Abstract
An important question in the cell cycle field is how cyclin-dependent kinases (cdks) target their substrates. We have studied the role of a conserved hydrophobic patch on the surface of cyclin A in substrate recognition by cyclin A-cdk2. This hydrophobic patch is ≈35Å away from the active site of cdk2 and contains the MRAIL sequence conserved among a number of mammalian cyclins. In the x-ray structure of cyclin A-cdk2-p27, this hydrophobic patch contacts the RNLFG sequence in p27 that is common to a number of substrates and inhibitors of mammalian cdks. We find that mutation of this hydrophobic patch on cyclin A eliminates binding to proteins containing RXL motifs without affecting binding to cdk2. This docking site is critical for cyclin A-cdk2 phosphorylation of substrates containing RXL motifs, but not for phosphorylation of histone H1. Impaired substrate binding by the cyclin is the cause of the defect in RXL substrate phosphorylation, because phosphorylation can be rescued by restoring a cyclin A–substrate interaction in a heterologous manner. In addition, the conserved hydrophobic patch is important for cyclin A function in cells, contributing to cyclin A’s ability to drive cells out of the G1 phase of the cell cycle. Thus, we define a mechanism by which cyclins can recruit substrates to cdks, and our results support the notion that a high local concentration of substrate provided by a protein–protein interaction distant from the active site is critical for phosphorylation by cdks.
Progression through the eukaryotic cell cycle is controlled in part by the ordered action of cyclin-dependent kinases (cdks) (reviewed in refs. 1–8). A number of studies have shown that cdks are active only after binding to a cyclin partner. Cyclin levels oscillate throughout the cell cycle and are restricted spatially within a cell, thus limiting cdk activity both temporally and spatially. Initially, the cyclin’s primary role was thought to be allosteric, changing the conformation of the cdk from an inactive to an active state. In recent years, however, cyclins have been shown to play a role in substrate selection, as different substrate preferences are observed for cdk2 bound to cyclin E or cyclin A and for cdc2 bound to cyclin A or cyclin B (9–11). In addition, a number of substrates have been shown to form complexes with cyclin-cdks, suggesting that high-affinity protein–protein interactions play a role in cdk substrate recognition (12–16). In this work we have investigated the mechanism of substrate recognition by cyclin A-cdk2.
Cyclin A associates with cdk2 during S-phase and cdc2 during G2 (17–20). Numerous observations suggest that cyclin A-cdk2 is involved in controlling DNA replication. First, cyclin A is synthesized at the onset of S-phase and is required for passage through S-phase (17–23). Second, cyclin A is localized at sites of DNA replication and can promote DNA replication in a cell-free system (24–26). Third, overexpression of cyclin A in cells accelerates exit from G1 phase (27, 28). Finally, a catalytically inactive mutant of cdk2 acts as a dominant negative, preventing exit from the G1 phase (29). It will be important to understand how cyclin A-cdk2 recognizes its substrates in order to understand its function in cell cycle progression.
We examined the crystal structure of cyclin A-cdk2 seeking clues for the role of cyclin A in substrate recognition (30). A hydrophobic patch on the surface of cyclin A, comprising residues conserved in many cyclins, is readily apparent (31). In general, hydrophobic interactions contribute the bulk of intermolecular binding energies (32). Therefore, we suspected that this hydrophobic patch may be involved in substrate recognition. Indeed, the crystal structure of cyclin A–cdk2 in complex with the inhibitory protein, p27, reveals that this hydrophobic patch on cyclin A contacts the RNLFG sequence in p27, termed a Cy or RXL motif (33, 34) that is conserved between both substrates and inhibitors of cyclin-cdks (33–37). We have made hydrophobic to alanine mutations in cyclin A based on this crystal structure to investigate the role of the hydrophobic patch in cyclin A-cdk2 function. We find that this hydrophobic patch mediates binding to RXL-containing proteins and that this binding is important for phosphorylation of a subset of substrates of cyclin A-cdk2. Restoring substrate binding to cyclin A in a heterologous manner is sufficient to rescue phosphorylation even with mutations in the hydrophobic patch. Thus, our results suggest a mechanism for substrate recruitment to cdk2 by cyclin A.
MATERIALS AND METHODS
Plasmids.
