Abstract
Histone acetylation and methylation are linked to a variety of nuclear activities, most notably transcriptional regulation. Both synergistic and antagonistic relationships between these two modifications have been reported in different systems. Here we show that the budding yeast histone H4 arginine 3 (R3) methyltransferase Hmt1p binds acetylated histones H3 and H4, and importantly, that acetylated H4 is a significantly better methylation substrate for Hmt1p. Kinetic studies show that acetylation at any of the four acetylatable lysine residues of histone H4 results in more efficient methylation. Among the four, K8 acetylation imposes the strongest effect on reducing KM, consistent with the observed acetylation-stimulated interaction. In vivo, hmt1! cells rescue the transcriptional defect caused by GCN5 deletion, indicating that one of the functions of Gcn5p is to neutralize the negative effect of Hmt1p. Mutating either K8 or R3 to alanine causes similar growth defects in selective histone and gcn5 mutant background, suggesting that these two residues function in the same pathway for optimal vegetative growth. Together, these results reveal functional connection between histone acetylation, methylation, and two of the responsible enzymes, Gcn5p and Hmt1p.
Keywords: histone, acetylation, methylation, GCN5, HMT1, RMT1, Saccharomyces cerevisiae
1. INTRODUCTION
Acetylation is one of the best-characterized post-translational modifications of histones. A variety of nuclear activities including transcriptional regulation, chromatin assembly, DNA repair, recombination, and mitosis, have been linked to histone acetylation (see [1] and references therein). Mechanistically, histone acetylation may reduce the affinity of histone-DNA interactions[2], changes the chromatin structures [3], and recruits or evicts selective histone- binding proteins [4, 5]. Furthermore, methylation, phosphorylation, and ubiquitination of histones in many cases are functionally tied to histone acetylation [6]. Both synergism and antagonism have been observed between different histone modifications.
In the budding yeast Saccharomyces cerevisiae, Gcn5p is a prototypical histone acetyltransferase (HAT) [7]. Histones H3, H4, and H2B are acetylated by Gcn5p [8–10], however the substrate specificity (histones and the target lysine residues) may vary depending on the assay conditions [11]. Gcn5p is the catalytic subunit of several chromatographically distinct complexes including SAGA [12], SALSA/SLIK [13, 14], and ADA [15], that have been linked to transcriptional regulation in yeast (see [16] for review). Recent works also linked Gcn5p and its HAT activity to DNA damage repair [17] and chromosome segregation [18]. In addition to Gcn5p, there are multiple HATs and histone deacetylases (HDACs) that target at different or overlapping histones and lysine residues for a variety of nuclear activities [19]. How these enzymes and selective acetylated histones appropriately perform their biological functions is an important yet largely unanswered question.
A prevailing hypothesis explaining the functional diversity of histone acetylation is that selective proteins are recruited to genomic domains with certain acetylated histones. The notion of modification-dependent recruitment of specific factors has been well-documented in protein phosphorylation [20], and there are an increasing number of examples for other modifications, such as methylation and sumoylation [21]. Similarly, acetylated histones interact with a conserved protein module, the bromodomain [4, 22, 23], which is found in many transcriptional regulators [24]. Recruitment of bromodomain-containing proteins is important for setting the boundary between euchromatin and heterochromatin [25]. However, the wide spectrum of nuclear functions linked to histone acetylation appears to exceed the number of proteins possessing the bromodomain, suggesting that other protein modules may be capable of binding to the acetylated histones. Using the tethered catalysis/yeast two-hybrid system designed to identify protein-protein interactions stimulated by post-translational modifications [26], several acetylated histone binding proteins were found, including Hmt1p (a.k.a. Rmt1p) for protein and histone H4 methylation [27–29], Cac1 (a.k.a. Rlf2) for chromatin assembly [30], and Exo84p that has been implicated in mRNA splicing [31, 32]. These results are in excellent agreement with the observations that histone acetylation functions beyond transcriptional regulation. In the current work, we focused on Hmt1p.
Hmt1p is a type I protein arginine methyltransferase that catalyzes asymmetric dimethylation [33, 34]. Known substrates for Hmt1p include hnRNP components [28, 29], several nucleolar proteins [35], and histone H4 [27]. Arginine 3 (R3) of histone H4 is the methylation target of Hmt1p [27]. Importantly, the Hmt1p methyltransferase activity is critical for rDNA silencing mediated by a histone deacetylase Sir2p [36]. Loss of Hmt1p methyltransferase activity results in hypomethylation of H4 at silent loci, and in impairment of Sir2p recruitment [36]. The transcriptional repression function of Hmt1p differs significantly from its mammalian homologue, PRMT1, in that the latter is important for transcriptional activation of multiple genes [37–39]. PRMT1-mediated histone H4 R3 methylation acts upstream of the p300 histone acetyltransferase in transcriptional activation [37]. How the yeast Hmt1p performs its transcriptional regulatory function remains an open question.
In this work, we present evidence that Hmt1p is an acetylated histone-binding protein, and that acetylated H4 is a better substrate for Hmt1p methyltransferase activity. Transcriptional defects caused by the gcn5 null allele can be suppressed by deleting HMT1. Furthermore, histone H4 K8 acetylation and R3 methylation appear to be part of a pathway that becomes essential for optimal mitotic growth in a background devoid of Gcn5p HAT activity and the predominant histone H3 acetylation site. These data reveal novel interactions between histone acetylation and methylation.
