Abstract
HLA class II-restricted antigen (Ag) processing and presentation are important for the activation of CD4+ T cells, which are the central orchestrating cells of immune responses. The majority of melanoma cells either expresses, or can be induced to express, HLA class II proteins. Thus, they are prime targets for immune mediated elimination by class II-restricted CD4+ T cells. We have previously shown that human melanoma cells lack an important enzyme, gamma interferon-inducible lysosomal thiol-reductase (GILT), capable of perturbing immune recognition of these tumors. Here, we show that GILT expression in human melanoma cells enhances Ag processing and presentation via HLA class II molecules. We also show that GILT expression influences the generation of active forms of cysteinyl proteases, cathepsins B, L and S, as well as an aspartyl protease cathepsin D in melanoma cells. Mechanistic studies revealed that GILT does not regulate acidic cathepsins at the transcriptional level; rather it colocalizes with the cathepsins and influences HLA class II Ag processing. GILT expression in melanoma cells also elevated HLA-DM molecules, which favor epitope loading onto class II in the endolysosomal compartments, enhancing CD4+ T cell recognition. These data suggest that GILT-expressing melanoma cells could prove to be very promising for direct antigen presentation and CD4+ T cell recognition, and may have direct implications for the design of cancer vaccines.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-008-0483-8) contains supplementary material, which is available to authorized users.
Keywords: Gamma-IFN-inducible lysosomal thiol reductase, Melanoma, HLA class II molecules, Cathepsins, HLA-DM, Antigen processing and presentation
Introduction
Melanoma is the rapidly growing cancer among US Caucasians [1–3]. While early, localized tumors are often treatable through wide excision, metastatic malignant tumors are largely refractory to cytotoxic chemotherapeutics and almost universally fatal [3–6]. Treatment through surgery, radiation, chemotherapy and gene therapy, though rapidly improving, are often ineffective [4–8]. The most promising option available to combat melanoma is immunotherapy, due to its potential specificity and lack of side effects [9–11]. One of the most promising outlets for this immunotherapy involves antigen (Ag) processing and direct presentation by tumor cells, inducing antitumor immune responses. The majority of melanoma cells express HLA classes I and II molecules necessary for tumor Ag presentation to T cells [12, 13]. However, the development of strong memory responses to tumor Ags also depends on processing and cross-presentation of shed tumor Ags via bystander antigen presenting cells (APCs) [14–16]. While dendritic cells, macrophages, and B cells express class II molecules and possess the machinery to optimally activate CD4+ T cells [17, 18], melanoma cells lack a thiol reductase, GILT, that disrupts antigen processing and tumor cell presentation of antigenic peptides [19]. Whether GILT expression in melanoma alters antigen processing, epitope generation and CD4+ T cell recognition via class II molecules remains unclear.
Crucial to the stimulation of immune response in humans is the ability to present epitopes on the surface of the cells that are recognizable by CD4+ T cells [19–21]. Epitopes are derived from exogenous and endogenous antigens (Ags), which are internalized by APCs as well as tumor cells and then are processed by intercellular proteases, specifically the cathepsins [22–25]. The cysteinyl proteases, cathepsins (Cat) B, S, and L, are crucial for most of the degradations leading to functional epitopes. Cat B has also been shown to upregulate the expression of class II proteins, thereby contributing Ag presentation via the HLA class II pathway. Cat D, an aspartyl protease, performs more specific degradations of Ags, splicing them at very specific points to yield short, functional peptides [25]. Once processed, the epitopes become complexed with the HLA class II molecules in the acidic endosomal/lysosomal compartments and appear on the cell surface [26]. HLA class II molecules are initially complexed with a glycoprotein known as the invariant chain (Ii), a molecule necessary for correct folding and function of the molecule as well as serving as a competitive inhibitor of the class II binding site [27, 28]. Epitopes cannot be loaded onto the class II molecules until proper cleavage of the Ii by one of the cysteinyl cathepsins, Cat S. After cleavage of the Ii, the class II associated Ii peptide (CLIP) remains in the class II binding groove, further preventing improper loading of peptides. A non-classical class II protein, HLA-DM, mediates the removal of CLIP and helps the loading of appropriate peptides into the class II binding groove [28, 29].
The processing of Ags by APCs is the main factor influencing autoimmunity, successful organ transplantation, and cancer memory responses [30–32]. In tumor cells, many of the enzymatic processes involved in Ag processing and presentation are altered. Many cancerous cells do not express measurable class II proteins, thus preventing the body from recognizing the tumors via the class II pathway, and allowing the cells to continue to proliferate and metastasize in some cases. Therefore, insights into the pathways, which govern class II Ag processing and presentation by melanoma may provide a very promising outlet for the treatment of cancer. One specific area within the realm of Ag processing that holds a great deal of promise is the role of the acidic proteases, such as cathepsins, and the chaperone molecule HLA-DM. The expression of these molecules differs in tumor cells, demonstrating that they play a role in the ability of tumors to evade the immune system. We have previously shown that GILT can process cysteinylated peptides by reducing disulfides, making the functional form of peptides readily accessible to class II proteins. In this study, we show that GILT expression in melanoma enhances class II Ag processing, upregulates the expression of HLA-DM molecules, and alters acidic proteases such as the cathepsins. This in conjunction with GILTs’ ability to break disulfide bonds in Ags may make GILT an excellent candidate for facilitating the direct presentation of class II-complexes by tumor cells for effective antitumor immune response.
Materials and methods
Cell lines
The human melanoma cell line 1359-mel which constitutively expresses HLA-DR4 (DRB1*0401) was cultured in complete RPMI containing 10% fetal bovine serum (FBS) (HyClone), 50 U/ml penicillin, 50 μg/ml streptomycin, and enriched with L-glutamate (Mediatech Inc., Herndon, VA, USA) [41, 42]. Human melanoma cells, J3 transfected with DR4 (J3.DR4), were cultured in complete IMDM medium containing 10% bovine growth serum (BGS) (HyClone), 50 U/ml penicillin, 50 μg/ml streptomycin [19]. Cells were grown in IMDM for 24 h in the absence or presence of actinomycin D (5 μg/ml, final concentration) (Sigma Chemicals).
T cell hybridomas line 2.18a, 1.21, 17.9 and 4027/99 specific for κ188–203, κ145–159, HSA64–76 and CII261–273, respectively, were cultured in complete RPMI 1640 with 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, and supplemented with L-glutamate and 50 μM β-mercaptoethanol (β-ME) [19, 34]. The IL-2 dependent cell line, HT-2 was cultured in RPMI 1640 with 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, 50 μM β-ME, and 20% Con A supernantant (T-STIM, Collaborative Biomedical Res., Bedford, MA) as a supplement [19, 34].
Antigens and peptides
Human IgG kappa (Igκ), human serum albumin (HSA) and type II collagen (CII) were purchased from Sigma. The human IgG immunodominant (κI) peptide κ188–203 (sequence KHKVYACEVTHQGLSS), subdominant (κII) peptide κ145–159 (sequence KVQWKVDNALQSGNS), HSA64–76 peptide (sequence VKLVNEVTEFAKTK) and CII261–273 (sequence AGFKGEQGPKGEP) were produced using Fmoc technology and an Applied Biosystems Synthesizer [34, 35]. Analysis of the peptide was performed using reverse phase HPLC purification and mass spectrometry, showing a peptide purity of >99%.