Mutants of cyclin A were generated using the Altered Sites system (Promega). Briefly, cyclin A(Δ1) (38) was subcloned from pBSK into the HindIII-BamHI sites of pALTER. Mutations, including addition of a hemagglutinin (HA) tag, were generated according to the manufacturer’s instructions. Cassettes containing mutations and flanked by unique restriction sites in cyclin A were sequenced entirely prior to subcloning. pCDNA3-Cyclin AHA was generated by subcloning the HindIII-EcoRI fragment from pALTER-CyAHA into the same sites in pCDNA3 (Invitrogen). pMycCyclin A was a gift from L. Zhu (Albert Einstein University, New York). pCMV-p107 (39), pT5T-Rbp3 (40), pCDNA3-p21HA (41), pGST-p27 (16), pGST-RbC (42), pCMV-cdk2, and pCMV-cdk2DN (29) have all been described previously.
Cell Culture, Transfections, Flow Cytometry, and Indirect Immunofluorescence.
Transfections into U2OS cells (American Type Culture Collection) were performed by calcium phosphate DNA coprecipitation. For biochemical experiments, 20 μg of DNA was used for a 10-cm plate: 10–15 μg from cyclin plasmid, 2–5 μg from cdk2 plasmid, and the rest from either plasmids expressing RXL proteins or vector. For immunofluorescence and cell cycle experiments, 22 μg of DNA was used per 10-cm plate: 2–5 μg of cyclin plasmid and 0.2–2 μg of cdk2 plasmid was used, with 2 μg of pCD20 in the case of cell cycle experiments, and the rest from vector. Fluorescence-activated cell sorter analysis of transfected cells was performed as described previously (29). All samples were analyzed for cyclin A expression by Western blotting. Only samples with comparable levels of wild-type and cyAhpm protein were included. For immunofluorescence, cells were fixed in 4% formaldehyde (Polysciences) in PBS, and permeabilized with 0.5% Triton-X-100 (Sigma) in PBS. Localization was determined by staining cells with mAb 12CA5 directed against the HA tag on cyclin A.
Protein Purification.
tub E2F1, glutathione S-transferase (GST)-p27, and GST-Rb-C were expressed in bacterial strain BL21 and expressed and purified as described previously by Dynlacht et al. (9), Vlach et al. (16), and Meyerson and Harlow (42), respectively. The “tub” tag on E2F1 refers to a three-amino acid epitope, EEF, derived from tubulin, which is recognized by the mAb YL1/2. p107 was purified from insect cell lysates as described previously (9), except with an additional wash with 0.5 M NaCl/HMGN [25 mM Hepes (pH 7.6), 12.5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40].
Immunoprecipitation, Western Blotting, and Kinase Assays.
Cells were lysed on ice in 0.8–1 ml of lysis buffer [50 mM Tris (pH 7.4), 250 mM NaCl, 5 mM EDTA (pH 8.0), 0.1% Nonidet P-40, 10% glycerol, 1 mM NaF, containing 0.1 mM Na3VO4, 0.5 mM DTT, and the following protease inhibitors: 5 μg/ml leupeptin, 5 μg/ml aprotinin, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride/HCl, and 4 mM phenylmethylsulfonyl fluoride] for 30 min with occasional vortexing and clarified by centrifugation at top speed in a microfuge for 10 min at 4°C. Immunoprecipitations and Western blots were performed as described previously (43). mAbs used for immunoprecipitations of HA-tagged cyclin A (12CA5), p107 (SD9, SD15), E2F-1 (KH1, KH95), and p21 (CP68) have all been described previously (33, 39, 44, 45). Antibodies used for Western blotting of cyclin A (12CA5, recognizing the HA tag, and 9E10, recognizing the myc tag), p107 (SD9), and E2F-1 (KH20) were described previously (39, 44–46). The following rabbit polyclonal antibodies from Santa Cruz Biotechnology were used in Western blots: M2, H432, H164, and C20, recognizing cdk2, cyclin A, p21, and E2F-1, respectively. For kinase assays, lysates were precleared with normal rabbit serum-protein A-Sepharose beads prior to immunoprecipitation with 12CA5. Kinase reactions were performed as described previously (9).
Phosphopeptide Mapping.
Phosphopeptides of p107 were prepared as described previously (47). Five hundred counts per minute of the resulting peptides were resolved on cellulose thin-layer plates in two dimensions. Electrophoresis was performed at pH 1.9 [88% formic acid:acetic acid:water, 25:78:897 (by volume)] for 40 min at 800 V followed by ascending chromatography in 1-butanol:acetic acid:pyridine:water, 75:15:50:60 (by volume).
RESULTS
The Conserved Hydrophobic Patch Is Important for Binding to RXLFG-Containing Proteins.
