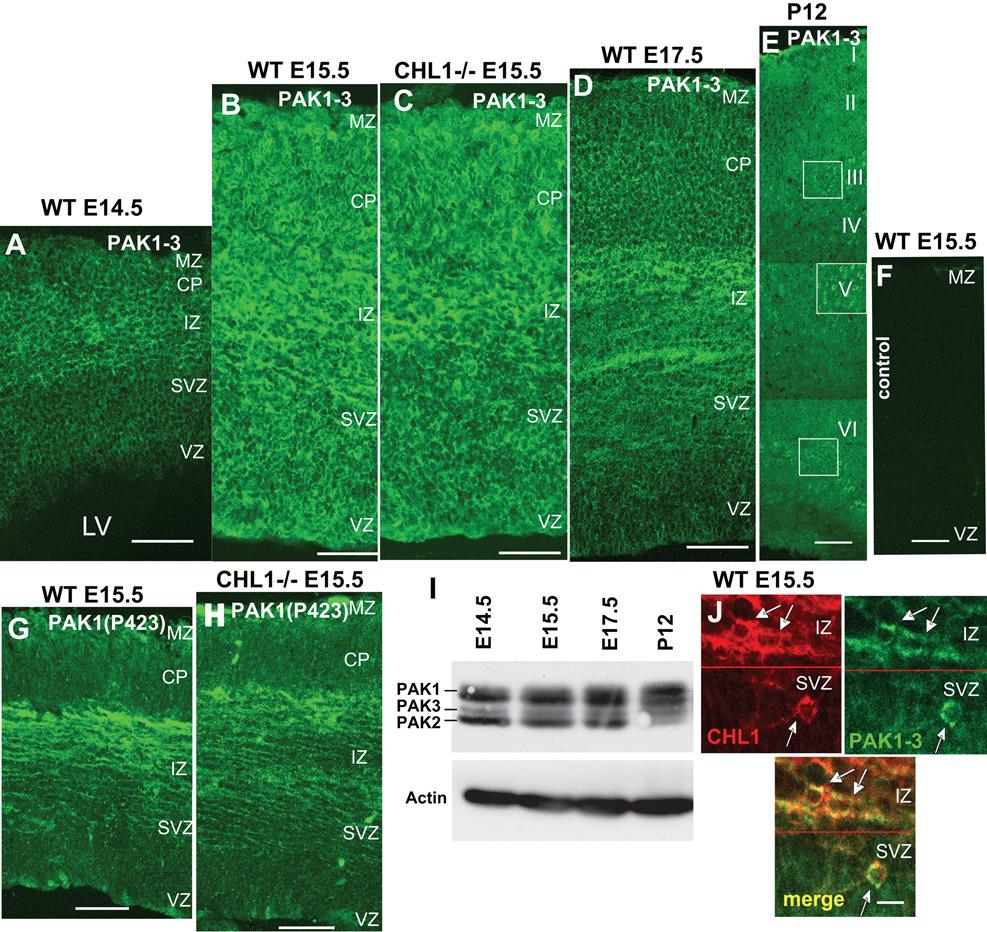
To evaluate the importance of PAK in the developing cerebral cortex, the expression of Group I PAK1-3 was examined by immunofluorescent staining in the developing neocortex of embryonic and postnatal mice (Fig. 1A – E) using polyclonal antibody PAK1(C-19), which also recognizes PAK 2 and 3. Cortical neurons undergo radial migration in the mouse neocortex from approximately E14–E16, differentiate as they reach their appropriate laminar positions, and continue maturation during early postnatal stages (Nadarajah et al., 2002). In wild type cortex at E14.5 high levels of PAK1-3 immunoreactivity were evident in the cortical plate (CP) and intermediate zone (IZ), containing neurons migrating by somal translocation and glial-guided mechanisms (Fig. 1A). Comparatively lower levels of PAK1-3 were seen in the ventricular (VZ) and subventricular (SVZ) zones, containing proliferating progenitors (Fig. 1A). In wild type cortex at E15.5 PAK1-3 expression occurred throughout the cortex, and was especially high in axonal fibers in the IZ (Fig. 1B). We scored the number of cells that were PAK1-3 immunopositive in different cortical layers of wild type E15.5 cortex counterstained with DAPI (not shown) in single confocal images. All DAPI-stained cells were PAK1-3 immunopositive in the CP/MZ (n=173), the IZ (n=193), and in VZ (n-183), thus PAK1-3 did not appear to be restricted to a particular layer or cell type. In homozygous CHL1 null mutants, the expression pattern of PAK1-3 in the neocortex was similar to wild type, as shown at E15.5 (Fig. 1C). PAK1-3 expression persisted in wild type embryos at E17.5, where it was strong in the IZ but reduced in the VZ and CP (Fig. 1D). In wild type postnatal cortex (P12) PAK1-3 cells in all layers of the CP (II–VI) with highest level of expression in pyramidal cell soma and processes in layers III and V, and somewhat lower levels in layer VI (Fig. 1E, boxes). Similar results were obtained in CHL1 null mutants (not shown). Control staining with non-immune IgG was negative at all stages (Fig. 1F). An antibody recognizing the activated forms of PAK1-3, phosphorylated at threonine 423 in PAK1 or equivalent sites in PAK2-3, was used to localize active PAK1-3 in wild type and CHL1 null embryonic cortex (E15.5) (Fig. 1G, H). Active phospho-PAK1-3 was most prominent in fibers and cells in the IZ, with less staining in the CP and VZ. The minimal differences in localization or intensity of phospho-PAK immunoreactivity in wild type versus CHL1-minus cortex suggested that PAK1-3 may not be a direct downstream effector of CHL1.