Transfection assays
Human melanoma J3 cells were transfected with HLA-DR4 (DRB1*0401) using retroviral vectors with linked drug selection markers for hygromycin and histidinol resistance [19]. To confirm DR4 surface expression, flow cytometric analysis with the specific monoclonal antibody, 359-F10, was used [36]. The J3.DR4 line is further transfected with GILT cDNA to generate J3.DR4.GILT [19, 33]. The 1359-mel cells, which innately express HLA-DR4 were also transfected with GILT as described [19, 33]. The expression of GILT was confirmed by western blotting [19].
Antigen presentation assays
Melanoma cells J3.DR4, J3.DR4.GILT, 1359-mel and 1359-mel.GILT were incubated with the whole antigen Igκ, HSA, or CII overnight at 37°C in culture media. Cells were washed, and co-cultured with the κ188–203, κ145–159, HSA64–76 or CII261–273 peptide-specific T cell hybridomas [2.18a for κ188–203, 1.21 for κ145–159, 17.9 for HSA64–76 and 4027/99 (kindly provided by Dr. Lars Fugger, Aarhus, Denmark) for CII261–273] for 24 h [19, 34]. T cell production of IL-2 was monitored by measuring 3H-thymidine (1 μCi/well) incorporation using HT-2 cells [19, 35]. All assays were repeated at least three times with the standard error for triplicate samples within a single experiment reported. Data were corrected for isotope counting efficiency and expressed as mean ccpm ± SEM.
Flow cytometric analysis
J3.DR4, J3.DR4.GILT, 1359-mel, 1359-mel.GILT and a B-cell line Frev were stained with mAbs directed against DR4 (359-F10) followed by a secondary antibody labeled with FITC as described [36]. Samples were analyzed on a FACScan using CellQuest software (BD Bioscience, Mountain View, CA). Background fluorescence was evaluated using an irrelevant isotype-matched mAb (IN-1) as described previously [36].
Western blot analysis
Total cell lysates obtained from melanoma cells J3·DR4, J3·GILT·DR4, 1359, 1359·GILT, and a B-cell line Frev were subjected to SDS-PAGE (10%) and analyzed by western blotting for cathepsins B, D, S and L (Santa Cruz Biotechnology, Santa Cruz, CA.), HLA-DR (L243), GILT (JSB-GILT), and HLA-DM (JSB-DM) as described [19, 33, 37].
Live confocal microscopic analysis
Melanoma cells 1359-mel.GILT were grown in 35-mm dishes, washed twice with PBS and stained with rabbit anti-cathepsin B and D, and costained with goat anti-GILT, followed by rhodamine-conjugated anti-rabbit IgG and fluorescein-conjugated anti-goat IgG antibodies. All antibodies were purchased from Santa Cruz Biotechnology. Samples were analyzed by a laser scanning confocal microscopy with a Leica TCS SP2 AOBS, 63x water immersion objective. Live images were acquired using excitation lasers 488 and 543. Overlay images were assembled, zoomed, and cropped using Leica software.
Reverse transcription-polymerase chain reaction analysis
Total RNA was isolated from melanoma cells, using RNAzol reagent (Biotex Labs, TX) according to the manufacturer’s protocol. Reverse transcription reaction was performed using poly-dT primer and Moloney murine leukemia virus reverse transcriptase (Applied Biosystem) in 25 μl reaction volume containing total RNA (2 μg), 1 × PCR buffer and 2 mM MgCl2, at 42°C for 15 min followed by 95°C for 5 min. PCR was carried out in a programmed thermal cycler (Applied Bio system). The primer sequences used to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were 5′-CCTACCCCCAATGTATCCGTTGTG-3′ (sense) and 5′-GGAGGAATGGGAGTTGCTGTTGAA-3′ (anti-sense) and for CatB mRNA were 5′-TCGGATGAGCTGGTCAACTATG-3′ (sense) and 5′-TCCAAGCTTCAGCAGGATAG-3′ (anti-sense); Cat D mRNA were 5′-TTCAGGGCGAGTACATGATCC-3′ (sense) and 5′-CCCTGTTGTTGTCACGGTCAA-3′ (anti-sense); Cat L mRNA were 5′-GACAGGGACTGGAAGAGAG-3′ (sense) and 5′-GTTTCCCTTCCCTGTATTC-3′(anti-sense); Cat S mRNA were 5′-GGGTACCTCATGTGACAAG-3′ (sense) and 5′-TCACTTCTTCACTGGTCATG-3′ (anti-sense); HLA-DMα mRNA were 5′-ATCCAGCAAATAGGGCCAAAACTT-3′ (sense) and 5′-GATGAGAACAATGCCCACGATGAT-3′ (anti-sense) and HLA-DMβ mRNA were 5′-AATAGCTTGGCGAATGTCCTCTCA-3′ (sense) and 5′-CATTGGGCTGGGCAGTCTTGTG-3′ (anti-sense). Thermal cycling parameters were 94°C for 3 min, followed by 40 cycles of amplifications at 94°C for 30 s, 60°C for 1 min, 72°C for 1 min, and 72°C for 5 min as the final elongation step as follows: 30 cycles of 1 min at 92°C. PCR products were visualized following electrophoresis in 1.5% agarose gel with ethidium bromide staining.
mRNA expression levels were also determined by real-time RT-PCR as described [38]. The quantitative real-time PCR was performed using IQTM SYBR Green Supermix in an iCycler (iCycler iQ Single–color Real Time PCR detection system; Bio-Rad, Hercules, CA). Relative levels of DM/cathepsins mRNA expression were normalized in all the samples analyzed with respect to the levels of GAPDH amplification.
Statistical analyses
Data from each experimental group were subjected to statistical analysis. Differences between experimental groups were analyzed for statistical significance using Student’s t-test. Values of P < 0.05 (*) were considered significant.