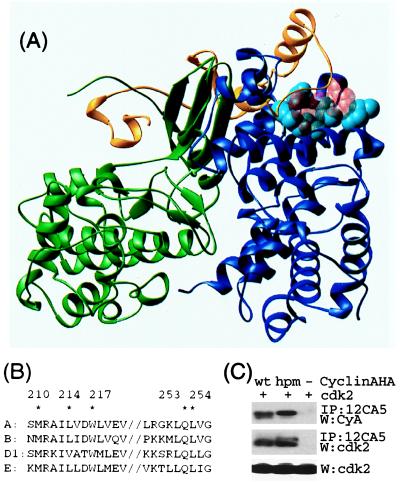
Many side chains contribute to the hydrophobic patch on cyclin A (30), including the residues M210, L214, W217, Q253, and L254 (Fig. 1). A number of mutants were constructed with three, four, or five simultaneous substitutions to alanine, and, for example, the mutants [M210A, L214A, W217A], [M210A, L214A, Q253A, L254A], and [M210A, L214A, W217A, Q253A, L254A] were particularly helpful in our initial experiments. Wild-type and mutant cyclin A variants were tagged with the HA epitope at the C terminus and transiently expressed in U2OS cells to test for protein stability and for high-affinity interaction with cdk2. Of the initial mutants, the triple mutant [M210A, L214A, W217A] appeared to have the properties that were desired for our studies. It was expressed at levels comparable to wild-type cyclin A and the mutations did not affect overall cyclin A integrity as it still bound to cdk2 in a cotransfection experiment (Fig. 1C). For convenience the [M210A, L214A, W217A] mutation is hereafter referred to as cyAhpm (cyclin A hydrophobic patch mutant).
Figure 1.
Structural basis for interactions with cyclin A. (A) Crystal structure of cyclin A (blue) -cdk2 (green) -p27 (yellow) (35). The residues mutated in this report are in light-blue space-filling representation. The RXL motif (R30, L32, and F33) involved important in p27–cyclin interactions is shown in transparent red space-filling representation. (B) The sequence in cyclin A that contacts p27 is conserved among a number of mammalian cyclins. Residues mutated in this report are marked with ∗. (C) The hydrophobic patch mutant binds cdk2 when immunoprecipitated from lysates of U2OS cells cotransfected with plasmids expressing HA-tagged cyclin A and cdk2. A representative experiment is shown. In a series of similar experiments, the amount of cdk2 present in 12CA5 immunoprecipitates is proportional to the amount of either HA-tagged cyclin A or cyAhpm.
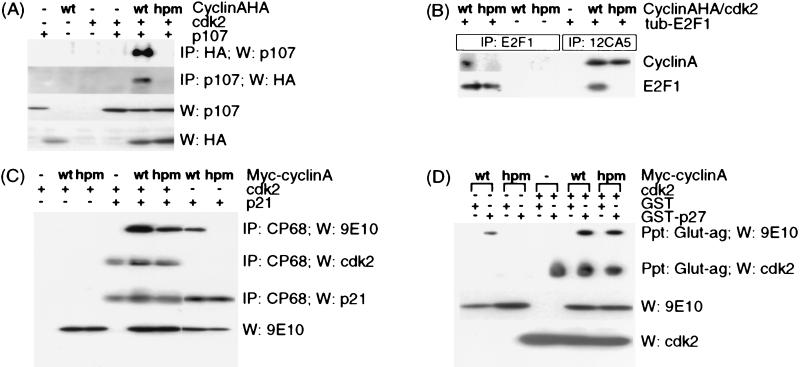
Because the x-ray structure of cyclin A-cdk2-p27 revealed that the hydrophobic patch on cyclin A contacts the RXL motif of p27 (35), we focused on interactions of CyAhpm with proteins containing similar motifs. Four candidate RXL motif proteins were tested: p107, E2F1, p21, and p27. CyAhpm is severely impaired for binding to p107 and E2F1 under all conditions assayed. To test binding to p107, plasmids encoding p107, cdk2, and either HA-tagged cyclin A or cyAhpm were cotransfected into U2OS cells. Immunoprecipitation with either a mixture of anti-p107 mAbs or the 12CA5 mAb recognizing the HA epitope tag on cyclin A revealed that mutation of the hydrophobic patch on cyclin A severely impairs binding to p107 (Fig. 2A). Even though the lysates contained similar amounts of each protein, and the 12CA5 immunoprecipitates contained similar amounts of cyclin A and cyAhpm as judged by Western blots (Fig. 2A and data not shown), p107 was found associated only with wild-type and not the mutant cyclin A. To test binding to E2F1, lysates from U2OS cells cotransfected with cdk2 and either cyclin A or cyAhpm and containing similar amounts of each protein were mixed with recombinant E2F1 purified from bacteria. Immunoprecipitation with either a mixture of mAbs recognizing E2F1 or with 12CA5 revealed that mutation of the hydrophobic patch on cyclin A severely impairs binding to E2F1 (Fig. 2B).