2. MATERIALS AND METHODS
2.1. Yeast strains and plasmids
All genetic methods were performed according to standard procedures [40]. Yeast transformation was done using the lithium acetate method [41]. The GST-Hmt1 plasmid was generously provided by S. Clarke (UCLA) [34]. The dot1! and hmt1! strains were a gift from J. Côté (Lava University).
To create yeast strains for RNA analyses and 3-AT tolerance tests, BY4742 (MATα his3! 1 leu2! 0 lys2! 0 ura3! 0), yMK1180 (isogenic to BY4742 except MATa dot1!), and yMK118 (isogenic to BY4742 except MATa hmt1!) were first transformed with the 1.75 kb Bam HI/Bam HI fragment from pJJ217 [42] that contained the entire HIS3 gene, resulting in yMK1185, yMK1186, and yMK1187, respectively. This procedure was to convert these strains to HIS+ so that the transcriptional status of HIS3 and cellular sensitivity to 3-AT could be tested. GCN5 was subsequently deleted from these three strains by using a gcn5! :: hisG-URA3-hisG fragment derived from pMK147 (following Xho I and Xba I digestion and gel-purification of the 4.6 kb fragment) to create yMK1188 (gcn5!), yMK1189 (dot1! gcn5!), and yMK1190 (hmt1! gcn!5!).
For histone mutants, pQQ18 (ARS1 CEN4 LEU2 HTA1-HTB1-HHT2-HHF2) [43] was subjected to mutagenesis (see [44] for detailed procedures; oligonucleotide sequences are available upon request) to replace selective lysine or arginine residues (Figure 4). JHY205 [43] (MATa his3! 1 leu20 ura3 ! 0 hta1-htb1! ::HPH hta2-htb2! ::NAT hht1-hhf1! ::KAN hhf2-hht2::NAT <pJH33 ARS1 CEN4 URA3 HTA1-HTB1-HHT2-HHF2>) was transformed with Ngo MIV-digested pMK284 F221A to replace the chromosomal copy of GCN5 with the F221A allele [44], resulting in yDA12. Desired histone mutant plasmids (pQQ18 derivatives) were transformed to either GCN5+ or gcn5 F221A strains. Leu+ Ura+ strains were grown overnight in YPD to saturation before plating to 5-FOA medium to assess viability.
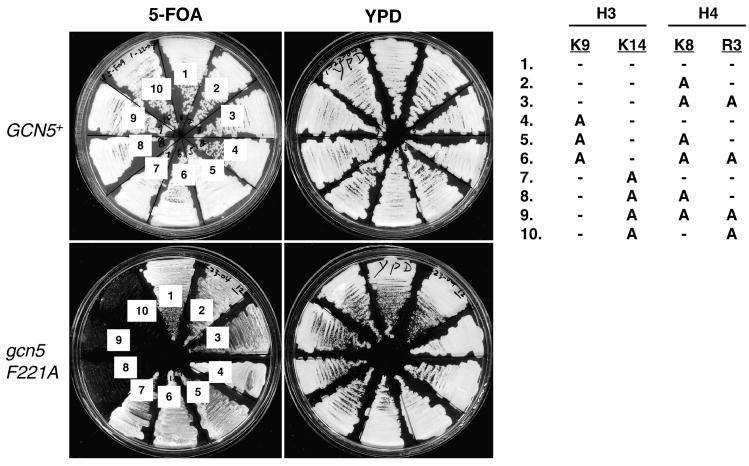
Figure 4. Genetic interactions between histone acetylation, methylation, and Gcn5p.
Histones H2A, H2B, H3, and H4 quadruple knockout strains bearing wildtype histone genes on a URA3 plasmid were transformed with a LEU2 plasmid containing wildtype or one of the indicated histone mutants. The transformants were streaked to 5-FOA plates to assess the effect of selective histone acetylation or methylation defective mutations on cellular growth. These histone mutants were tested in GCN5+ and gcn5− background. Numbers 1–10 were assigned to each GCN5+ and gcn5 F221A strains. The corresponding alanine substitution of selective lysine or arginine residues are listed on the right. “–” signs indicate un-changed residues.
2.2. Protein expression, purification, and biochemical assays
Induction and purification of the recombinant Hmt1p were according to Gary et al [34]. Bacterially expressed yeast histones H3 and H4 were a kind gift of K. Luger (Colorado State University, Fort Collins). Synthetic histone H4 peptides were purchased from the Upstate Biotechnology Inc. 3H-S-adensyl-methionine (SAM) was purchased through Amersham (15 Ci/mmol). Purification of core histones from yeast was based on Edmondson et al [5]. Methods for recombinant Gcn5p production and in vitro histone acetylation were as previously described [8] except that 1 μg of recombinant histone H3 or H4, or approximately 10 μg of yeast core histones were first treated with 50 ng of His-tagged Gcn5p in 20 μl reactions containing 50 mM Tris-HCl, pH 8.0, 10% glycerol (v/v), 1 mM EDTA and 1 mM unlabelled acetyl coenzyme A. For mock acetylation reaction, 1 mM of coenzyme A was substituted for acetyl coA. The acetylation reactions were conducted at 30°C for 30 minutes, immediately followed by GST pulldown or in vitro methylation reactions.
For GST pulldown assays, approximately 1 μg of GST-Hmt1p was added to acetylation or mock-acetylation reactions that had been brought to 200 μl with the acetylation buffer without the cofactor. The reactions were gently rocked at 4°C for overnight, followed by addition of 5 μl of reduced glutathione beads (1:1 slurry). The binding reaction was continued at 4°C for an additional hour. The matrix was pelleted (14,000 rpm for 15 seconds at room temperature), and washed twice with 500 μl of acetylation buffer. 20 μl of 1X SDS-PAGE loading dye was added to the beads, which were then boiled for 5 minutes. The supernatant was loaded to 15% SDS-PAGE for resolution and Coomassie Blue staining.