Results
GILT expression in human melanoma enhances HLA-DR4-restricted Ag processing and epitope presentation
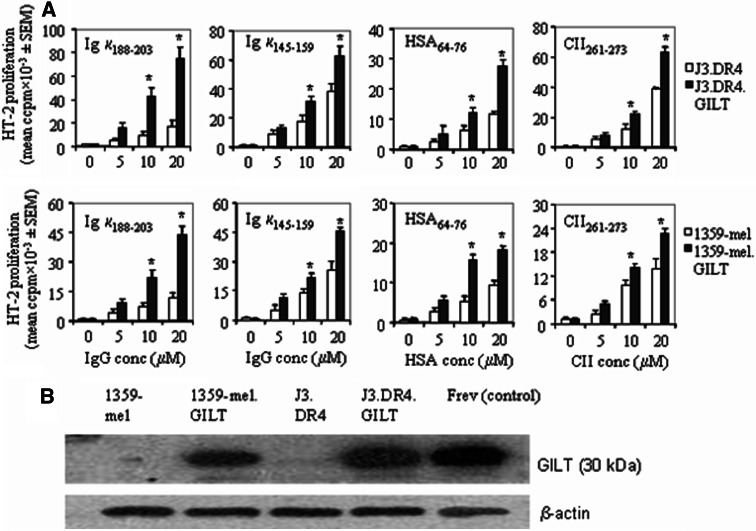
Recent evidence suggests that presentation of antigenic peptides by HLA class II molecules to CD4+ T cells is critical to the generation of anti-tumor immunity [39–41]. CD4+ T cells recognize their specific Ag in the context of class II molecules. In contrast to HLA class I, class II molecules have a more restricted distribution pattern, and are predominantly expressed in professional APCs. Tumors such as melanoma, also express class II molecules, and could be potential targets for CD4+ T cells [39–42]. We have previously shown that melanoma cells lack GILT, and are unable to optimally process disulfide or cysteine containing Ags/peptides to stimulate CD4+ T cells [19, 43]. Here, we have investigated whether GILT expression in melanoma enhances the processing and generation of epitopes from Ags regardless of disulfides or cysteines in Ags/epitopes. Using a number of model Ags such as human IgG, human serum albumin (HSA), and type II collagen (CII), we found that GILT influences Ag processing and CD4+ T cell recognition (Fig. 1a). Data showed that GILT expression in melanoma cells significantly enhanced κ188–203, κ145–159, HSA64–76 and CII261–273 epitope presentation when the cells were incubated with 10–20 μM of Ag (P < 0.05). While GILT may influence cysteine containing κ188–203 epitope presentation because of its reductase activity, κ145–159, HSA64–76 and CII261–273 epitopes do not contain any cysteine residues. Thus, GILT expression may modulate Ag processing and epitope generation in tumor cells. In parallel experiments, we performed western blot analysis which confirmed that J3.DR4 and 1359-mel cells lacked GILT and that those transfected with GILT cDNA (J3.DR4.GILT and 1359-mel.GILT) expressed high levels of GILT (Fig. 1b). Enhanced Ag processing and presentation was also observed in another melanoma cell line (Sk-mel-31) when transfected with GILT (19, data not shown). Taken together, these data suggest that GILT plays an important role in the class II pathway enhancing Ag processing and presentation.
Fig. 1.
Role of GILT in antigen processing and immune recognition of human melanoma cells in the context of HLA class II molecules. Melanoma cell lines J3.DR4, J3.DR4.GILT, 1359-mel, 1359-mel.GILT were incubated with either Igk, HSA or CII overnight, followed by coculture with 2.18a, 1.21, 17.9 and 4027/99 T cell hybridomas specific for κ188–203, κ145–159, HSA64–76 and CII261–273 epitopes, respectively. Cell supernatants were assayed for measurement of IL-2 using the HT-2 cell line as described in methods. a Processing of Igk, HSA and CII antigens by J3.DR4 ± GILT and 1359-mel ± GILT cells, and functional presentation of κ188–203, κ145–159, HSA64–76 and CII261–273 epitopes to CD4+ T cells. Data are representative of at least three separate experiments and expressed as mean ccpm ± SEM of triplicate counts. *Indicates that the data are statistically significant. b Expression of GILT in J3.DR4 ± GILT and 1359-mel ± GILT cells. Cell lysates from melanoma cells 1359-mel and J3.DR4, and those transfected with GILT cDNA 1359-mel.GILT and J3.DR4.GILT, were analyzed by western blotting using GILT-specific antibody (JSB-GILT). The wild-type B-cell line Frev, which constitutively expresses GILT, was used as a control. The blot was reprobed with β-actin to confirm equal loading of proteins from the lysates. Data are representative of at least three separate experiments
GILT expression minimally alters HLA class II protein levels in human melanoma cells
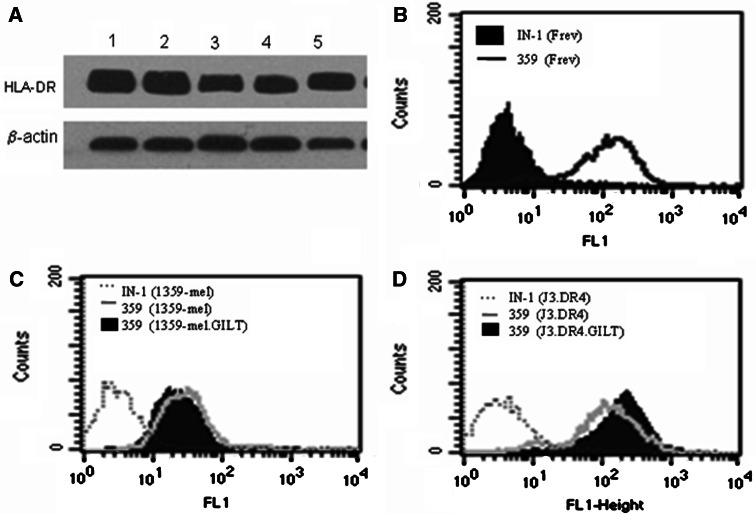
To investigate whether GILT expression in melanoma cells altered HLA protein expression and enhanced epitope presentation, we first measured steady-state class II protein expression in cells plus or minus GILT by western blot analysis. Data showed that GILT expression did not significantly alter class II protein levels in melanoma cells 1359-mel, 1359-mel.GILT, J3.DR4 and J3.DR4.GILT (Fig. 2a). We then measured cell surface class II protein expression by flow cytometric analysis and expressed as percentages (%) and mean fluorescence intensities (MFI). The B-cell line Frev, which constitutively expresses HLA-DR4 was used as a control (Fig. 2b). Data showed that the cell surface DR4 expression on 1359-mel cells was not significantly altered upon GILT expression [1359 = 87.2% (MFI:72.6) versus 1359.GILT = 84.6% (MFI:71.1)] (Fig. 2c). In J3.DR4.GILT cells, a slight increase in cell surface class II molecules was detected when compared with those of J3.DR4 cells [J3 = 95.4% (MFI:92.5) vs. J3.GILT = 99.1% (MFI:101.2)] (Fig. 2d). These data suggest that GILT expression in melanoma cells only minimally affect HLA class II protein expression on the cell surface.
Fig. 2.