Figure 2.
The hydrophobic patch on cyclin A recruits RXL proteins. (A) Binding to p107 was assayed by immunoprecipitation and Western blotting of p107 or HA-tagged cyclin A in lysates from transfected U2OS cells. (B) Binding to E2F1 was assayed by immunoprecipitation and Western blotting of E2F1 or HA-tagged cyclin A. Lysates from U2OS cells transfected with either cyclin A- or cyAhpm-cdk2 were mixed with recombinant E2F1 before immunoprecipitation. (C) Binding to p21 was detected by Western blotting for Myc-tagged cyclin A or cdk2 after immunoprecipitation of p21 from transfected U2OS cells. (D) Binding to p27 was assayed by glutathione-agarose precipitation of GST or GST-p27 and Western blotting for Myc-tagged cyclin A or cdk2. Lysates from U2OS cells transfected with either cyclin A or cyAhpm and/or cdk2 were mixed with recombinant GST or GST-p27 before precipitation.
p21 and p27 fall into a second class of cyclin A-binding proteins. It has been shown previously that both p21 and p27 can bind cyclin A alone, cdk2 alone, and cyclin A–cdk2 complexes (16, 33), and we find that both p21 and p27 fail to associate with cyAhpm, but do associate with cyAhpm–cdk2 complexes. A previously described version of cyclin A with a Myc tag at the N terminus was used in these experiments to facilitate detection by Western blotting (36, 48). To test binding to p21, plasmids encoding p21, cdk2, and either cyclin A or cyAhpm were cotransfected into U2OS cells, and lysates were immunoprecipitated with CP68, which recognizes the C terminus of p21 (33). CyAhpm associates with p21 only when in complex with cdk2, suggesting that the hydrophobic patch on the surface of cyclin A is responsible for p21 association with cyclin A alone (Fig. 2C). In agreement with previous studies of p21 and p27 mutants (16, 33), cdk2 could recruit p21 to the cyclin A mutant. To test binding to p27, lysates from transfected U2OS cells were mixed with either recombinant GST or GST-p27 purified from bacteria, and precipitated with glutathione-agarose beads. The results for p27 were similar to the results for p21 (Fig. 2D).
Substrate Binding by Cyclin A Plays a Major Role in Recognition by cdk2.
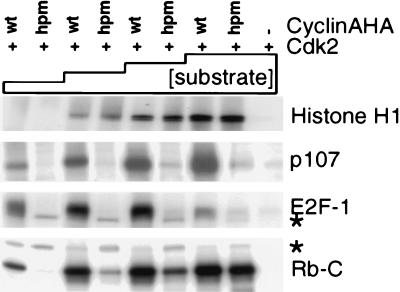
We investigated the role of the conserved hydrophobic patch on cyclin A in substrate phosphorylation by cyclin A–cdk2. Cyclin A–cdk2 and cyAhpm–cdk2 complexes were compared for their ability to phosphorylate a variety of substrates. Kinase complexes from transfected cell lysates were immunoprecipitated through the HA tag on cyclin A with the mAb 12CA5, the precipitates were divided equally, and kinase activity toward each substrate was tested over a broad range of substrate concentration. For each experiment, the same kinase preparation was used for several substrates in parallel, always including histone H1 as a control. The levels of histone H1 phosphorylation were identical for each preparation of cyAhpm–cdk2 and cyclin A–cdk2 complexes. However, cyAhpm–cdk2 is specifically defective for phosphorylating E2F1, p107, and Rb-C terminus, suggesting that the hydrophobic patch is involved in substrate recruitment to cdk2 (Fig. 3). Interestingly, a significantly higher substrate concentration is required for phosphorylation of histone H1, which does not tightly associate with cyclin A–cdk2. To test further the correlation between binding to and phosphorylation of RXL proteins, four additional mutants were studied: [M210A, L214A], [W217A], [E220A], and [Q253A, L254A]. Although all four mutants could activate cdk2 to phosphorylate histone H1, those that did not bind p27 or p107 were impaired for phosphorylating p107 (Table 1).
Figure 3.
The hydrophobic patch on cyclin A is important for phosphorylation of a subset of substrates. HA-tagged cyclin A–cdk2 or cyAhpm–cdk2 complexes immunoprecipitated from transfected U2OS cells were incubated with [γ-32P]ATP and the following amounts of the following substrates: histone H1, 100 ng, 250 ng, 1 μg, and 2 μg; p107, 10 ng, 50 ng, 200 ng, and 500 ng; E2F1, 10 ng, 50 ng, 200 ng, and 500 ng; Rb-C, 10 ng, 50 ng, 200 ng, and 1 μg. Autophosphorylated cyclin A is indicated with ∗.