A typical methylation reaction (20 μl) (for non-kinetic studies) contained approximately 3 μM histone or H4 peptide substrates, 3.3 μM of 3H-SAM, and about 50 ng of recombinant GST- Hmt1p in 50 mM Tris-HCl, pH 8.0, 10% glycerol (v/v), and 0.1 mM EDTA. Reactions were carried out at 30°C for 30 minutes before P-81 filter assays to assess the incorporation (see below). Alternatively, 15% SDS-PAGE was used to resolve histones for fluorography. Acetylation and methylation reactions were done in the same buffer. Thus, methylation of Gcn5p-acetylated histones was conducted by directly adding radioactive SAM and GST-Hmt1p to the (mock) acetylation reactions after the 30-minute acetylation reaction had been completed. Methylation was extended for 30 minutes before SDS-PAGE loading dye was added to stop the reaction.
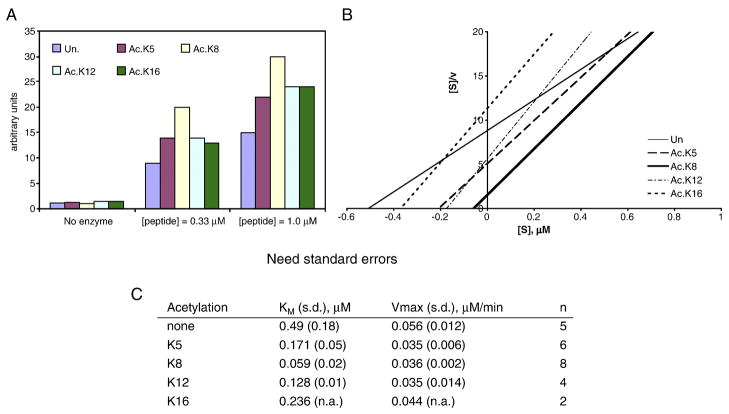
Kinetic studies of H4 peptide methylation was conducted in the following way. Each 10-μl reaction contained 50 mM Tris-HCl, pH 8.0, 10% glycerol, 0.1 mM EDTA, app. 25 ng of GST- Hmt1p, varying concentrations of peptide (ranging from 0.01 to 10 μM), and 3.3 μM 3H-SAM. All ingredients except 3H-SAM were mixed and made into 9-μl aliquots. The mixture was capped and pre-warmed at 30°C for 30 seconds. 1 μl of 33 μM 3H-SAM was then added to each tube to start the reaction. Reactions were carried out for exactly 3 minutes. Four 2-μl aliquots from each reaction were then spotted to Whatman P-81 paper discs [45]. After all sample discs were air- dried, they were washed for 10 minutes in generous amount of 20 mM NaHCO3, pH9.2 under gentle shaking. The wash was repeated twice, followed by brief 95% ethanol wash and air dry. Each disc was then submerged in 2 ml of scintillation fluid and shook gently overnight at room temperature before scintillation counting. The initial rate of methylation from each concentration of the peptide was calculated, and KM and Vmax were derived from the Hanes/Wolffe [S]/v vs. [S] plot {[S]/v = (1/Vmax)·[S] + (KM/Vmax)}. Average numbers of KM and Vmax are presented in Figure 2C.
Figure 2. Lysine 8 acetylation stimulates H4 methylation.
Unacetylated and monoacetylated histone H4 peptides (residues 1–18) were used for in vitro methylation. A. Synthetic H4 peptides were effective substrates for methylation. Unacetylated and monoacetylated peptides with acetyllysine at positions 5, 8, 12, or 16 were methylated by Hmt1p for 30 min. Relative efficiency of 3H-methyl incorporation was quantified. Un: unacetylated peptide. Shown are representative of two independent experiments. B and C. Kinetic studies of Hmt1p mediated methylation of synthetic H4 peptides. A representative Hanes/Wolffe plot ([S]/v vs. [S]) from single experiments is shown in panel B. Plots were best fit with r2 between 0.970 and 0.999. Panel C summarizes results from multiple experiments.
2.3. RNA preparation and RT-PCR
Yeast cells were grown in appropriate selective media till culture density reached 0.5 OD600/ml. Cells were then collected by centrifugation (5,000 × g, 5 min, 4°C) and transferred to synthetic minimal medium supplemented with required nutrients and 40 mM 3-AT for HIS3 induction. Cell cultures were further incubated at 37°C for 2 to 3 h before harvesting for RNA preparation. Procedures for RNA preparation were described previously [44]. 10 μg of total RNA was treated with 10 units of DNaseI (Roche) in 100 μl (50mM Tris-HCl, pH7.5, 5mM MgCl2), and incubated at 37°C for 1 hour. cDNA was synthesized following the instruction of ImpronII reverse transcriptase kit (GE Life Sciences) using 30 ng of poly (dT) primer. Semi-quantitative PCR reactions were conducted in 50 mM KCl, 10 mM Tris-HCl (pH 9.0, 25°C), 1% Triton X-100, 2 mM MgCl2, 0.1 mM each dNTP, 0.5 μM each primer, and 1.25 U Taq DNA polymerase (Promega), and appropriately diluted DNA templates. PCR parameters were (94°C, 4 min; 50°C, 4 min; 72°C, 30 sec) for 2 cycles; (94°C, 45 sec; 50°C, 45 sec; 72°C, 30 sec) for 24 cycles; and 72°C, 3 min. PCR products were resolved in polyacrylamide gels followed by ethidium bromide staining.