Expression of HLA class II proteins in human melanoma cells plus or minus GILT. a Western blot analysis of lysates (90 μg) from J3.DR4 (lane 1), J3.DR4.GILT (lane 2), 1359-mel (lane 3) and 1359-mel.GILT (lane 4) cells was performed using HLA-DR-specific antibody (L243) to measure steady-state class II protein expression. The wild-type B-cell line Frev (50 μg), which constitutively expresses high levels of class II-DR, was used as a control (lane 5). b–d Flow cytometric analysis of cell surface protein (HLA-DR4) expression on melanoma ± GILT. Cells were also stained with antibody against DR4 (359-F10) plus FITC-labeled secondary antibody and isotype control (IN-1). Data are representative of at least four separate experiments
GILT expression alters acidic cysteine proteases in human melanoma cells
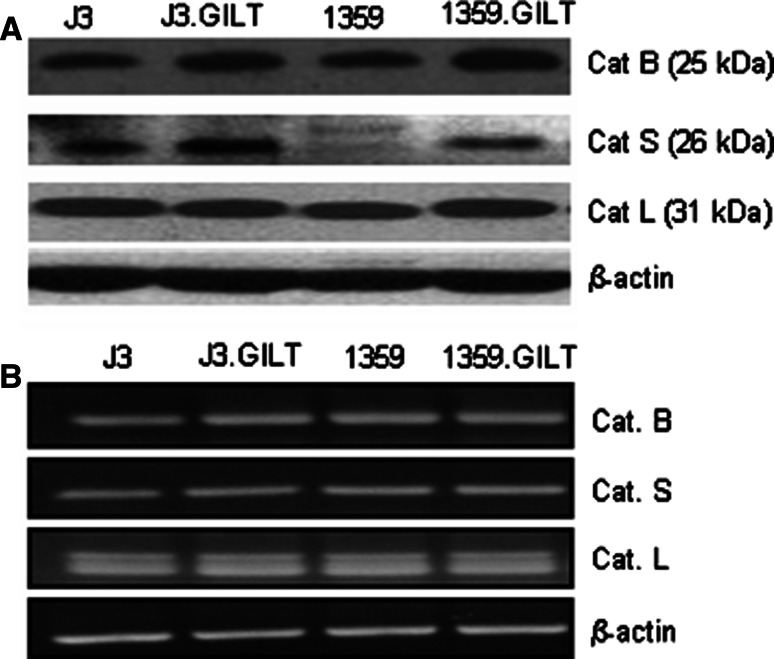
Because class II protein expression in human melanoma cells remained unchanged, we tested whether GILT expression altered intracellular acidic proteases that process Ags and generate epitopes for class II-mediated presentation. We examined cysteine proteases, cathepsins B, L and S. Cat B, active form 25 to 31 kDa, is crucial to the processing of Ags and has also been shown to influence class II Ag presentation [44, 45]. Cat S, active form (25–31 kDa), is crucial to one of the most important steps along the class II pathway, degradation of the Ii chain [44–46]. Without the removal of the Ii from the binding site on class II molecules, no peptides can be loaded onto the molecule for transport to the cell surface and subsequent T cell recognition. Cat L, active form (25–35 kDa), also participates in the cleavage of the Ii. It is believed that Cat S and Cat L perform the same function but in different types of cells [44–46]. Cat S in bone marrow-derived cells and Cat L in medullary thymic epithelial cells. However, it is also possible that each has a specific role in a specific stage of Ii degradation. All of these proteases are necessary for proper functioning of the class II pathway. Western blot analysis showed that GILT enhanced [J3 = 0.91 vs. J3.GILT = 1.2; 1359 = 0.92 vs 1359.GILT = 1.43, (P < 0.05)] the expression of the active forms of the cysteinyl cathepsins, B (25 kDa) as analyzed by densitometry and normalized with β-actin (Fig. 3a). GILT also enhanced [(J3 = 0.76 vs. J3.GILT = 0.97; 1359 = 0.87 vs. 1359.GILT = 1.2 (P < 0.05)] the expression of the active forms of the cysteinyl cathepsins, and S (26 kDa) (Fig. 3a, second row). GILT expression also slightly increased the active forms of Cat L (31 kDa) in 1359-mel.GILT compared to those in 1359-mel cells [1359 = 0.91 vs. 1359.GILT = 0.96, (P < 0.1)], but did not alter mature Cat L levels in J3.DR4/J3.DR4.GILT cells (J3 = 0.91 vs. J3.GILT = 0.91) (Fig. 3a, third row). Up-regulation of Cat B and S was also observed in melanoma cells Sk-mel-31 when transfected with GILT (19, data not shown). These data suggest that the presence of GILT results in increased levels of the catalytically active form of the cysteinyl cathepsins within melanoma cells that might enhance Ag processing and epitope presentation via class II molecules. RT-PCR analysis showed that Cat B, S and L mRNA levels remained unchanged in melanoma ± GILT cells (Fig. 3b). Quantitative PCR analyses confirmed that cathepsins mRNA levels were not significantly altered in cells ± GILT (supplementary Fig. 1), suggesting that GILT expression may not influence transcriptional regulation of cysteinyl cathepsins. Treatment of 1359.GILT cells with the transcriptional inhibitor actinomycin D (5 μg/ml for 24 h period) did not alter Cat B and Cat S protein levels (data not shown), further suggesting that GILT did not affect transcriptional regulation of cathepsins, rather GILT interacted with the proteins and peptide processing.
Fig. 3.
Influence of GILT on the expression of active forms of cysteinyl cathepsins B, S and L in human melanoma cells. a Cell lysates obtained from the melanoma cell lines J3.DR4, J3.DR4.GILT, 1359-mel and 1359-mel.GILT cells were analyzed by Western blotting for the expression of the cysteinyl proteases Cat B, Cat S and Cat L. Each lane was loaded with 60 μg of protein from the cell lysates. The membrane was reprobed with β-actin to confirm equal loading of proteins from the lysates. Data showed that GILT enhanced the expression of the active forms cysteinyl cathepsins B and S, and are representative of at least three separate experiments. b Analysis of Cat B, S and L mRNA by RT-PCR. Total RNA extracted from J3.DR4, J3.DR4.GILT, 1359-mel and 1359-mel.GILT cells were reverse-transcribed, amplified, and separated by electrophoresis. Specific products were detected in these cells by RT-PCR using the Cat B, S, and L primers as described in the “Materials and methods”. The amplification of housekeeping gene β-actin was included as a control
GILT expression alters acidic aspartyl proteases in human melanoma cells
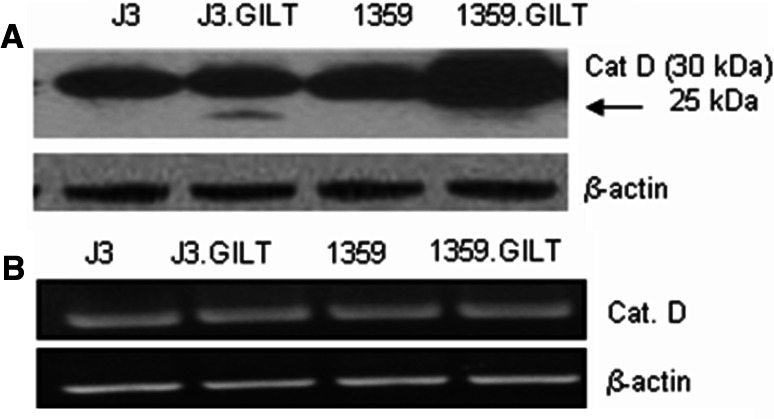
Cat D, active form (25–30 kDa), is another intracellular acidic aspartyl protease crucial to Ag processing [44, 47]. Once the Ag has been uptaken by APC, Cat D processes the Ag into usable peptides through selective cleavage. Without Cat D, the functional epitopes necessary for T cell stimulation would remain buried within the complex structure of the large Ag. Cat D, along with other intracellular proteases, cleaves Ags into small peptide fragments for loading onto the class II molecules. Only appropriately sized peptides can be loaded onto the HLA class II binding groove. Here, data showed that an increased expression of the active form of aspartyl protease Cat D (25 kDa) is generated in GILT-expressing melanoma cells (Fig. 4a). 1359-mel.GILT cells expressed high levels of active Cat D (30 kDa) compared to those found in 1359-mel cells [1359 = 1.18 versus 1359.GILT = 1.6, (P < 0.05)]. J3.DR4.GILT cells also expressed slightly higher levels of active Cat D protein [J3 = 1.10 vs. J3.GILT = 1.14 (P < 0.1)], but it was not statistically significant. However, data showed another active form of Cat D (25 kDa) in both J3.DR4 and 1359-mel cells expressing GILT (Fig. 4a, arrow). Elevation of active forms of cathepsins was also detected in Sk-mel-31 cells (data not shown), suggesting that GILT enhances the processing of Cat D. These data also suggest that the generation of active forms of Cat D (25 and 30 kDa) in conjunction with the ability of GILT to break the disulfide bonds holding Ags together may lead to more efficient processing of Ags. We also performed RT-PCR and quantitative PCR analysis of Cat D mRNA. Data showed that the Cat D mRNA levels in melanoma cells ± GILT remained unchanged (Fig. 4b, supplementary Fig. 1). Treatment of 1359-mel.GILT cells with or without actinomycin D did not show a significant effect on Cat D protein expression (data not shown), suggesting that GILT may influence posttranslational regulation of acidic cysteinyl and aspartyl cathepsins, thereby modulating Ag processing and presentation.