Figure 1. PAK1-3 expression in the Developing Mouse Neocortex.
(A – E), Coronal sections of wild type embryos at E14.5, E15.5, E17.5 (A,B, D), wild type (WT) P12 mice (E), and CHL1 null mutant E15.5 embryos (C) were stained with antibody to PAK1, which also recognizes PAK 2 and 3. Neuronal soma in layers II, V, and VI are indicated in boxes.
(F), Control staining with nonimmune IgG in WT (E15.5) cortex.
(G, H), Staining for active PAK1-3 in E15.5 cortex from wild type and CHL1 null mutant mice with antibody recognizing phospho-Thr423 in PAK1 or equivalent sites in PAK2-3 showed equivalent patterns with highest levels in the IZ.
(I), Immunoblot of cortical lysates (60 µg) from E14.5, E17.5, and P12 wild type brains for PAK1-3 with reprobing for actin as loading control.
(J), Staining for CHL1 (red) and PAK1-3 (green) in wild type (E15.5) cortex showing colocalization (merge) in cortical neurons located in the IZ/SVZ (arrows). Red line indicates the IZ and SVZ boundary.
I–VI, layers of the cortex; LV, lateral ventricle; VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate; MZ, marginal zone.
Scale bars: (A–H) 100 m; (I) 10 µm.
Immunoblotting was used to demonstrate that the PAK1(C-19) antibody was specific for PAK1-3 in embryonic and postnatal cortex (Fig. 1I). PAK1 (upper band) was prominently expressed in cortex from E14.5 to E17.5, as well as at postnatal and adult stages. Postnatal expression of PAK1 is consistent with its known function in memory in the adult brain (Hayashi et al., 2004, Zhang et al., 2005). PAK2,3 isoforms were also expressed at embryonic through adult stages. The identification of PAK1-3 isoforms on Western blots was based on established band patterns (Hayashi et al., 2002);(Hayashi et al., 2004). However, four newly recognized splice variants of PAK 3 have been identified in adult mouse brain (Kreis et al., 2008), rendering assignment tentative in the adult. In addition PAK1-3 may be modified by phosphorylation, which can alter electrophoretic mobility. The developmental expression pattern of PAK isoforms which we observed was in general accord with studies (Hayashi et al., 2002, Zhong et al., 2003, Causeret et al., 2009) that used different PAK antibodies.
Double immunostaining for CHL1 and PAK1-3 in wild type embryos at E15.5 showed colocalization of CHL1 and PAK1-3 in soma and processes of cells in the caudal neocortex, where CHL1 is known to be abundant (Fig. 1J). Highest levels of CHL1 expression were seen in the IZ, with lower levels in the CP, SVZ, and VZ, in agreement with our published study (Demyanenko et al., 2004). Cells were scored for CHL1 immunoreactivity in the IZ/CP and VZ/SVZ from single confocal images. All cells in the IZ/CP (111/111) and 3% of cells in the VZ /SVZ (9/261) were immunopositive for CHL1. Furthermore, all of the CHL1-labeled cells in the IZ and SVZ were found to also express PAK1-3, as shown by double labeling (Fig. 1J, merge).
PAK1-3 and CHL1 cooperate to regulate morphology of embryonic cortical neurons
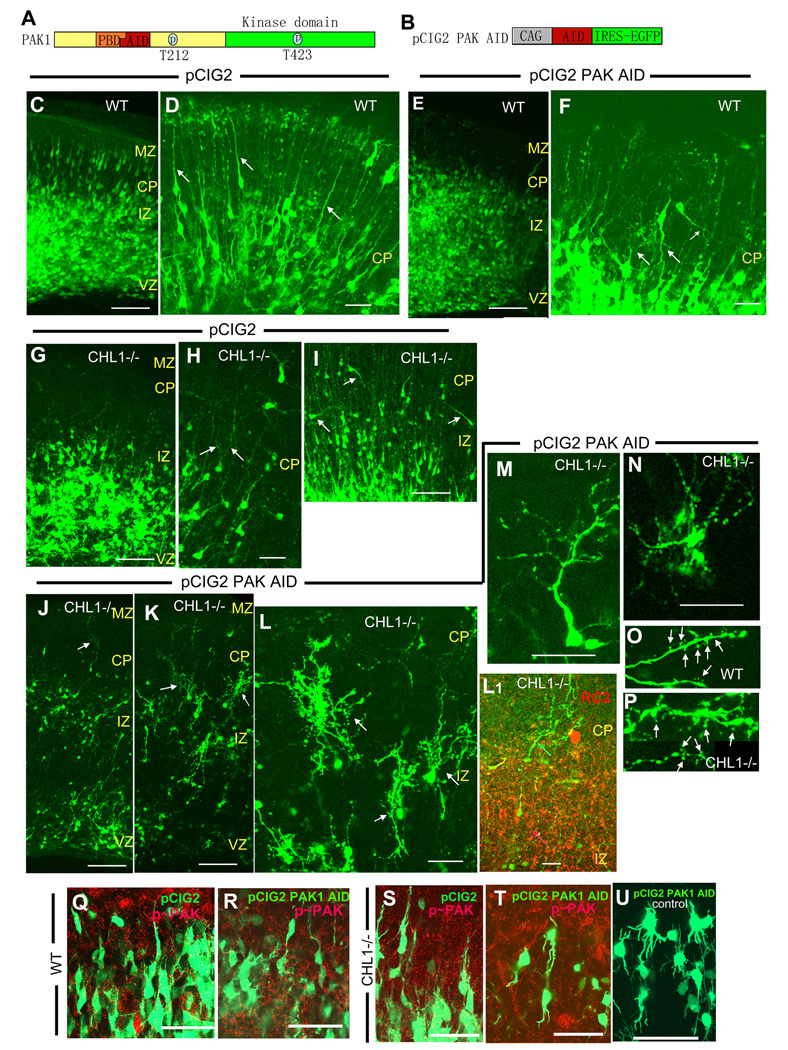
PAK1 contains a p21-binding domain (PBD) for Cdc42 and Rac1, an auto-inhibitory domain (AID), and a serine/threonine-specific kinase domain, as well as sites for inhibitory Thr212 phosphorylation and Thr423 autophosphorylation (Fig. 2A). PAK controls a variety of cellular responses including motility by both kinase-dependent and kinase-independent functions (Frost et al., 1998). Kinase-dependent functions of PAK1-3 can be inhibited by expression of the auto-inhibitory domain (AID) of PAK1, comprising amino acids 83–149, which acts in a dominant negative mechanism to inhibit the catalytic activity of PAK 1-3 isoforms (Zhao et al., 1998). To study a potential role of PAK1-3 kinase in morphological differentiation or neuronal positioning in the developing neocortex, PAK kinase was acutely suppressed by electroporation-mediated expression of the dominant negative PAK1 AID from the plasmid pCIG2 PAK AID, which co-expresses PAK1 AID and enhanced green fluorescent protein (EGFP) polycistronically from an internal ribosome entry site (IRES) (Fig. 2B). Embryos (E14.5) from timed pregnant wild type and CHL1 null mutant mice were removed from the uterus and electroporated into the lateral ventricles with pCIG2 (control) or pCIG2 PAK AID plasmids (2µg/µl). Slices were prepared from the caudal part of the cortex and maintained in organotypic culture for 2–4 days in vitro. The pCIG2 plasmid provided strong expression of EGFP allowing detection of cells in the VZ, IZ, CP and marginal zone (MZ) by 2–4 div.
Figure 2. Effect of PAK1 AID Expression on Neuronal Morphology and Positioning in Wild Type and CHL1 Null Mutant Cortex.
(A), Diagram of PAK1, showing the p21-binding domain (PBD), auto-inhibitory domain (AID), N-terminal regulatory domain (yellow), kinase domain (green), as well as sites of phosphorylation at T212 and T423.
(B), Diagram of pCIG2 PAK1 AID plasmid containing the auto-inhibitory domain under the chicken β-actin promoter with a CMV early-immediate enhancer. The plasmid insert is followed by an IRES-EGFP sequence for coexpression of EGFP.
(C,D), GFP-expressing cells after electroporation of control pCIG2 plasmid into wild type embryos at E14.5, and culturing of slices for 3 div. (D), High magnification of wild type cortical plate (CP) showing GFP-labeled cells with one leading process (arrows).
(E,F), pCIG2 PAK1 AID was electroporated into wild type embryos at E14.5 and cortical slices were incubated for 3 div. (F), High magnification of wild type CP shows cells with misoriented and/or branching processes (arrows).
(G,H,I), GFP-expressing cells after control pCIG2 plasmid was electroporated into CHL1-minus embryos at E14.5, and cortical slices incubated for 3 div. (H), High magnification showing cells with branched or wavy leading processes (arrows). (I), Neurons in IZ and CP with inverted or misoriented leading processes (arrows).
(J–O) pCIG2 PAK1 AID was electroporated into CHL1-minus embryos at E14.5, and caudal cortical slices incubated for 3 div. (J), arrow shows wavy apical dendrite in the CP. (K), arrows show highly branched GFP-labeled cells. (L), Higher magnification of K shows details of highly branched cells in the upper IZ and CP (arrows). (L1), Radial glial processes (red arrow) were labeled with RC2 antibody (red) and GFP-expressing highly branched cells (green arrow) were labeled with GFP antibodies (green) in CHL1-minus cortex (E15.5). Colocalization was not evident in the merged image shown.
(M), High magnification of GFP-labeled cell expressing pCIG2 PAK1 AID with densely branched processes off a single leading process/apical dendrite.
(N), High magnification of GFP-labeled cell expressing pCIG2 PAK1 AID with multiple branched processes emerging from the soma.
(O,P), Dendritic spines (arrows) on GFP-labeled wild type and CHL1-minus neuronal processes expressing PAK1 AID.
(Q–U) pCIG2 or pCIG2-PAK1 AID plasmids were electroporated into wild type or CHL1-minus embryos at E14.5, and cortical slices incubated for 3 days in vitro (div), then immunostained for activated, phospho-PAK1 (p~PAK). Localization of phospho-PAK1 (red) in GFP-positive neurons (green) was evident as punctate labeling (yellow) in single confocal images. Control staining with no primary antibody in (U) for pCIG2-PAK1AID plasmid in CHL1-minus neurons.
VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate; MZ, marginal zone. Scale bars: (C,E,G,I,J,K) 50 µm; (D,F,H,L–P) 30 µm; (Q–U) 25 µm.
After 3 div, electroporation of either pCIG2 or pCIG2 PAK1 AID plasmids into wild type embryonic brains (E14.5) resulted in EGFP-positive cells in all layers of the developing neocortex with maximal levels in the VZ and IZ (Fig. 2 C, E). Neurons labeled at E14.5 include later migrating progenitors of pyramidal cells destined for layers II–IV (Anton et al., 1999). To evaluate an effect on neuronal positioning, the percent of EGFP-positive cells was quantified in each of 3 bins (VZ/SVZ, IZ, and CP/MZ) and analyzed for significant mean differences (Fig. 3 A–C). Results showed that expression of PAK1 AID in wild type neurons tended to decrease the percent of labeled neurons in the CP/MZ with a corresponding increase in the VZ/SVZ, but these differences were not statistically significant at 3 div (Fig. 3 A–C) or 4 div (not shown). In contrast, expression of PAK1 AID in CHL1-deficient neurons resulted in a statistically significant decrease in the percent of labeled neurons in the CP/MZ (Fig. 3 A–C) as evaluated by either the t-test or 1-way ANOVA. Expression of pCIG2-PAK1 AID did not further affect neuronal positioning in any of the cortical layers in either CHL1-minus or wild type caudal cortex compared to the respective genotypes expressing pCIG2 (Fig. 3 A–C).
Figure 3. Analysis of Neuronal Positioning in Wild Type and CHL1 Null Mutant Cortex and Influence of PAK1 AID.
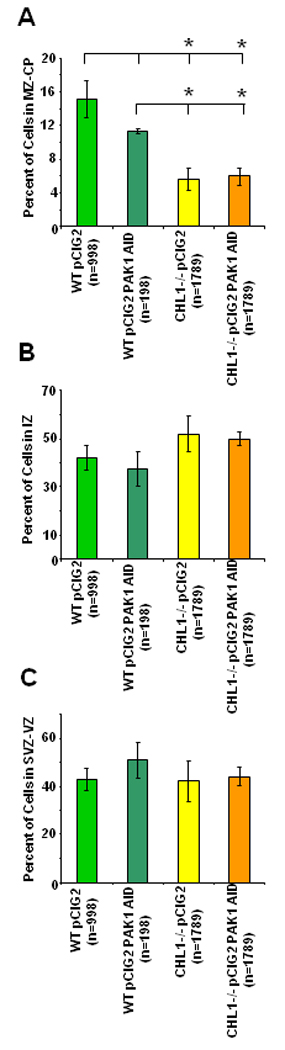
Wild type or CHL1-deficient cortex (E14.5) was electroporated with pCIG2 or pCIG2 PAK1 AID plasmids, and caudal cortical slices incubated for 3 div, as shown in Fig. 2. GFP-positive neurons (n, number of cells) were scored in multiple confocal images in bins corresponding to the VZ/SVZ, IZ, CP/MZ. The percent of cells in each bin was calculated (A, MZ-CP; B, IZ; C, SVZ-VZ), and group means ± SEM compared for significant differences in a pairwise manner by the t-test (p< 0.05) as indicated by asterisks.
Most wild type neurons in the upper IZ and CP expressing pCIG2 showed one unbranched leading process that was directed perpendicularly to the pial surface and often reached the MZ (Fig. 2 D, arrows). Expression of pCIG2-PAK1 AID in wild type neurons affected their morphology, causing some misorientation and branching of leading processes (Fig. 2 F, arrows). Cells were scored from multiple confocal images for misoriented leading processes or the presence of more than one branch from the leading process. Approximately 57% of wild type neurons expressing pCIG2-PAK1AID (n=47 total) showed process misorientation with respect to the pial surface or displayed multiple branches, compared to only 4.6% of wild type cells expressing the control pCIG2 (n=108 total).
Most CHL1-minus neurons expressing pCIG2 or pCIG2-PAK1 AID were located in the VZ and IZ, with some neurons present in the CP and MZ (Fig. 2 G,J). In agreement with our previous findings (Demyanenko et al., 2004), pCIG2-expressing CHL1-minus neurons (Fig. 2G) showed a somewhat lower laminar positioning compared to pCIG2-expressing wild type neurons (Fig. 2C). Quantification of pCIG2-expressing neurons showed a significantly decreased percent of CHL1-minus neurons in the MZ/CP compared to wild type (Fig. 3 A), but no change in the percent of cells in the IZ (Fig. 3 B) or SVZ/VZ (Fig. 3 C). CHL1-minus neurons expressing pCIG2 often displayed abnormal leading processes that were slightly branched (Fig. 2 H, arrows) or misoriented (Fig. 2 I, arrows), particularly in the lower CP. These features in electroporated slices were in accord with the morphology of pyramidal cells in layers V/VI in caudal cortical areas of unlabeled CHL1-minus mice in vivo (Demyanenko et al., 2004).