3. RESULTS
3.1. Hmt1p binds acetylated histones H3 and H4 preferentially
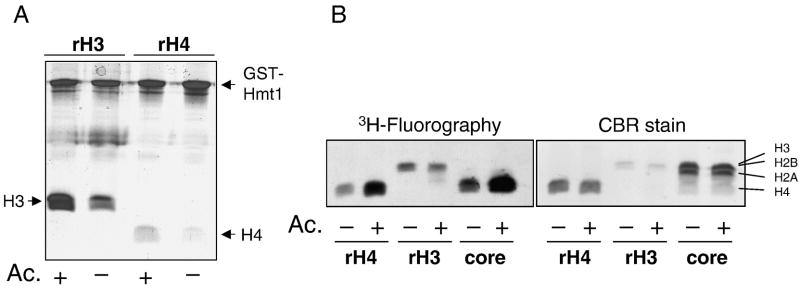
Hmt1p was found to be an acetylated histones H3 and H4 binding protein in two screens with the tethered catalysis/yeast two-hybrid system. To elaborate on this acetylation-dependent interaction, we conducted GST pulldown assays. Hmt1p was expressed in E. coli as a glutathione S transferase (GST) fusion protein. For histone ligand, bacterially expressed recombinant H3 and H4 (rH3 and rH4) were first subjected to acetylation reaction by Gcn5p in vitro, or to a mock acetylation reaction in which acetyl coenzyme A was replaced with coenzyme A. Recombinant Gcn5p acetylates yeast H3 at K14 and H4 at K8 [8]. Acetylated and mock acetylated histones were then incubated with GST-Hmt1p, and immobilized by glutathione beads. GST-Hmt1p and the trapped histones H3 and H4 were resolved by SDS-PAGE and quantified by Coomassie Blue staining. In agreement with the tethered catalysis/yeast two- hybrid results, pretreatment of H3 and H4 with Gcn5p and acetyl coA resulted in enhanced retention by Hmt1p (Figure 1A, compare lanes 1, 3, with 2, 4), whereas much weaker binding was seen in the mock acetylated histones. We thus conclude that Hmt1p is a bona fide acetylated histone H3- and H4-binding protein.
Figure 1. Hmt1p is an acetylated histone binding protein.
A. GST pulldown assays. Bacterially expressed recombinant histones H3 and H4 (rH3 and rH4) were treated with Gcn5p in the presence of acetyl coA or coA, followed by incubation with GST-Hmt1p and then glutathione beads. Proteins bound to the beads were resolved by SDS-PAGE and stained by Coomassie Blue (CBR). B. Methylation of acetylated and mock acetylated histones by Hmt1p. Recombinant H3 and H4, as well as core histones purified from yeast were acetylated by Gcn5p. Hmt1p catalyzed methylation was then conducted using radiolabelled S-adenosyl methionine as the methyl donor. Final methylation reactions were resolved and subjected to Coomassie Blue staining (right panel) and fluorography (left panel).
3.2. Acetylation enhances Hmt1p-mediated H4 methylation
Côté and colleagues showed that recombinant Hmt1p methylates R3 of histone H4, whereas no appreciable H3 methylation by Hmt1p was detected [27]. As acetylation effectively facilitates the interaction between the underlying histone H3 or H4 with Hmt1p, it is possible that the methylation efficiency of histone H3 or H4 would be enhanced by acetylation as well. To see if this was the case, we subjected the acetylated and mock acetylated H3 and H4 to Hmt1p methylation reactions. In addition, purified yeast core histone mixture were also pre-treated with Gcn5p with or without acetyl coA prior to incubation with Hmt1p and 3H-S-adenosyl methionine (SAM). The methylation status of histones was monitored by following the incorporation of the radioactive methyl group to histones. Results shown in Figure 1B clearly demonstrate that acetylated histone H4 was a significantly better methylation substrate for Hmt1p (compare lanes 1, 2). However, histone H3 was only radiolabelled weakly in an acetylation-independent manner (lanes 3 and 4), consistent with the previous finding that Hmt1p only methylates H4 among the four core histones [27]. Interestingly, core histones purified from yeast also showed acetylation- stimulated methylation by Hmt1p (lanes 5, 6). Under this condition, H3 methylation remained negligible. Neither H2A nor H2B was methylated. It is important to note that histone H4 purified from yeast, even without prior treatment with Gcn5p, was a better substrate when compared with its recombinant counterpart (lanes 1 and 5). This result was not surprising because yeast histones are known to be hyperacetylated in vivo [46]. Thus, compared with the bacterially expressed, acetylation-free recombinant H4, the yeast histone H4 justifiably exhibited a significantly better methylation efficiency even without pretreatment with Gcn5p and acetyl coA.
There are four acetylatable lysine residues in the amino terminal domain of H4: K5, K8, K12, and K16. Gcn5p acetylates yeast H4 at K8 [8], while other enzymes such as p300/CBP acetylate all four residues with roughly equivalent efficiency. To see which of these four acetylatable lysine residues played a more potent role in facilitating the binding and methylation by Hmt1p, we used synthetic peptides corresponding to the first 18 amino acids of H4 for in vitro methylation. One of these peptides was unmodified (Un), whereas each of the other four peptides contained one acetyllysine residue (i.e. Ac.K5, 8, 12, or 16) and were otherwise identical to others. These peptides would help delineate the contribution made by these acetyllysine residues in stimulating the action of Hmt1p.