Fig. 4.
Influence of GILT on the expression of the active forms of aspartyl protease, cathepsin D in human melanoma cells. a Cell lysates obtained from the melanoma cell lines J3.DR4, J3.DR4.GILT, 1359-mel and 1359-mel.GILT were analyzed by western blotting for the expression of the aspartyl protease Cat D. Each lane was loaded with 60 μg of protein from the cell lysates. Melanoma cells containing GILT expressed the active forms of Cat D (arrow). GILT-expressing cells also altered another active form of Cat D (dark band, 30 kDa). β-actin served as a loading control. Results shown are representative of at least three separate experiments. b Analysis of Cat D mRNA by RT-PCR. Total RNA extracted from J3.DR4, J3.DR4.GILT, 1359-mel and 1359-mel.GILT cells were reverse-transcribed, amplified, and separated by electrophoresis. Specific products were detected in these cells by RT-PCR using the Cat D primers as described in the methods. The amplification of β-actin was included as a control
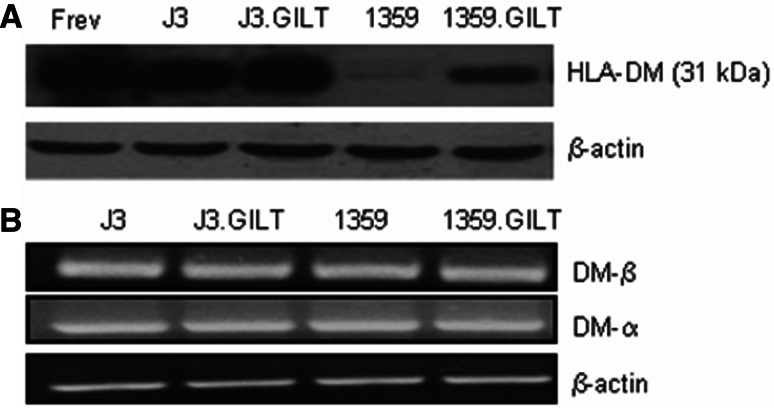
GILT expression modulates the peptide editor HLA-DM in human melanoma cells
Sustained anti-tumor immunity is dependent on the activation of CD4+ T cells, which recognize class II-restricted tumor peptides. Ag processing and presentation by the class II pathway is dependent on the key immune components such as the chaperon molecule HLA-DM, which catalyzes the removal of the CLIP fragment from the binding groove of class II molecules [29]. While DM-dependent or independent peptide presentation by class II proteins has been documented, DM expression mainly favors peptide loading, enhancing Ag presentation. Interestingly, we found that GILT expression up-regulated (J3 = 1.2 vs. J3.GILT = 1.8; 1359 = 1.91 vs. 1359.GILT = 2.06) DM molecules in melanoma cells J3.DR4.GILT and 1359-mel.GILT (Fig. 5a). These data suggest that high levels of DM in GILT-positive cells may influence epitope loading and presentation by the class II pathway. GILT expression did not alter DM mRNA levels in melanoma cells in the presence or absence of GILT (Fig. 5b). Quantitative PCR analyses also showed that DM mRNA levels were not significantly altered in melanoma cells ± GILT (data not shown), further suggesting that GILT expression does not regulate DM molecules at the transcriptional level. Although another non-classical HLA accessory molecule, DO, may influence epitope presentation by the class II pathway, we did not observe any detectable DO expression in melanoma cells ± GILT (data not shown). Taken together, these data suggest that Ag processing and epitope selection are possibly modulated by GILT and DM in melanoma cells, where GILT expression may favorably regulate dominant epitope presentation enhancing CD4+ T cell recognition.
Fig. 5.
Influence of GILT on the expression of the peptide editor HLA-DM in human melanoma cells. Cell lysates obtained from the melanoma cell lines J3.DR4, J3.DR4.GILT, 1359-mel and 1359-mel.GILT were analyzed by western blotting for the expression of HLA-DM. Each lane was loaded with 60 μg of protein from the cell lysates. The B-cell line Frev constitutively expresses DM molecules and was used as a control. Data are representative of at lest three separate experiments, and clearly showed that GILT expression in melanoma cells significantly elevated HLA-DM proteins. b RNA isolated from melanoma cells J3.DR4, J3.DR4.GILT, 1359-mel and 1359-mel.GILT were analyzed by RT-PCR for DM α and DM β mRNA. The amplification of β-actin was included as a control
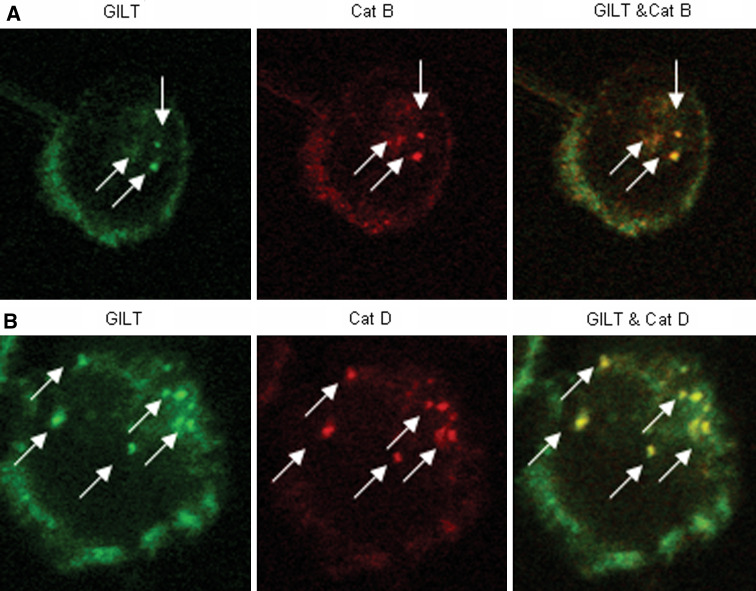
GILT colocalizes with Cat B and Cat D in human melanoma cells
There is increasing evidence that cysteinyl and aspartyl cathepsins are involved in proteolytic steps required for HLA class II-restricted antigen presentation. Thus, GILT may cooperate with cysteinyl and aspartyl cathepsins to enhance cathepsin protein processing, thereby up-regulating active forms of Cat B and Cat D, which are critically involved in antigen processing. The intracellular localization of GILT and Cat B/D in melanoma cells was examined by live confocal microscopy (Fig. 6). Data showed that GILT was colocalized with Cat B and D (arrows) in the endolysosomal compartments of melanoma cells. This data suggests that the colocalization of GILT and cathepsins may influence Ag processing and epitope generation for presentation via HLA class II molecules.