Table 1.
Phosphorylation of p107 correlates with binding to RXL-containing proteins
| Mutant | Histone H1 kinase | p107 Kinase | p107 Binding | p27 Binding |
|---|---|---|---|---|
| M210A, L214A | + | − | − | − |
| W217A | + | − | − | − |
| E220A | + | + | + | + |
| Q253A, L254A | + | − | − | − |
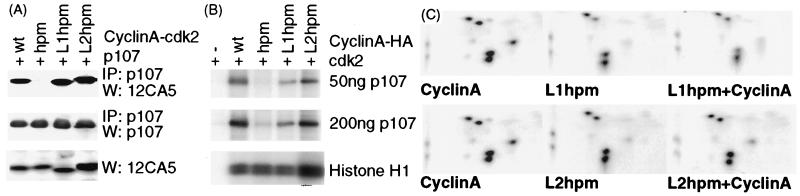
There are two simple explanations for how substrate binding to cyclin A could contribute to recognition by cdk2. First, binding to cyclin A could increase the local concentration of substrate relative to the kinase active site. Second, binding to the hydrophobic patch on cyclin A could correctly position the substrate with respect to the active site of cdk2. It would be possible to evaluate the relative importance of cyclin A–substrate interactions in cdk2 recognition if substrate binding to cyAhpm could be restored in a heterologous manner. One substrate, p107, contains several independent protein interaction domains (49). In addition to its RXL motif, p107 contains the distinct pocket domain, originally identified for its ability to bind to viral oncoproteins. These viral proteins all contain a pocket-binding motif, LXCXE, and a peptide derived from the papilloma virus E7 protein containing the sequence LYCYEQL has been shown to bind to pocket domains (50). Thus, to restore binding to p107, we made two different mutants of cyAhpm, adding the sequence LYCYEXφ (where φ is hydrophobic). The sites of insertion were selected based on three criteria: (i) they were dispensable for cdk binding (38, 51); (ii) they were likely to be solvent accessible, based on the extreme proteolytic sensitivity of the N-terminal 173 residues of cyclin A in vitro (30); and (iii) a minimum number of mutations would be needed to change the sequence to LYCYEXφ. L1hpm contains, in addition to the hydrophobic patch mutations, the sequence YCY substituted for QEDQ between L26 and E31, giving the sequence ALYCYENI at residues 25–33 of the cyclin A sequence. L2hpm contains the sequence YCYEQ substituted for KD between L75 and L78, resulting in the sequence PLYCYEQL. Both binding to and phosphorylation of p107 are restored by the LYCYEXφ insertions in cyAhpm (Fig. 4 A and B).
Figure 4.
Cyclin A binding is important for cdk2 phosphorylation of an RXL substrate. (A) Binding to p107 by LXCXE-hpm mutants of cyclin A was tested by immunoprecipitation of p107 and Western blotting of HA-tagged cyclin A from lysates of transfected U2OS cells. (B) L1hpm– and L2hpm–cdk2 complexes were tested as in Fig. 3 for their ability to phosphorylate the indicated amounts of p107 or 500 ng of histone H1. (C) Phosphopeptide maps of p107 phosphorylated by wild-type cyclin A-, L1hpm- and L2hpm-cdk2 in B.
As a first step toward addressing the importance of the location of the substrate-binding site on cyclin A, phosphopeptide maps were compared for p107 phosphorylated by the cyclin A–, L1hpm–, and L2hpm–cdk2 complexes. Insufficient 32P was incorporated into p107 by cyAhpm–cdk2 complexes to compare this to the other kinases. The phosphopeptide maps generated by each of the three complexes are identical, although the degree of phosphorylation of one peptide is different (Fig. 4C). Although we do not know the identity of the phosphorylation sites or where they are located with respect to the RRLFG or pocket regions of p107, the simplest interpretation of this result is that the major role of substrate binding by cyclin A is not to precisely position the phosphoacceptor site in the active site of cdk2, but to increase the local concentration of substrate (Fig. 5).
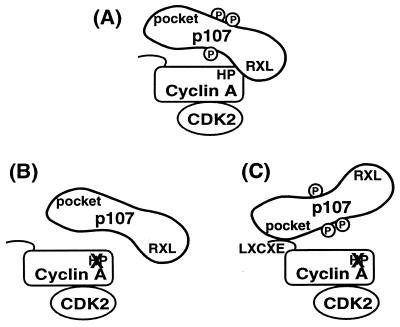
Figure 5.