Using 3H-SAM as the methyl donor, we first compared the efficiency of H4 peptide radiolabeling when both the enzyme and H4 peptides were fixed at constant concentrations. Figure 2A shows that, similar to using the intact H4 histone protein, all five H4 synthetic peptides were positively methylated by Hmt1p. However, the monoacetylated peptides exhibited 50–100% increase of the methylation efficiency. Intriguingly, the Ac.K8 peptide appeared to elicit the strongest stimulation, suggesting that modification at this residue may create the most effective substrate for Hmt1p. To more rigorously and quantitatively examine how acetylation enhanced methylation, we used kinetic studies to obtain key parameters for Hmt1p action on different H4 species, including the affinity (as reflected by the Michaelis Constant, KM), and maximal reaction rate (Vmax). Figure 2B depicts a representative Hanes/Wolffe [S]/v vs. [S] plot (where [S] is the substrate peptide concentration, and v is the initial reaction rate). Figure 2C summarizes the results from multiple experiments (n ranging from 2 to 8). Consistent with Figure 2A, the Ac.K8 peptide showed the lowest KM value (0.059 ± 0.02 μM), suggesting that this peptide had the highest affinity for the enzyme. The other three monoacetylated peptides also showed moderate reduction in KM compared with the unmodified control, indicating that acetylation at any of the four lysine residues can effectively enhance the binding of Hmt1p. These results were in excellent agreement with our tethered catalysis/yeast two-hybrid test results in that mutating single lysine residues failed to quantitatively diminish the two-hybrid interaction with the Hmt1p prey, yet mutating the tethered HAT, Gcn5p, eliminated the interactions completely (DG and MHK, data not shown). It is also interesting that the maximal velocity (Vmax) of all four acetylated peptides was somewhat lower than the unmodified counterpart. However, the mild reduction in the reaction rate was effectively compensated for by the increase in affinity. Thus, the overall efficiency of methylation turned out to be positively influenced by acetylation. Together, these kinetic studies verified the biochemical and yeast two-hybrid test results, and suggested that K8 acetylation had the strongest effect in facilitating the binding and methylation of H4.
3.3. Antagonism between Hmt1p and Gcn5p
The acetylation-stimulated binding and methylation of H4 by Hmt1p suggested functional connection between these two histone modifications. Since there are more than one HAT in yeast, to understand the in vivo connection between these two modification, we began our analyses by first examining the genetic relationship between Gcn5p and Hmt1p. To this end, we assessed transcriptional regulation of a model gene, and investigated overall cellular growth of strains bearing selective mutations in histones and/or Gcn5p.
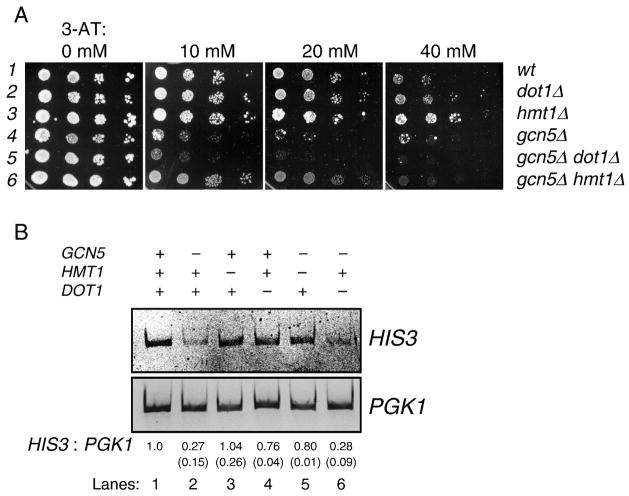
One of the best characterized target genes of Gcn5p is HIS3. Eliminating the Gcn5p HAT activity results in defects of HIS3 activation [47], which in turn causes cellular hypersensitivity to 3-amino-1,2,4-triazole (3-AT). If Hmt1p and Gcn5p interacted functionally, antagonistic or synergistic phenotypes may result from mutations that target both enzymes. To test this possibility, GCN5 and HMT1 were deleted. In addition, DOT1, which encodes a histone H3 K79 methyltransferase [48], was also knocked out as a comparison. Yeast cells bearing single or double knockouts of these genes were grown for HIS3 expression tests, including semi-quantitative reverse transcription PCR (RT-PCR) and cellular growth in medium containing varying concentrations of 3-AT.
Figure 3A shows that deleting GCN5 rendered cells hypersensitive to 3-AT (row 2), whereas deleting the histone H3 K79 methyltransferase DOT1 gene did not have an obvious effect. Interestingly, deleting HMT1 appeared to enable yeast cells to sustain higher concentrations of 3-AT (compare rows and 3 and 1 on 20- and 40-mM 3-AT plates), suggesting that Hmt1p plays a negative role in the cellular response to 3-AT toxicity. The genetic interaction between Gcn5p and Hmt1p was further revealed by the observation that the gcn5! hmt1! double knockout cells exhibited significantly higher resistance to 3-AT than did the gcn5! strain (compare rows 6 and 4). In contrast, deleting DOT1 in the gcn5! background did not alter the degree of 3-AT hypersensitivity. These results suggest functional differentiation of these two histone methyltransferases, and more importantly, that Hmt1p and Gcn5p might play opposing roles in containing the 3-AT toxicity.
Figure 3. Gcn5p antagonizes Hmt1p for HIS3 activation.