Fig. 6.
GILT colocalizes with cysteinyl and aspartyl cathepsins in the endolysosomal compartments of melanoma cells. 1359-mel.GILT cells were stained with either rabbit anti-cathepsin B or D, and costained with goat anti-GILT, followed by rhodamine-conjugated anti-rabbit IgG and FITC-conjugated anti-goat IgG antibodies. Live images were acquired by using a Leica TCS SP2 AOBS confocal system and processed with Leica software as described in the methods. Colocalization of GILT (green) with Cat B (a, red), Cat D (b, red) is indicated by yellow staining. Confocal images are representative of three separate experiments
Discussion
Professional APCs such as dendritic cells, macrophages, and B cells express high levels of GILT, whereas melanoma cells express either no GILT or very low levels of the enzyme [19]. We show this to be correlated to these cells’ ability to display functional class II peptide complexes on their cell surface, necessary for T cell recognition. While professional APCs are able to process Ags and stimulate T cells, malignant tumor cells often lack the proper machinery required to generate functional class II peptide complexes necessary for optimum CD4+ T cell responses [17, 19, 43]. Unraveling the machinery of the class II Ag presentation pathway in melanoma may provide promising targets for immunotherapy. In this study, we showed that GILT expression in melanoma cells enhanced class II-restricted antigen processing and presentation. Our data also showed an increase in the expression of the active forms of both cysteinyl and aspartyl cathepsins, B, S and D, in GILT-expressing melanoma cells. GILT expression also altered HLA-DM protein expression in melanoma cells. While the cathepsins and HLA-DM protein expressions were altered by GILT, mRNA levels were not significantly changed as demonstrated by PCR analysis. Confocal microscopic analysis showed that GILT colocalized with cystenyl and aspartyl cathepsins in melanoma cells, suggesting that GILT may influence the processing of cathepsins modulating Ag presentation. Up-regulation of active cathepsins, in conjunction with GILTs ability to break the disulfide bonds in Ags, may lead to more efficient processing of tumor Ags and CD4+ T cell responses [48, 49]. Investigating the role of GILT in direct Ag presentation by melanoma as well as other tumors may offer opportunities for developing new modalities of cancer immunotherapy.
Both exogenous and endogenous Ags including tumor Ags can get access to the endosomal/lysosomal compartments for processing and loading onto class II molecules. The predominant forms of cysteinyl and aspartyl proteases found in human melanoma were inactive proforms (data not shown). The elevation of active forms of Cat B and Cat D in melanoma by GILT is extremely important to the processing of IgG, HSA and CII Ags and the subsequent presentation of said Ags via class II molecules. The active forms of cathepsins B and S are enhanced by the presence of the GILT enzyme. This increase in the expression of the active forms of these molecules corresponds with an increase in the efficiency of Ag processing and presentation. This also suggests that there will be more functional epitopes available for class II loading and subsequent delivery to the cell’s surface. The ultimate result is an increased CD4+ T cell response. We also used GILT siRNA to determine whether the up-regulation of cathepsins observed, could be reversed by abolishing GILT expression in melanoma cells. Unfortunately, the commercially available GILT siRNA (Santa Cruz) did not inhibit GILT expression to a significant level that could be used to dissect cathepsins alteration by GILT in melanoma cells. But our studies with the melanoma cell lines plus or minus GILT confirmed that GILT alters cathepsins, which influence class II antigen processing. While melanoma cells express a variety of self Ags including tyrosinase, gp-100, and Mart-1 [12–15, 19], they seldom induce effective antitumor immune response. Enhanced Ag presentation by GILT-expressing cells could be exploited through immunotherapy, by allowing the in vitro generation of T cells specific against Ags expressed by melanoma.
Like the cysteinyl proteases, Cat Ds active form is enhanced by the presence of GILT with striking differences between the cells ± GILT. Utilizing site-specific editing, Cat D ensures that a functional epitope is generated from the protein. We also found that GILT expression in melanoma modulated the proforms of cathepsins B, L, S and D (data not shown). Confocal microscopy showed that GILT colocalizes with cysteinyl and aspartyl cathepsins, suggesting that the interaction of GILT and cathepsins may positively regulate class II antigen processing and presentation. Professional APC such as B cells express GILT [19] and with the up-regulation of Cat B and D, cells can efficiently generate antigenic peptides from ovalbumin or hen egg lysozyme that could be functionally presented to T cells [24, 26, 44]. Besides, experiments using splenocytes and macrophages prepared from Cat D-deficient mice showed that Cat D is dispensable for processing of a number of exogenous and endogenous antigens [17, 30, 31]. Thus the association of GILT and cathepsins may play a pivotal role in the HLA class II-restricted Ag processing and presentation. Another important component of the class II pathway, HLA-DM, surveys the class II DR complex to see if a correct epitope has arrived in the binding groove [28, 29]. Otherwise, DM replaces the incorrect peptide with a more suitable one [49, 50]. Peptides with higher binding affinities are usually correct and vice versa. Thus, DM acts as an enzyme that accelerates the forward and reverse reaction from unstable to stable class II-peptide complexes for immune recognition. The presence of GILT enhanced the expression of DM molecules, thus increasing the efficiency of Ag presentation and CD4+ T cell recognition. As one of the main weapons of tumor cells is its novel ability to evade the body’s defenses, any treatment option, which can ensure that more of the necessary defense signals reach the appropriate receptor cells represents a promising avenue for exploration.
Our results show that when GILT is inserted into malignant melanoma cells, there is an increase in Ag processing and epitope presentation in the context of class II molecules. We also show that the insertion of GILT leads to a greater population of the active forms of the cathepsins as well as the peptide editor DM. GILT may help processing of acidic cathepsins as they colocalize in the endocytic compartments thereby influencing Ag processing and presentation via the HLA class II pathway. This study suggests that the regulation of DM as well as acidic proteases by GILT could lead to “self-help”, with cells acting as their own defense against cancer. If Ag presentation can be enhanced within tumor cells, in conjunction with the presence of more functional peptides, then, cancerous cells have a greater chance of being recognized by T cells. This strategy would utilize the body’s own defense thus increasing the likelihood of prolonged immunity to reemerging tumor cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 GILT expression did not alter cathepsins mRNA levels in melanoma cells. RNA isolated from melanoma cells 1359-mel and 1359-mel.GILT were analyzed by real time RT-PCR for cathepsins B, D, L and S mRNA. Relative levels of Cat mRNA expression were normalized in all the samples analyzed with respect to the levels of GAPDH amplification. Cat B, D, L and S mRNA expression levels were presented as fold increase over control. (TIF 98 kb)
Acknowledgments
We are grateful to Dr. Janice Blum (Indiana University, Indianapolis) for providing us with cell lines, antibodies and reagents. We also thank Dr. P. Cresswell (Yale University) and Patrick W. O’Donnell (Indiana University) for GILT-expressing melanoma cells. This work was supported by grants from the Leukemia and Lymphoma Society (No. 3024), ACS-IRG (No. 85241), Hollings Cancer Center Seed Grant (GC-3319-05-4498CM) to A.H., and the NIH grant DE12603 (S.V.R).