Expression of pCIG2-PAK1 AID in CHL1-minus neurons caused a striking change in morphology, typified by wavy (Fig. 2 J, arrow) or misoriented leading processes that were profusely branched particularly in the lower CP and IZ (Fig. 2 K,L, arrows). Higher magnification of these cells showed two classes of approximately equal proportion: one class had densely branched processes emanating from a single leading process (Fig. 2 M), while another class had densely branched processes that emerged from the cell soma at multiple points (Fig. 2N). These highly branched EGFP-labeled cells were of neuronal origin, as their processes bore dendritic spines characteristic of wild type neurons (Fig. 2 O, P). These EGFP-labeled cells were not radial glia or astrocytes, which do not have spines, and they did not co-express RC2, a marker localized to radial glial processes (Fig. 2, L1). Furthermore, other studies have demonstrated that 2–3 days after electroporation of E14.5 embryos into the lateral ventricles, GFP-labeled cells within the CP and IZ are neurons (Yokota et al., 2007).
An antibody recognizing the activated forms of PAK1-3 was used to verify that PAK1-AID expression inhibited PAK1-3 activity in wild type and CHL1-minus cortical slices (E14.5) after 3 days in culture. The phospho-PAK antibody used detects endogenous activated PAK1, PAK2, and PAK3 only when phosphorylated at Thr423, Thr402, and Thr421, respectively. In wild type (Fig. 2 Q) and CHL1-minus neurons (Fig. 2 S) expressing pCIG2, puncate staining of active, phospho-PAK1-3 was present in the cytoplasm of neuronal soma and processes, including the leading process, and appeared similar in both genotypes. This pattern of staining has also been observed in neurons in developing mouse neocortex (Causeret et al., 2009) and in primary cultured cortical neurons from E16.5 mice (Hayashi et al., 2002, Hayashi et al., 2007). The similar intensity of phospho-PAK staining in wild type and mutant neurons suggested that PAK might not be a direct downstream effector of CHL1 but a component of a parallel pathway. In pCIG2-PAK1AID–expressing wild type (Fig. 2 R) and CHL1-minus neurons (Fig. 2 T) staining for active PAK1-3 was much reduced, indicting inactivation of PAK1-3. Control staining without phospho-PAK antibodies was negative for all samples, as shown in CHL1-minus neurons expressing pCIG2-PAK1AID (Fig. 2 U).
To quantitatively compare the morphological changes, the number of nodes, terminals, and total length of processes of EGFP-labeled neurons in the upper IZ and CP were compared in cortical slices after 3 div (Fig. 4 A–C). Electroporation of PAK1 AID in wild type cortical neurons caused a small increase (4–6 fold, t-test; p<0.05) in the number of branch points (nodes), terminals (ends), and total length of neuronal processes per cell, compared to wild type embryos electroporated with pCIG2 (Fig. 4). CHL1 null mutant cells electroporated with pCIG2 also showed a small, significant increase in the number of nodes and terminals per neuron compared to wild type cells containing pCIG2 (2–3.6 fold), although total process length was not different. Electroporation of PAK1 AID into CHL1 null mutants caused large increases in the number of nodes and terminals of leading processes/apical dendrites compared to CHL1 mutants with pCIG2 alone (6–11 fold), as well as an increase in total process length. The increase in nodes, terminals, and process length of EGFP-labeled neurons in PAK1 AID-expressing CHL1 null mutants was also significantly greater than in wild type cells with or without PAK1 AID expression (Fig. 4). These results indicate that CHL1 and PAK1-3 cooperate in regulating morphological development of the leading process/apical dendrite of embryonic cortical neurons.
Figure 4. Analysis of Neuronal Morphology in Wild Type and CHL1 Null Mutant Cortex and Influence of PAK1 AID Expression.