A. Cellular growth in the presence of varying concentrations of 3-AT. The concentrations of 3-AT in synthetic complete medium minus histidine (SC –His) are listed above each panel, and the relevant genotypes are on the right. Log-phase cells in rich medium were serially diluted and spotted to the 3-AT plates. Cells were grown at 30°C for three to five days. B. HIS3 transcriptional defect caused by deleting GCN5 can be rescued by HMT1 knockout. Semi-quantitative reverse transcription-PCR (RT- PCR) was used to compare the relative amount of HIS3 transcripts, using PGK1 as the control. Log-phase yeast cells were transferred to minimal medium containing 3-AT to induce HIS3 transcription. The ratio of HIS3/PGK1 of each sample was quantified and normalized to the wildtype (lane 1) strain. Quantification data were from two independent experiments and the errors are shown in parentheses under the mean.
Alteration in cellular resistance to 3-AT may result from changes in HIS3 expression or other mechanisms such as modulation of 3-AT flux (see Discussion for details). To link the restoration of 3-AT resistance in the gcn5! hmt1! cells to transcriptional regulation, semi- quantitative RT-PCR was conducted to compare the transcription status of HIS3 and PGK1, a GCN5-independent control gene that encodes the housekeeping 3-phosphoglycerol kinase. Log-phase yeast cells were starved for amino acids to activate amino acid biosynthesis genes, including HIS3. Total RNA was then quantified by RT-PCR. Figure 3B shows that gcn5! cells were defective in activating HIS3, and that neither hmt1! nor dot1! cells exhibited significant changes in HIS3 expression (109% and 76% of the wildtype level). The gcn5! dot1! strain exhibited a similar degree of reduction as that of gcn5! (compare lanes 2, 6), indicating again that Dot1p was not an important factor for HIS3 activation. In contrast, the HIS3 transcriptional defect caused by GCN5 deletion (down to 27% of the wildtype level) was effectively restored by knocking out the HMT1 gene (80%; lane 5). The restoration of HIS3 transcriptional activity by HMT1 deletion strongly suggests that one of the functions of Gcn5p is to antagonize a negative role of Hmt1p. When Gcn5p is absent, the transcriptional repression function of Hmt1p prevails, hence diminishing the expression of HIS3.
Given the biochemical and genetic connection between Gcn5p and Hmt1p, it is tempting to speculate that these two proteins may reside in the same loci. We have used an HA-tagged Hmt1p for chromatin immunoprecipitation assays but could not conclusively identify stabile association of Hmt1p with either the HIS3 gene or the HMLα silent locus (supplemental Figure 1) even though Hmt1p is predicted (but has not been substantiated) to be detectable at the silent loci [36]. Other in vivo approaches might be needed to examine this hypothesis.
3.4. Histone H4 K8 and R3 are in the same genetic pathway
Gcn5p is one of several histone acetyltransferases in yeast and controls about 10% of yeast genes [49, 50]. In vivo studies suggest that H3 is the major acetylation target for Gcn5p and that it has relatively minor contribution for H4 acetylation. Rather, the major histone H4 HAT activity is brought about by the NuA4 complex in which Esa1p is the catalytic subunit [12, 51, 52]. Although Figure 3 demonstrates that Gcn5p and Hmt1p are functionally linked to each other in controlling HIS3 activation and cellular growth in response to 3-AT, it is possible that histone acetylation, a result of balancing actions of multiple HATs and deacetylases, and H4 R3 methylation together control more cellular/nuclear activities. To further examine the possible connection between histone acetylation and H4 R3 methylation, we asked whether normal cell growth depended on the capacity of cells to maintain the full spectrum of H3/H4 acetylation, and H4 R3 methylation. To this end, we used a yeast strain in which the chromosomal histone genes were deleted. Cell viability was supported by a plasmid bearing all four core histone genes. Two versions of the histone plasmid were available, one with the URA3 marker and wildtype histone genes, and the other with the LEU2 marker and histone genes bearing selective mutations (see below). Yeast cells that had lost the URA3 plasmid but maintained the LEU2 plasmid were selected by the drug 5-fluoroorotic acid (5-FOA) that was converted to a lethal toxin by Ura3p [53]. Thus, the LEU2 plasmid provided the sole source of histones in cells resistant to 5-FOA, and the cellular growth under influence of these mutant histones could then be assessed. In addition, we also replaced GCN5 with the F221A catalytically inactive allele [47] to examine the essentiality of the HAT activity of Gcn5p when certain histone mutations were combined.
GCN5+ and gcn5 F221A strains bearing the URA3 plasmid and wildtype histone genes were transformed with the LEU2 construct bearing alanine mutations on the acetylatable residues K8 (H4), K9 (H3), and K14 (H3), as well as the methylatable R3 of H4. The transformant cells were grown in the YPD rich medium to allow for spontaneous plasmid loss, followed by streaking to either YPD or 5-FOA plates. Figure 4 shows that eliminating the modifications at H3 K9, K14, H4 K8, or R3 did not cause appreciable impairment of cellular growth in the presence of wildtype GCN5. In most cases, replacing the functional Gcn5p with the F221A mutant did not generate obvious phenotypes either, consistent with the reports that Gcn5p and the SAGA complex control about 10% of yeast genes that are mostly related to stress responses [49, 50]. However, gcn5 F221A H3 K14A cells could not efficiently form colonies on the 5-FOA plate if either H4 K8 or R3 was substituted by alanine (streaks 8-10, bottom panel). This result indicated that the Gcn5p HAT activity was critical for optimal cellular growth when selective histone acetylation or methylation sites were impaired. Mutating K9 of H3, which is the acetylation target for a functionally distinct HAT, Hat1p, had no effect in any combinations tested thus far. The identical phenotype caused by histone H4 R3A (i.e. lack of methylation) and K8A (i.e. lack of acetylation) mutations strongly suggested that these two modifications functioned in the same pathway, a notion consonant with the biochemical and yeast two-hybrid interaction data (Figures 1 and 2) that histone R3 methylation is closely linked to H4 acetylation, in particular at K8.