References
- 1.Gloster HM, Jr, Neal K. Skin cancer in skin of color. J Am Acad Dermatol. 2006;55:741–760. doi: 10.1016/j.jaad.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 2.O’Day S, Boasberg P. Management of metastatic melanoma. Surg Oncol Clin N Am. 2006;15:419–437. doi: 10.1016/j.soc.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Ross M. New American joint commission on cancer staging system for melanoma: prognostic impact and future directions. Surg Oncol Clin N Am. 2006;15:341–352. doi: 10.1016/j.soc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Francis SO, Mahlberg MJ, Johnson KR, Ming ME, Dellavalle RP. Melanoma chemoprevention. J Am Acad Dermatol. 2006;55:849–861. doi: 10.1016/j.jaad.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Hersey P, Zhuang L, Zhang XD. Current strategies in overcoming resistance of cancer cells to apoptosis melanoma as a model. Int Rev Cytol. 2006;251:131–158. doi: 10.1016/S0074-7696(06)51004-6. [DOI] [PubMed] [Google Scholar]
- 6.Queirolo P, Acquati M. Targeted therapies in melanoma. Cancer Treat Rev. 2006;32:524–531. doi: 10.1016/j.ctrv.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Burmeister BH, Mark Smithers B, Burmeister E, Baumann K, Davis S, Krawitz H, Johnson C, Spry N. A prospective phase II study of adjuvant postoperative radiation therapy following nodal surgery in malignant melanoma-Trans Tasman Radiation Oncology Group (TROG) Study 96.06. Radiother Oncol. 2006;81:136–142. doi: 10.1016/j.radonc.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Young SE, Martinez SR, Essner R. The role of surgery in treatment of stage IV melanoma. J Surg Oncol. 2006;94:344–351. doi: 10.1002/jso.20303. [DOI] [PubMed] [Google Scholar]
- 9.Saleh F, Renno W, Klepacek I, Ibrahim G, Asfar S, Dashti H, Romero P, Dashti A, Behbehani A. Melanoma immunotherapy: past, present, and future. Curr Pharm Des. 2005;11:3461–3473. doi: 10.2174/138161205774414529. [DOI] [PubMed] [Google Scholar]
- 10.Lizee G, Radvanyi LG, Overwijk WW, Hwu P. Immunosuppression in melanoma immunotherapy: potential opportunities for intervention. Clin Cancer Res. 2006;12:2359s–2365s. doi: 10.1158/1078-0432.CCR-05-2537. [DOI] [PubMed] [Google Scholar]
- 11.van der Bruggen P, Van den Eynde BJ. Processing and presentation of tumor antigens and vaccination strategies. Curr Opin Immunol. 2006;18:98–104. doi: 10.1016/j.coi.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Storkus WJ, Zarour HM. Melanoma antigens recognised by CD8+ and CD4+ T cells. Forum (Genova) 2000;10:256–270. [PubMed] [Google Scholar]
- 13.Phan GQ, Touloukian CE, Yang JC, Restifo NP, Sherry RM, Hwu P, Topalian SL, Schwartzentruber DJ, Seipp CA, Freezer LJ, Morton KE, Mavroukakis SA, White DE, Rosenberg SA. Immunization of patients with metastatic melanoma using both class I- and class II-restricted peptides from melanoma-associated antigens. J Immunother. 2003;26:349–356. doi: 10.1097/00002371-200307000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang RF. Enhancing antitumor immune responses: intracellular peptide delivery and identification of MHC class II-restricted tumor antigens. Immunol Rev. 2002;188:65–80. doi: 10.1034/j.1600-065X.2002.18807.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang RF, Zeng G, Johnston SF, Voo K, Ying H. T cell-mediated immune responses in melanoma: implications for immunotherapy. Crit Rev Oncol Hematol. 2002;43:1–11. doi: 10.1016/S1040-8428(01)00186-X. [DOI] [PubMed] [Google Scholar]
- 16.Poehlein CH, Ruttinger D, Ma J, Hu HM, Urba WJ, Fox BA. Immunotherapy for melanoma: the good, the bad, and the future. Curr Oncol Rep. 2005;7:383–392. doi: 10.1007/s11912-005-0066-1. [DOI] [PubMed] [Google Scholar]
- 17.Marks MS, Theos AC, Raposo G. Melanosomes and MHC class II antigen-processing compartments: a tinted view of intracellular trafficking and immunity. Immunol Res. 2003;27:409–426. doi: 10.1385/IR:27:2-3:409. [DOI] [PubMed] [Google Scholar]
- 18.Haque A, Blum JS. New insights in antigen processing and epitope selection: development of novel immunotherapeutic strategies for cancer, autoimmunity and infectious diseases. J Biol Regul Homeost Agents. 2005;19:93–104. [PubMed] [Google Scholar]
- 19.Haque MA, Li P, Jackson SK, Zarour HM, Hawes JW, Phan UT, Maric M, Cresswell P, Blum JS. Absence of gamma-interferon-inducible lysosomal thiol reductase in melanomas disrupts T cell recognition of select immunodominant epitopes. J Exp Med. 2002;195:1267–1277. doi: 10.1084/jem.20011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busch R, Rinderknecht CH, Roh S, Lee AW, Harding JJ, Burster T, Hornell TM, Mellins ED. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–260. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 21.Mandic M, Castelli F, Janjic B, Almunia C, Andrade P, Gillet D, Brusic V, Kirkwood JM, Maillere B, Zarour HM. One NY-ESO-1-derived epitope that promiscuously binds to multiple HLA-DR and HLA-DP4 molecules and stimulates autologous CD4+ T cells from patients with NY-ESO-1-expressing melanoma. J Immunol. 2005;174:1751–1759. doi: 10.4049/jimmunol.174.3.1751. [DOI] [PubMed] [Google Scholar]
- 22.Turk V, Turk B, Guncar G, Turk D, Kos J. Lysosomal cathepsins: structure, role in antigen processing and presentation, and cancer. Adv Enzyme Regul. 2002;42:285–303. doi: 10.1016/S0065-2571(01)00034-6. [DOI] [PubMed] [Google Scholar]
- 23.Moss CX, Villadangos JA, Watts C. Destructive potential of the aspartyl protease cathepsin D in MHC class II-restricted antigen processing. Eur J Immunol. 2005;35:3442–3451. doi: 10.1002/eji.200535320. [DOI] [PubMed] [Google Scholar]
- 24.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 25.Rudensky A, Beers C. Lysosomal cysteine proteases and antigen presentation. Ernst Schering Res Found Workshop. 2006;56:81–95. doi: 10.1007/3-540-37673-9_5. [DOI] [PubMed] [Google Scholar]
- 26.Bryant PW, Lennon-Dumenil AM, Fiebiger E, Lagaudriere-Gesbert C, Ploegh HL. Proteolysis and antigen presentation by MHC class II molecules. Adv Immunol. 2002;80:71–114. doi: 10.1016/S0065-2776(02)80013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon AM, Stanley BJ, Matthews EE, Dawson JP, Engelman DM. Invariant chain transmembrane domain trimerization: a step in MHC class II assembly. Biochemistry. 2006;45:5228–5234. doi: 10.1021/bi052112e. [DOI] [PubMed] [Google Scholar]