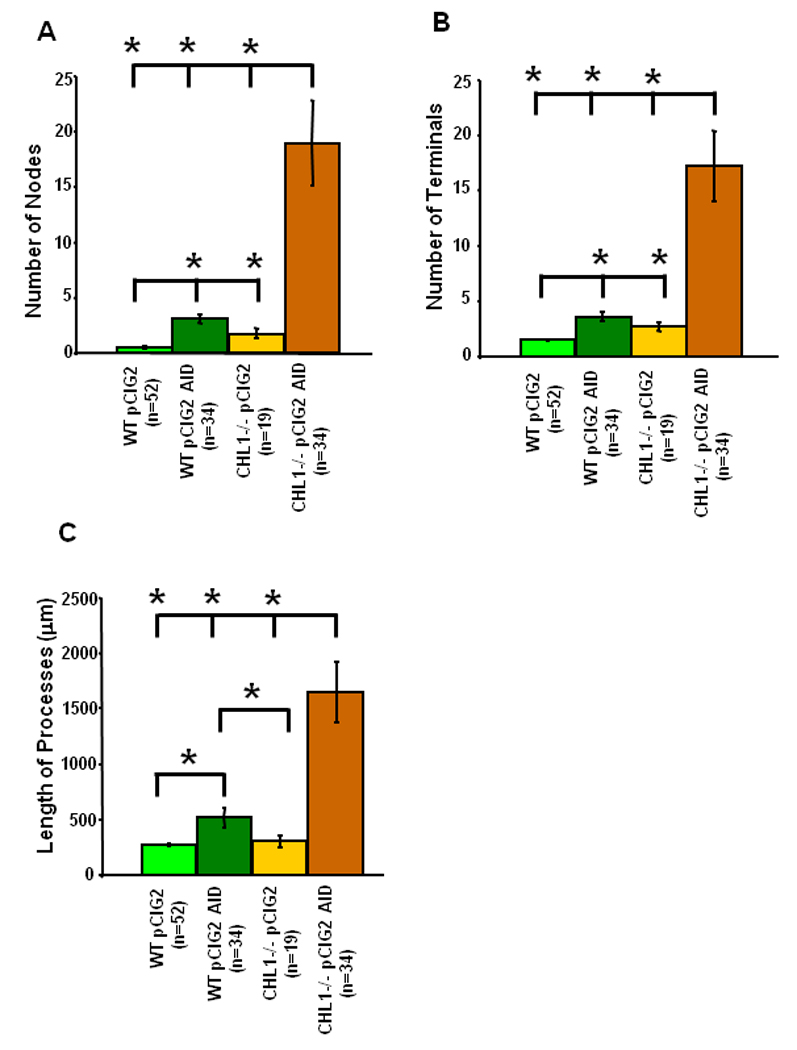
Wild type and CHL1 null mutant cortex were electroporated at E14.5 with control pCIG2 or pCIG2 PAK1 AID plasmids, and caudal cortical slices incubated for 3 div. The number of nodes (branch points) (A), terminals (ends) (B), and total length of processes (C) were measured for each neuron (n, number of neurons) from confocal images using Neurolucida software and means ± SEM calculated. Pairwise comparison of means was determined using the t-test with significant differences at p<0.05, as indicated by asterisks.
Discussion
Here, we identify a novel cooperation between CHL1 and PAK1-3 in regulating the morphology of neuronal precursors during stages of radial migration and early differentiation in mouse embryonic cortex. Group I PAK1-3 isoforms were prominently expressed in neuronal precursors in the IZ and CP of the developing mouse neocortex during stages corresponding to migration and differentiation. PAK1-3 colocalized with CHL1 in the soma and processes of embryonic cortical neurons in the caudal region of the neocortex, where CHL1 functions in neuronal positioning and dendritic projection (Demyanenko et al., 2004). Inactivation of PAK1-3 kinase in wild type embryos (E14.5) induced by expressing PAK1 AID caused small but significant increases in neuronal branching, as well as some misorientation of leading processes. In CHL1 null mutant mice many cortical neurons have misoriented apical dendrites and shortened leading processes, as well as neuronal positioning defects (Demyanenko et al., 2004). Here we show that loss of CHL1 coupled with inactivation of PAK1-3 by PAK1-AID induced profuse branching of the leading process/apical dendrite, as well as elaboration of processes from the soma to form multipolar-like cells in the IZ and CP. This phenotype was most likely cell autonomous in neurons, because radial glia and Cajal-Retzius cells are unaffected by loss of CHL1 (Demyanenko et al., 2004). PAK1-3 activation did not appear to be downstream of CHL1, as phosphorylated, activated PAK1-3 immunoreactivity appeared similar in wild type and CHL1-minus cortical neurons. Thus CHL1 and Group I PAKs may function independently to suppress process branching of neuronal precursors in the developing neocortex during migration and differentiation.
A role for Group I PAKs in regulating neuronal process elaboration is supported by studies in which premature expression of PAK3 in immature, migratory interneurons induces inappropriate outgrowth of neurites, causing cells to arrest in the basal ganglia (Cobos et al., 2007). Our studies are in general accord with another report in which downregulation of PAK1 by shRNA expression in wild type E14.5 mouse cortical neurons induced elaboration of protrusions from the soma, as well as disorganization of apical extensions (Causeret et al., 2009). However, these cells did not exhibit the densely branched morphology of neurons lacking CHL1. In contrast to our findings, knockdown of PAK1 decreased neuronal migration through the IZ and lower CP, when PAK1-specific shRNA was electroporated at E14.5 and allowed to proceed in utero until P0 (Causeret et al., 2009). Although we did not observe a statistically significant effect of PAK1 AID expression on cortical positioning of wild type neurons after electroporation at E14.5 after 3 div in organotypic slice cultures, there was a decreasing trend in the percent of cells in the MZ and CP and a corresponding increase in the SVZ and VZ. This trend might have become significant at longer incubation times, which was precluded by the limitations of our organotypic cultures. Another consideration is that PAK inhibition by PAK1 hnRNA and PAK1 AID could have different effects, since PAK1 AID inhibits kinase-dependent functions but not necessarily scaffolding functions. Scaffolding functions of PAK might contribute to migration, for example by recruitment of signaling intermediates to the plasma membrane (Sundberg-Smith et al., 2005, Higuchi et al., 2008).