4. DISCUSSION
In this work, genetic and functional relationship between Gcn5p, Hmt1p, histone acetylation and methylation was examined. H4 acetylation enhances the association and methylation by Hmt1p in vitro, suggesting that the functions of histone methylation and acetylation are connected in certain cellular functions. Indeed, alanine substitution of either K8 or R3 results in identical cellular growth defects when Gcn5p and one of its most prominent substrates, K14 of H3, are mutated (Figure 4). It is rather interesting that the observed defective growth requires that both Gcn5p and H3 K14 are mutated. These results are reminiscent of a report by Dent and colleagues [54] in that combining multiple histone K-to-A or K-to-Q mutations with GCN5 deletion causes severe growth defects, indicating that Gcn5p functions beyond acetylating the known lysine residues of histones H3. K14 of histone H3 appears to be a common substrate for many histone acetyltransferases [51]. Changing this lysine to alanine can potentially hinder the in vivo functions of multiple HATs. In other words, in a background where the predominant H3 acetylation is blocked and the enzymatic activity of Gcn5p is further ablated, histone H4 K8 and R3 assume critical mitotic growth functions. These results point to the functional redundancy among chromatin and histone modifying enzymes.
That Hmt1p binds both acetylated H3 and H4 but only methylates the latter suggests an intriguing possibility that acetylated H3 may facilitate the recruitment of Hmt1p to methylate R3 of H4. We envision that the acetylated H3 and H4 ligands for Hmt1p may reside within the same nucleosomes, or in different nucleosomes but at the same genetic loci. Alternatively, since the predominant H4 acetyltransferase is Esa1p in the NuA4 complex, it is also possible that in loci where H4 is not the best binding and methylation substrate for Hmt1p, the Gcn5p-acetylated H3 plays an important role in recruiting Hmt1p for certain molecular functions.
Deleting HMT1 restores the transcriptional defects of HIS3 in gcn5! cells, suggesting that Gcn5p counteracts an inhibitory role of Hmt1p. Interestingly, HIS3 transcription appears to be normal or slightly reduced in GCN5+ hmt1! cells (Figure 3B). However, these cells exhibit higher resistance to 3-AT (Figure 3A), a competitive inhibitor of the HIS3 gene product, Imidazoleglycerol-phosphate dehydratase, that catalyzes the sixth step of histidine biosynthesis [55]. The disconnection between the steady-state HIS3 transcript level and cellular resistance to 3-AT seen in hmt1! cells can be due to several possibilities. Firstly, since the Imidazoleglycerol- phosphate dehydratase (i.e. His3p) is competitively inhibited by 3-AT, conditions that increase the substrate (D-erythro-imidazole-glycerol-phosphate) abundance can effectively dampen the toxicity of 3-AT. That is, if Hmt1p also inhibits the expression/activity of any of the enzymes upstream of His3p (i.e. His1p, His4p, His6p, and His7p), deleting HMT1 likely will raise 3-AT resistance without triggering significant HIS3 upregulation. Secondly, Hmt1p methylates multiple proteins, including several that affect mRNA metabolism [28, 56] and nucleolar functions [35]. Thus, translation of histidine biosynthesis enzymes may be controlled by Hmt1p, resulting in changes in cellular tolerance to 3-AT (at a post-transcription level) when HMT1 is deleted. Lastly, 3-AT is transported by Atr1p [57]. If Hmt1p affects the function or expression of ATR1, it is conceivable that yeast cells will exhibit different tolerance to 3-AT as well.
With respect to transcriptional regulation, the potential antagonism between Hmt1p and Gcn5p in HIS3 transcription suggests a previously unknown relationship between two different histone modifications. If acetylation plays a critical role in recruiting Hmt1p, and that Gcn5p is responsible for histone H3 hyperacetylation in vivo, how can gcn5! hmt1! cells express HIS3 efficiently when the Gcn5p co-activator has already been deleted? We envision two possible scenarios. Firstly, histone H4 is acetylated in a Gcn5p-independent manner at HIS3 and many other loci [58]. Hmt1p may thus be recruited to the HIS3 locus via direct interactions with acetylated H4, and possibly deposits the R3 methylation mark. Normally, neutralizing the negative effect of Hmt1p is part of the transcriptional activation process. Deleting HMT1 in a GCN5+ background thus does not necessarily increase HIS3 transcription. However, in the absence of Gcn5p, the inhibitory action of Hmt1p becomes so prominent that it contributes significantly to the transcriptional defects associated with gcn5− cells. Deleting HMT1 can thus allow cells to activate transcription even when Gcn5p is absent. Secondly, it has been shown that the methyltransferase activity of Hmt1p is required for recruiting a histone deacetylase to silent loci for transcriptional silencing [36]. We thus suggest that Hmt1p recruits a histone deacetylase to the promoter region by interacting with histones acetylated by Gcn5p, NuA4, and/or other HATs. Recruitment of histone deacetylases results in turnover of acetyllysine in each round of transcriptional initiation, allowing for fast control of transcription. This hypothetic situation is similar to the cyclic H2B ubiquitylation/deubiquitylation that is important for full-fledge transcription [59, 60]. When GCN5 is deleted, the balance between histone acetylation and deacetylation is skewed, thus reducing HIS3 transcription. By deleting HMT1 in gcn5− cells, other transcriptional co-activators can then provide the needed activities to activate HIS3.