- 28.Stern LJ, Potolicchio I, Santambrogio L. MHC class II compartment subtypes: structure and function. Curr Opin Immunol. 2006;18:64–69. doi: 10.1016/j.coi.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Sant AJ, Chaves FA, Jenks SA, Richards KA, Menges P, Weaver JM, Lazarski CA. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II: peptide complexes. Immunol Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 30.Overwijk WW, Restful NP. Autoimmunity and the immunotherapy of cancer: targeting the “self” to destroy the “other”. Crit Rev Immunol. 2000;20:433–450. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, Maekawa Y, Hanba J, Dainichi T, Nashed BF, Hisaeda H, Sakai T, Asao T, Himeno K, Good RA, Katunuma N. Lysosomal cathepsin B plays an important role in antigen processing, while cathepsin D is involved in degradation of the invariant chain in ovalbumin-immunized mice. Immunology. 2000;100:13–20. doi: 10.1046/j.1365-2567.2000.00000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 2005;307:1630–1634. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 33.O’Donnell PW, Haque A, Klemsz MJ, Kaplan MH, Blum JS. Cutting edge: induction of the antigen-processing enzyme IFN-gamma-inducible lysosomal thiol reductase in melanoma cells Is STAT1-dependent but CIITA-independent. J Immunol. 2004;173:731–735. doi: 10.4049/jimmunol.173.2.731. [DOI] [PubMed] [Google Scholar]
- 34.Pathak SS, Blum JS. Endocytic recycling is required for the presentation of an exogenous peptide via MHC class II molecules. Traffic. 2000;1:561–569. doi: 10.1034/j.1600-0854.2000.010706.x. [DOI] [PubMed] [Google Scholar]
- 35.Haque MA, Hawes JW, Blum JS. Cysteinylation of MHC class II ligands: peptide endocytosis and reduction within APC influences T cell recognition. J Immunol. 2001;166:4543–4551. doi: 10.4049/jimmunol.166.7.4543. [DOI] [PubMed] [Google Scholar]
- 36.Hiraiwa A, Yamanaka K, Kwok WW, Mickelson EM, Masewicz S, Hansen JA, Radka SF, Nepom GT. Structural requirements for recognition of the HLA-Dw14 class II epitope: a key HLA determinant associated with rheumatoid arthritis. Proc Natl Acad Sci USA. 1990;87:8051. doi: 10.1073/pnas.87.20.8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haque A, Das A, Hajiaghamohseni LM, Younger A, Banik NL, Ray SK. Induction of apoptosis and immune response by all-trans retinoic acid plus interferon-gamma in human glioblastoma T98G and U87MG cells. Cancer Immunol Immunother. 2007;56:615–625. doi: 10.1007/s00262-006-0219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundaram K, Nishimura R, Senn J, Youssef RF, London SD, Reddy SV. RANK ligand signaling modulates matrix metalloproteinase-9 gene expression during osteoclast differentiation. Exp Cell Res. 2007;313:168–178. doi: 10.1016/j.yexcr.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM. NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res. 2000;60:4946–49452. [PubMed] [Google Scholar]
- 40.Dissanayake SK, Thompson JA, Bosch JJ, Clements VK, Chen PW, Ksander BR, Ostrand-Rosenberg S. Activation of tumor-specific CD4(+) T lymphocytes by major histocompatibility complex class II tumor cell vaccines: a novel cell-based immunotherapy. Cancer Res. 2004;64:1867–1874. doi: 10.1158/0008-5472.CAN-03-2634. [DOI] [PubMed] [Google Scholar]
- 41.Godefroy E, Scotto L, Souleimanian NE, Ritter G, Old LJ, Jotereau F, Valmori D, Ayyoub M. Identification of two Melan-A CD4+ T cell epitopes presented by frequently expressed MHC class II alleles. Clin Immunol. 2006;121:54–62. doi: 10.1016/j.clim.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Brady MS, Lee F, Petrie H, Eckels DD, Lee JS. CD4(+) T cells kill HLA-class-II-antigen-positive melanoma cells presenting peptide in vitro. Cancer Immunol Immunother. 2000;48:621–626. doi: 10.1007/s002620050010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P, Haque MA, Blum JS. Role of disulfide bonds in regulating antigen processing and epitope selection. J Immunol. 2002;169:2444–2450. doi: 10.4049/jimmunol.169.5.2444. [DOI] [PubMed] [Google Scholar]
- 44.Lennon-Dumenil AM, Bakker AH, Wolf-Bryant P, Ploegh HL, Lagaudriere-Gesbert C. A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol. 2002;14:15–21. doi: 10.1016/S0952-7915(01)00293-X. [DOI] [PubMed] [Google Scholar]
- 45.Riese RJ, Chapman HA. Cathepsins and compartmentalization in antigen presentation. Curr Opin Immunol. 2000;12:107–113. doi: 10.1016/S0952-7915(99)00058-8. [DOI] [PubMed] [Google Scholar]
- 46.Hsing LC, Rudensky AY. The lysosomal cysteine proteases in MHC class II antigen presentation. Immunol Rev. 2005;207:229–241. doi: 10.1111/j.0105-2896.2005.00310.x. [DOI] [PubMed] [Google Scholar]
- 47.Moss CX, Villadangos JA, Watts C. Destructive potential of the aspartyl protease cathepsin D in MHC class II-restricted antigen processing. Eur J Immunol. 2005;35:3442–3451. doi: 10.1002/eji.200535320. [DOI] [PubMed] [Google Scholar]
- 48.Ostrand-Rosenberg S. CD4+ T lymphocytes: a critical component of antitumor immunity. Cancer Invest. 2005;23:413–419. [PubMed] [Google Scholar]
- 49.Weber DA, Dao CT, Jun J, Wigal JL, Jensen PE. Transmembrane domain-mediated colocalization of HLA-DM and HLA-DR is required for optimal HLA-DM catalytic activity. J Immunol. 2001;167:5167–5174. doi: 10.4049/jimmunol.167.9.5167. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson MJ, Moradi B, Seth NP, Xing X, Cuny GD, Stein RL, Wucherpfennig KW. Small molecules that enhance the catalytic efficiency of HLA-DM. J Immunol. 2006;176:4208–4220. doi: 10.4049/jimmunol.176.7.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 GILT expression did not alter cathepsins mRNA levels in melanoma cells. RNA isolated from melanoma cells 1359-mel and 1359-mel.GILT were analyzed by real time RT-PCR for cathepsins B, D, L and S mRNA. Relative levels of Cat mRNA expression were normalized in all the samples analyzed with respect to the levels of GAPDH amplification. Cat B, D, L and S mRNA expression levels were presented as fold increase over control. (TIF 98 kb)