A model for cyclin A-cdk2 recognition of a substrate, p107. (A) The RXL motif in p107 normally contacts the hydrophobic patch in cyclin A. (B) Mutation of the hydrophobic patch on cyclin A dramatically reduces binding to and phosphorylation of p107. (C) Binding between p107 and a hydrophobic patch mutant of cyclin A can be restored by addition of the p107 pocket-binding sequence, LXCXE, to cyclin A. Restoration of binding is sufficient to rescue phosphorylation. Thus, a specific trajectory between substrate binding and the kinase active site is not necessary for phosphorylation, and the purpose of substrate binding is to increase the local concentration available to the kinase.
Cyclin A Function in Cells.
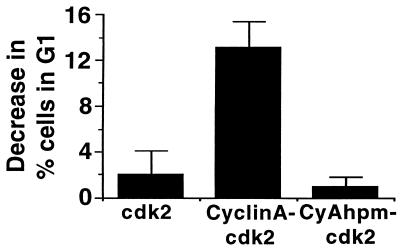
Overexpression of cyclin A in cultured cells results in a decrease in the G1 phase population (27, 28) that reflects a shortened G1 phase. We compared the effects of cyclin A-cdk2 and cyAhpm-cdk2 on the cell cycle distribution by cotransfecting the cell surface marker CD20 and analyzing the CD20-positive population by fluorescence-activated cell sorting. There is a dramatic decrease in the G1 population of cells expressing cyclin A-cdk2. However, cyAhpm-cdk2 does not affect the cell cycle profile differently from cdk2 alone (Fig. 6). Similar effects on the cell cycle profile were observed previously for two different cyclin E mutants, one which cannot phosphorylate pRb and another which binds cdk2 but fails to activate its catalytic activity (15, 52). Thus, our results suggest a role for substrate recognition by the hydrophobic patch on cyclin A in cells.
Figure 6.
Decrease in G1 population of U2OS cells transiently expressing either cdk2 alone, cyclin A-cdk2, or cyAhpm-cdk2. The results are the mean of at least three independent experiments, and the error bars indicate SD from the mean. The baseline G1 fraction in vector-transfected cells is 32.8%.
There are multiple ways in which cyclins are thought to regulate cdk activity. Although cyclins both activate the kinase and recruit substrates, they also limit cdk activity to the subcellular compartments in which they are localized. To ensure that the defect in cyclin A’s ability to accelerate G1 exit was not due to mislocalization, we examined subcellular localization of cyclin A by indirect immunofluorescence with 12CA5. Importantly, both cyclin A and cyAhpm localize to the nucleus to the same extent when cotransfected with either cdk2 or the catalytically inactive cdk2dn (data not shown), consistent with previous studies (53, 54). Because the cell cycle experiments and the key biochemistry described above were carried out either under these conditions or in vitro, subcellular localization is unlikely to account for the differences in behavior described above for wild-type cyclin A- and cyAhpm-cdk2.
DISCUSSION
A number of previous studies have shown that the RXL motifs on the substrates p107 and E2F-1 and the inhibitors p21 and p27 are important for cyclin binding (16, 33, 34, 36, 37, 55). Our work extends these studies by showing that the RXL contact site on cyclin A is important for cyclin A-cdk2 function. We have demonstrated that the hydrophobic patch on cyclin A serves as a docking site, important for tight physical interaction with and phosphorylation of RXL substrates by cyclin A-cdk2.
Cyclin A and Substrate Selection.
The residues that influence substrate binding by cyclin A are conserved in a number of mammalian cyclins (Fig. 1A) (31), and these same residues may also serve as binding sites on D-, E-, and B-type cyclins. These cyclins can all bind p21 family members (16, 33, 34, 56). However, because numerous different cyclins have the same amino acids found in the hydrophobic patch on cyclin A, it is likely that additional factors dictate substrate specificity. Indeed, there are differences among the abilities of different cyclins to bind to some RXL proteins. For example, cyclin A-cdk2 and cyclin A-cdc2 bind and efficiently phosphorylate p107, but cyclin B–cdk2 and –cdc2 complexes do not (10, 11). Thus, while the conserved hydrophobic patch serves as a docking site, its accessibility to different proteins may be limited by surrounding side chains on the surface of a given cyclin or by other sites that have not yet been described. Such additional constraints could determine which substrates have access to which cyclins. Identification of such specificity determinants will await the crystal structures of the other cyclins, alone and in complex with other RXL proteins.