The crystal structure of Hmt1p has been solved at 2.9 Å resolution [61]. There seems to be no obvious structural similarity between Hmt1p and the bromodomain [62]. Since Hmt1p interacts with both acetylated H3 and H4, it probably does not have very stringent sequence preference for acetyllysine interaction. On the other hand, this flexibility of binding appears to be at odds with its selectivity on choosing H4 R3 as the methylation target. Future structural studies of Hmt1p in the presence of acetylated histone H4 will provide an insight into this novel mode of acetyllysine interaction.
Supplementary Material
Acknowledgments
We thank Steve Clarke, Karolin Luger, and Jacques Côté for their generous supply of materials. The authors thank Bill Henry, Steve Triezenberg, Yang Liu, and David Almy for discussions and suggestions during the development of this project. This work was supported by an NIH grant (GM62282) to MHK. HAB is a recipient of a Study-Abroad Scholarship by the German National Academic Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cary PD, Crane-Robinson C, Bradbury EM, Dixon GH. Effect of acetylation on the binding of N-terminal peptides of histone H4 to DNA. Eur J Biochem. 1982;127:137–143. doi: 10.1111/j.1432-1033.1982.tb06847.x. [DOI] [PubMed] [Google Scholar]
- 3.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 4.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 5.Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 6.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 7.Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 8.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 9.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 10.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 11.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 13.Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterner DE, Belotserkovskaya R, Berger SL. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci U S A. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, 3rd, Berger SL, Workman JL. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 17.Teng Y, Yu Y, Waters R. The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene. J Mol Biol. 2002;316:489–499. doi: 10.1006/jmbi.2001.5383. [DOI] [PubMed] [Google Scholar]
- 18.Vernarecci S, Ornaghi P, Bagu A, Cundari E, Ballario P, Filetici P. Gcn5p plays an important role in centromere kinetochore function in budding yeast. Mol Cell Biol. 2008;28:988–996. doi: 10.1128/MCB.01366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol. 2002;3:177–186. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- 21.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 23.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. Embo J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winston F, Allis CD. The bromodomain: a chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 25.Ladurner AG, Inouye C, Jain R, Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell. 2003;11:365–376. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 26.Guo D, Hazbun TR, Xu XJ, Ng SL, Fields S, Kuo MH. A tethered catalysis, two- hybrid system to identify protein-protein interactions requiring post-translational modifications. Nat Biotechnol. 2004;22:888–892. doi: 10.1038/nbt985. [DOI] [PubMed] [Google Scholar]
- 27.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of Telomeric Silencing-1 Is a Chromatin-specific Histone H3 Methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 28.Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA- binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 31.Guo W, Grant A, Novick P. Exo84p is an exocyst protein essential for secretion. J Biol Chem. 1999;274:23558–23564. doi: 10.1074/jbc.274.33.23558. [DOI] [PubMed] [Google Scholar]
- 32.Awasthi S, Palmer R, Castro M, Mobarak CD, Ruby SW. New roles for the Snp1 and Exo84 proteins in yeast pre-mRNA splicing. J Biol Chem. 2001;276:31004–31015. doi: 10.1074/jbc.M100022200. [DOI] [PubMed] [Google Scholar]
- 33.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 34.Gary JD, Lin WJ, Yang MC, Herschman HR, Clarke S. The predominant protein- arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- 35.Xu C, Henry PA, Setya A, Henry MF. In vivo analysis of nucleolar proteins modified by the yeast arginine methyltransferase Hmt1/Rmt1p. RNA. 2003;9:746–759. doi: 10.1261/rna.5020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu MC, Lamming DW, Eskin JA, Sinclair DA, Silver PA. The role of protein arginine methylation in the formation of silent chromatin. Genes Dev. 2006;20:3249–3254. doi: 10.1101/gad.1495206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 39.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, Allis CD. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 40.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 41.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 43.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Xu X, Singh-Rodriguez S, Zhao Y, Kuo MH. Histone H3 Ser10 phosphorylation-independent function of Snf1 and Reg1 proteins rescues a gcn5- mutant in HIS3 expression. Mol Cell Biol. 2005;25:10566–10579. doi: 10.1128/MCB.25.23.10566-10579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horiuchi K, Fujimoto D. Use of phosph-cellulose paper disks for the assay of histone acetyltransferase. Anal Biochem. 1975;69:491–496. doi: 10.1016/0003-2697(75)90151-7. [DOI] [PubMed] [Google Scholar]
- 46.Waterborg JH. Steady-state levels of histone acetylation in Saccharomyces cerevisiae. J Biol Chem. 2000;275:13007–13011. doi: 10.1074/jbc.275.17.13007. [DOI] [PubMed] [Google Scholar]
- 47.Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 49.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 50.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 51.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 53.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. Embo J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alifano P, Fani R, Lio P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 57.Kanazawa S, Driscoll M, Struhl K. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol Cell Biol. 1988;8:664–673. doi: 10.1128/mcb.8.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuo MH, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- 59.Wyce A, Henry KW, Berger SL. H2B ubiquitylation and de-ubiquitylation in gene activation. Novartis Found Symp. 2004;259:63–73. discussion 73–67, 163–169. [PubMed] [Google Scholar]
- 60.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat Struct Biol. 2000;7:1165–1171. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 62.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.