Our results suggest that CHL1 and PAK1-3 function in independent, possibly compensatory pathways to suppress neuronal branching and regulate morphological development of the leading process/apical dendrite, since attenuation of both leads to a much greater change in dendritic morphology than attenuation of each one separately. In the developing neocortex the secreted protein Semaphorin3A (Sema3A) attracts apical dendrites/leading processes of cortical neurons to the pial surface (Polleux et al., 1998), and inhibits branching of cortical neuronal processes in vitro (Dent et al., 2004). Loss of Sema3A or its receptor Neuropilin-1 induces branching and misorientation of the leading process in developing mouse cortex, impairing radial migration (Chen et al., 2008). CHL1 is known to bind Neuropilin-1 and mediate Sema3A-induced neuronal responses such as growth cone collapse (Wright et al., 2007). Thus, CHL1 may be an upstream component of the Sema3A/Neuropilin-1 signaling pathway, which suppresses branching of the apical dendrite/leading process during migration. In a parallel pathway, PAK1-3 might also suppress branching by maintaining integrin-dependent adhesion of migrating neurons to radial glia, as it is a well-established integrin signaling intermediate in various cell types (Slack-Davis et al., 2003, Guo et al., 2007). In the developing neocortex, α3β1 integrin regulates attachment of the leading process to radial glia, facilitating radial migration of cortical neurons (Anton et al., 1999, Sanada et al., 2004). By functioning in independent pathways, CHL1 and PAK1-3 may coordinately suppress neuronal process elaboration, until the neuron attains its appropriate laminar position, detaches from radial glial guides, and elaborates its dendritic arbor.
Acknowledgments
This work was supported by NIH grants 5R01NS049109-04 and 5 P30 NS045892-06 to the UNC at Chapel Hill Neuroscience Center for support of the Confocal Microscopy Facility. We thank Dr. Eva Anton and Dr. Leslie V. Parise (UNC at Chapel Hill) for helpful discussion.
Abbreviations
- CHL1
close homolog of L1
- CP
cortical plate
- GFP
green fluorescent protein
- IZ
intermediate zone
- MZ
marginal zone
- PAK
p21-activated kinase (PAK1-3) family of serine/threonine kinases
- SVZ
subventricular zone
- VZ
ventricular zone
- wild type
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anton ES, Kreidberg JA, Rakic P. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron. 1999;22:277–289. doi: 10.1016/s0896-6273(00)81089-2. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Buhusi M, Midkiff BR, Gates AM, Richter M, Schachner M, Maness PF. Close homolog of L1 is an enhancer of integrin-mediated cell migration. J Biol Chem. 2003;278:25024–25031. doi: 10.1074/jbc.M303084200. [DOI] [PubMed] [Google Scholar]
- Causeret F, Terao M, Jacobs T, Nishimura YV, Yanagawa Y, Obata K, Hoshino M, Nikolic M. The p21-activated kinase is required for neuronal migration in the cerebral cortex. Cereb Cortex. 2009;19:861–875. doi: 10.1093/cercor/bhn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Sima J, Jin M, Wang KY, Xue XJ, Zheng W, Ding YQ, Yuan XB. Semaphorin-3A guides radial migration of cortical neurons during development. Nat Neurosci. 2008;11:36–44. doi: 10.1038/nn2018. [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Schachner M, Anton E, Schmid R, Feng G, Sanes J, Maness PF. Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron. 2004;44:423–437. doi: 10.1016/j.neuron.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Dent EW, Barnes AM, Tang F, Kalil K. Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J Neurosci. 2004;24:3002–3012. doi: 10.1523/JNEUROSCI.4963-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- Frost JA, Khokhlatchev A, Stippec S, White MA, Cobb MH. Differential effects of PAK1-activating mutations reveal activity- dependent and - independent effects on cytoskeletal regulation. J Biol Chem. 1998;273:28191–28198. doi: 10.1074/jbc.273.43.28191. [DOI] [PubMed] [Google Scholar]
- Guo D, Tan YC, Wang D, Madhusoodanan KS, Zheng Y, Maack T, Zhang JJ, Huang XY. A Rac-cGMP signaling pathway. Cell. 2007;128:341–355. doi: 10.1016/j.cell.2006.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, Schuurmans C, Guillemot F, Polleux F. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ohshima T, Hashimoto M, Mikoshiba K. Pak1 regulates dendritic branching and spine formation. Dev Neurobiol. 2007;67:655–669. doi: 10.1002/dneu.20363. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ohshima T, Mikoshiba K. Pak1 is involved in dendrite initiation as a downstream effector of Rac1 in cortical neurons. Mol Cell Neurosci. 2002;20:579–594. doi: 10.1006/mcne.2002.1144. [DOI] [PubMed] [Google Scholar]
- Hayashi ML, Choi SY, Rao BS, Jung HY, Lee HK, Zhang D, Chattarji S, Kirkwood A, Tonegawa S. Altered cortical synaptic morphology and impaired memory consolidation in forebrain- specific dominant-negative PAK transgenic mice. Neuron. 2004;42:773–787. doi: 10.1016/j.neuron.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Onishi K, Kikuchi C, Gotoh Y. Scaffolding function of PAK in the PDK1-Akt pathway. Nat Cell Biol. 2008;10:1356–1364. doi: 10.1038/ncb1795. [DOI] [PubMed] [Google Scholar]