Our data using a heterologous method of establishing cyclin–substrate interactions suggest that cyclin binding is of major importance in substrate selection, but the actual stereochemistry of how the substrate binds to the cyclin may be less important (Fig. 5). This conclusion is supported by a number of previous observations. First, numerous sites on a given substrate are phosphorylated by cdks (47). Thus, there may not be a rigid path between the substrate binding site on a cyclin and the catalytic site on a cdk. Second, phosphoacceptor sequences predicted from studies of peptide substrates closely resemble those found in physiological substrates, showing that the local sequence around the phosphorylated residue plays a major role in determining which sites are recognized by cdk2 (57). Third, some cdk substrates associate tightly with cyclins (12–14). The importance of tight cyclin–substrate interactions is supported by the result that transfer of the RXL motif from E2F1 to the closely related protein E2F4 is sufficient to allow phosphorylation by cyclin A-cdk2, even though E2F4 is not normally a substrate (37). In addition, a mutation in cyclin E which abolishes pRb binding also eliminates pRb phosphorylation (52). Finally, peptides containing the RXL motif block phosphorylation of RXL-containing substrates by cyclin–cdk complexes (33, 34). These results underscore the importance of a high local concentration of substrate.
When both cyclin A and cdk2 are overexpressed, the only defect we observe for cyAhpm–cdk2 complexes is binding to and phosphorylation of RXL substrates. CyAhpm–cdk2 complexes remain inert and do not affect the cell cycle profile whereas wild–type cyclin A-cdk2 complexes phosphorylate their substrates and drive exit from G1, suggesting that the hydrophobic patch on cyclin A recruits substrates involved in cell cycle progression.
Cyclin A as an Integrator.
A new view of the role of cyclins is emerging, with cyclins acting somewhat analogously to the adapter, anchoring, or scaffold proteins in other signal transduction pathways (for review, see ref. 58). Adapter proteins used in receptor tyrosine kinase signaling pathways have numerous protein-docking sites, with an SH2 or PTB domain to bind the receptor itself, and additional motifs to connect the kinase to other components of the signaling pathway (59, 60). Similarly, the scaffolding protein ste5p brings together consecutive members of the mitogen-activated protein kinase pathway so that signals pass rapidly and in the correct order from one kinase to the next (61, 62). Src family kinases have evolved an even simpler mechanism for recruiting substrates, containing SH2 and SH3 domains in addition to the kinase domain, which directly recruit tightly associated substrates (63). Similarly, cyclin A tethers substrates to cdk2.
Cyclin A coordinates multiple functions of cdk2. It is an activator, a timer, and a substrate recruiter. In addition, cyclin A restricts kinase activity to the correct subcellular compartment. Interestingly, for the substrate E2F1, nuclear localization may be coupled to cyclin A binding, with the same short region of sequence containing both its RXL motif and its sequences required for nuclear localization (64). Given the importance of cyclin A’s recognizing the correct substrate in the correct subcellular location, it seems reasonable to suggest that proper localization of substrates could be coordinated with cyclin binding. Our data indicate that a number of functions are integrated at a single site on the surface of cyclin A.
Acknowledgments
We thank M. Classon, S. Salama, and J. Zhao for helpful and stimulating discussions. We thank B. Amati for GST-p27, A. Dutta for pCDNA3-p21HA, E. Lees for pBSKCyclinA(Δ1), L. Zhu for pMyc-cyclin A, and B. Dynlacht for the p107 baculovirus. We are grateful to N. Pavletich for providing us with coordinates for the structures of cyclin A-cdk2 and cyclin A-cdk2-p27. We thank A. Amon, S. Boulton, D. Fingar, and P. Murray for critical reading of the manuscript and B. Kennedy for help with immunofluorescence. We thank D. Dombkowsky for assistance with flow cytometry. We are grateful to N. Dyson and members of the Harlow and Dyson laboratories for continuous advice and encouragement. B.A.S. is a Foundation for Advanced Cancer Studies Fellow of the Life Sciences Research Foundation. This work was supported by grants from the National Institutes of Health (to E.H.).
ABBREVIATIONS
- HA
hemagglutinin
- GST
glutathione S-transferase
References
- 1.Hunt T. Curr Opin Cell Biol. 1989;1:268–274. doi: 10.1016/0955-0674(89)90099-9. [DOI] [PubMed] [Google Scholar]
- 2.Nurse P. Nature (London) 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 3.Norbury C, Nurse P. Annu Rev Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 4.Pines J, Hunter T. Ciba Found Symp. 1992;170:187–196. [PubMed] [Google Scholar]
- 5.Nasmyth K. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 6.Sherr C J. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 7.Morgan D O. Nature (London) 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 8.Morgan D O. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 9.Dynlacht B D, Flores O, Lees J A, Harlow E. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 10.Pan Z Q, Amin A, Hurwitz J. J Biol Chem. 1993;268:20443–20451. [PubMed] [Google Scholar]
- 11.Peeper D S, Parker L L, Ewen M E, Toebes M, Hall F L, Xu M, Zantema A, van der Eb A J, Piwnica-Worms H. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 13.Ewen M E, Faha B, Harlow E, Livingston D M. Science. 1992;255:85–87. doi: 10.1126/science.1532457. [DOI] [PubMed] [Google Scholar]
- 14.Faha B, Ewen M E, Tsai L H, Livingston D M, Harlow E. Science. 1992;255:87–90. doi: 10.1126/science.1532458. [DOI] [PubMed] [Google Scholar]
- 15.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 16.Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5334–5544. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pines J, Hunter T. Nature (London) 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 19.Rosenblatt J, Gu Y, Morgan D O. Proc Natl Acad Sci USA. 1992;89:2824–2828. doi: 10.1073/pnas.89.7.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai L H, Harlow E, Meyerson M. Nature (London) 1991;353:174–177. doi: 10.1038/353174a0. [DOI] [PubMed] [Google Scholar]
- 21.Girard F, Strausfeld U, Fernandez A, Lamb N J. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 22.Zindy F, Lamas E, Chenivesse X, Sobczak J, Wang J, Fesquet D, Henglein B, Brechot C. Biochem Biophys Res Commun. 1992;182:1144–1154. doi: 10.1016/0006-291x(92)91851-g. [DOI] [PubMed] [Google Scholar]
- 23.Henglein B, Chenivesse X, Wang J, Eick D, Brechot C. Proc Natl Acad Sci USA. 1994;91:5490–5494. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Urso G, Marraccino R L, Marshak D R, Roberts J M. Science. 1990;250:786–791. doi: 10.1126/science.2173140. [DOI] [PubMed] [Google Scholar]
- 25.Cardoso M C, Leonhardt H, Nadal-Ginard B. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 26.Krude T, Jackman M, Pines J, Laskey R A. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 27.Resnitzky D, Hengst L, Reed S I. Mol Cell Biol. 1995;15:4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg A R, Zindy F, Le Deist F, Mouly H, Metezeau P, Brechot C, Lamas E. Oncogene. 1995;10:1501–1509. [PubMed] [Google Scholar]
- 29.van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 30.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Nature (London) 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 31.Brown N R, Noble M E, Endicott J A, Garman E F, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson L N. Structure. 1995;3:1235–1247. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 32.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G., Jr Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Nature (London) 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 36.Zhu L, Harlow E, Dynlacht B D. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 37.Dynlacht B D, Moberg K, Lees J A, Harlow E, Zhu L. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lees E M, Harlow E. Mol Cell Biol. 1993;13:1194–1201. doi: 10.1128/mcb.13.2.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 40.Huber H E, Edwards G, Goodhart P J, Patrick D R, Huang P S, Ivey-Hoyle M, Barnett S F, Oliff A, Heimbrook D C. Proc Natl Acad Sci USA. 1993;90:3525–3529. doi: 10.1073/pnas.90.8.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 42.Meyerson M, Harlow E. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Q J, Bautista C, Edwards G M, Defeo-Jones D, Jones R E, Harlow E. Mol Cell Biol. 1991;11:5792–5799. doi: 10.1128/mcb.11.11.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helin K, Wu C L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 46.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lees J A, Buchkovich K J, Marshak D R, Anderson C W, Harlow E. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heald R, McLoughlin M, McKeon F. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 49.Zhu L, Enders G, Lees J A, Beijersbergen R L, Bernards R, Harlow E. EMBO J. 1995;14:1904–1913. doi: 10.1002/j.1460-2075.1995.tb07182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyson N, Guida P, Munger K, Harlow E. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi H, Stewart E, Poon R, Adamczewski J P, Gannon J, Hunt T. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly B L, Wolfe K G, Roberts J M. Proc Natl Acad Sci USA. 1998;95:2535–2540. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maridor G, Gallant P, Golsteyn R, Nigg E A. J Cell Sci. 1993;106:535–544. doi: 10.1242/jcs.106.2.535. [DOI] [PubMed] [Google Scholar]
- 54.Pines J, Hunter T. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin J, Reichner C, Wu X, Levine A J. Mol Cell Biol. 1996;16:1786–1793. doi: 10.1128/mcb.16.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Songyang Z, Blechner S, Hoagland N, Hoekstra M F, Piwnica-Worms H, Cantley L C. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 58.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 59.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 60.Clark S G, Stern M J, Horvitz H R. Nature (London) 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- 61.Choi K Y, Satterberg B, Lyons D M, Elion E A. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 62.Marcus S, Polverino A, Barr M, Wigler M. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reynolds A B, Kanner S B, Wang H C, Parsons J T. Mol Cell Biol. 1989;9:3951–3958. doi: 10.1128/mcb.9.9.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]