- Jaffer ZM, Chernoff J. p21-activated kinases: three more join the Pak. Int J Biochem Cell Biol. 2002;34:713–717. doi: 10.1016/s1357-2725(01)00158-3. [DOI] [PubMed] [Google Scholar]
- Kreis P, Rousseau V, Thevenot E, Combeau G, Barnier JV. The four mammalian splice variants encoded by the p21-activated kinase 3 gene have different biological properties. J Neurochem. 2008;106:1184–1197. doi: 10.1111/j.1471-4159.2008.05474.x. [DOI] [PubMed] [Google Scholar]
- Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, Harrison SC. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- Montag-Sallaz M, Schachner M, Montag D. Misguided axonal projections, neural cell adhesion molecule 180 mRNA upregulation, and altered behavior in mice deficient for the close homolog of L1. Mol Cell Biol. 2002;22:7967–7981. doi: 10.1128/MCB.22.22.7967-7981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Alifragis P, Wong RO, Parnavelas JG. Ventricle-directed migration in the developing cerebral cortex. Nat Neurosci. 2002;5:218–224. doi: 10.1038/nn813. [DOI] [PubMed] [Google Scholar]
- Nikolic M. The Pak1 kinase: an important regulator of neuronal morphology and function in the developing forebrain. Mol Neurobiol. 2008;37:187–202. doi: 10.1007/s12035-008-8032-1. [DOI] [PubMed] [Google Scholar]
- Nikolic M, Chou MM, Lu W, Mayer BJ, Tsai LH. The p35/Cdk5 kinase is a neuron-specific Rac effector that inhibits Pak1 activity. Nature. 1998;395:194–198. doi: 10.1038/26034. [DOI] [PubMed] [Google Scholar]
- Nikonenko AG, Sun M, Lepsveridze E, Apostolova I, Petrova I, Irintchev A, Dityatev A, Schachner M. Enhanced perisomatic inhibition and impaired long-term potentiation in the CA1 region of juvenile CHL1-deficient mice. Eur J Neurosci. 2006;23:1839–1852. doi: 10.1111/j.1460-9568.2006.04710.x. [DOI] [PubMed] [Google Scholar]
- Polleux F, Ghosh A. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE. 2002:L9. doi: 10.1126/stke.2002.136.pl9. 2002. [DOI] [PubMed] [Google Scholar]
- Polleux F, Giger RJ, Ginty DD, Kolodkin AL, Ghosh A. Patterning of cortical efferent projections by semaphorin-neuropilin interactions. Science. 1998;282:1904–1906. doi: 10.1126/science.282.5395.1904. [DOI] [PubMed] [Google Scholar]
- Rashid T, Banerjee M, Nikolic M. Phosphorylation of Pak1 by the p35/Cdk5 kinase affects neuronal morphology. J Biol Chem. 2001;276:49043–49052. doi: 10.1074/jbc.M105599200. [DOI] [PubMed] [Google Scholar]
- Sakakibara A, Horwitz AF. Mechanism of polarized protrusion formation on neuronal precursors migrating in the developing chicken cerebellum. J Cell Sci. 2006;119:3583–3592. doi: 10.1242/jcs.03080. [DOI] [PubMed] [Google Scholar]
- Sanada K, Gupta A, Tsai LH. Disabled-1-regulated adhesion of migrating neurons to radial glial fiber contributes to neuronal positioning during early corticogenesis. Neuron. 2004;42:197–211. doi: 10.1016/s0896-6273(04)00222-3. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Maness PF. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr Opin Neurobiol. 2008 doi: 10.1016/j.conb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid RS, Midkiff BR, Kedar VP, Maness PF. Adhesion molecule L1 stimulates neuronal migration through Vav2-Pak1 signaling. Neuroreport. 2004;15:2791–2794. [PubMed] [Google Scholar]
- Sells MA, Knaus UG, Bagrodia S, Ambrose DM, Bokoch GM, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- Slack-Davis JK, Eblen ST, Zecevic M, Boerner SA, Tarcsafalvi A, Diaz HB, Marshall MS, Weber MJ, Parsons JT, Catling AD. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J Cell Biol. 2003;162:281–291. doi: 10.1083/jcb.200212141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg-Smith LJ, Doherty JT, Mack CP, Taylor JM. Adhesion stimulates direct PAK1/ERK2 association and leads to ERK-dependent PAK1 Thr212 phosphorylation. J Biol Chem. 2005;280:2055–2064. doi: 10.1074/jbc.M406013200. [DOI] [PubMed] [Google Scholar]
- Swartz M, Eberhart J, Mastick GS, Krull CE. Sparking new frontiers: using in vivo electroporation for genetic manipulations. Dev Biol. 2001;233:13–21. doi: 10.1006/dbio.2001.0181. [DOI] [PubMed] [Google Scholar]
- Wright AG, Demyanenko GP, Powell A, Schachner M, Enriquez-Barreto L, Tran TS, Polleux F, Maness PF. Close homolog of L1 and neuropilin 1 mediate guidance of thalamocortical axons at the ventral telencephalon. J Neurosci. 2007;27:13667–13679. doi: 10.1523/JNEUROSCI.2888-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Ring C, Cheung R, Pevny L, Anton ES. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron. 2007;54:429–445. doi: 10.1016/j.neuron.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke FT, King CC, Bohl BP, Bokoch GM. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem. 1999;274:32565–32573. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZS, Manser E, Chen XQ, Chong C, Leung T, Lim L. A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong JL, Banerjee MD, Nikolic M. Pak1 and its T212 phosphorylated form accumulate in neurones and epithelial cells of the developing rodent. Dev Dyn. 2003;228:121–127. doi: 10.1002/dvdy.10351. [DOI] [PubMed] [Google Scholar]