Abstract
The ceramide synthase (CerS) enzymes are key regulators of ceramide homeostasis. CerS1 is central to regulating C18 ceramide which has been shown to be important in cancer and the response to chemotherapeutic drugs. Previous work indicated that some drugs induced a novel and specific translocation of CerS1 from the endoplasmic reticulum to the Golgi apparatus. We now show that diverse stresses such as UV light, DTT, as well as drugs with different mechanisms of action induce CerS1 translocation. The stresses cause a specific cleavage of the CerS1 enzyme, and the cleavage is dependent on the action of the proteasome. Inhibition of proteasome function inhibits stress-induced CerS1 translocation, indicating that this proteolytic cleavage precedes the translocation. Modulation of protein kinase C activity shows that it plays a central role in regulating CerS1 translocation. Analysis of the C-terminus of the CerS1 protein shows that several KxKxx motifs are not involved in regulating stress induced translocation. The study suggests that diverse stresses initiate responses through different signaling pathways, which ultimately converge to regulate CerS1 localization. The data provide an increasingly detailed understanding of the regulation of this important enzyme in normal and stressed cells, and support the idea that it is uniquely regulated with respect to the other CerS enzymes.
Keywords: sphingolipids, cisplatin, stress
Introduction
Ceramide is an important bioactive sphingolipid which functions in many eukaryotic cellular processes including the response to cellular stress, cancer and cell death [1–3]. Ceramide often appears to work in opposition to another bioactive sphingolipid, sphingosine-1-phosphate (S-1-P), and the dynamic balance between these two molecules is a critical factor in regulating these functions [4]. Because of their central role in cell signaling, and as potential drug targets for cancer and other human diseases, there is a great deal of interest in these molecules, and thus there is a corresponding level of interest in the regulation of the enzymes that are responsible for their synthesis and degradation [3, 5, 6]. Six ceramide synthase enzymes (CerS) are responsible for the de novo synthesis of ceramide in mammalian cells [7], but little is yet known about the regulation of these enzymes which are central to structural and regulatory functions in all cells.
Each of the CerS enzymes is the product of a unique gene, and the six CerS each preferentially use fatty acyl-CoAs of different chain lengths, along with the long chain base sphinganine, to synthesize ceramides with correspondingly different chain lengths [8–12]. The different CerS enzymes have unique patterns of tissue expression, implying transcriptional regulation in different tissues [9, 13, 14]. Of the CerS enzymes, CerS1 is clearly structurally and functionally different from the others [7]. It has been shown that C18 ceramide, the primary product of CerS1, is reduced in head and neck cancer [15–17]. In addition, we have shown that CerS1 preferentially sensitizes cells to chemotherapeutic drugs [18], that it has a short half life, and that the steady state protein level is regulated by ubiquitination/proteasomal mediated turnover [19].
In an earlier study we discovered that CerS1 translocates from the endoplasmic reticulum (ER) to the Golgi apparatus in response to drug stress [18]. The translocation is robust, and is inducible by treatment with either the chemotherapeutic drug cisplatin or doxorubicin. The stress-induced ER to Golgi translocation is specific to CerS1, as neither the CerS4 nor the CerS5 enzyme translocate from the ER to the Golgi apparatus in response to the drugs. Moreover, ectopic expression of sphingosine kinase 1 blocked the translocation of CerS1 [18]. Studies of other sphingolipid metabolizing enzymes also have revealed that movement between intracellular compartments is a central theme in the regulation of sphingolipid biosynthesis and degradation [20, 21].
In this study we demonstrate that CerS1 translocates to the Golgi apparatus in response to diverse types of stresses representing different mechanisms of action. Following stress, CerS1 is processed into a C-terminal fragment encompassing about half of the protein. The stress-induced translocation of CerS1 is dependent on proteasome function, and is regulated by protein kinase C (PKC). Overall, this study reveals underlying molecular details of the fate of this important enzyme in response to stress, and its role in ceramide homeostasis.
Materials and Methods
Reagents
Cell culture reagents, Lipofectamine 2000, PureLink plasmid purification kit, goat anti-rabbit Alexa Fluor 488 antibody, goat anti-mouse Alexa Fluor 568, To-Pro 3-iodide and Prolong gold anti-fade reagent were from Invitrogen Corp., Carlsbad, CA. Fetal bovine serum (FBS) was from Atlanta Biologicals, Lawrenceville, GA. Mouse monoclonal anti-Golgi GM130 antibody (Ab) was purchased from BD Biosciences–Pharmingen, San Diego, CA. Mammalian expression vectors were from Stratagene, La Jolla, CA and DNA cloning enzymes were from Promega Corp., Madison, WI. Anti-FLAG M2 mouse monoclonal antibody (mAb), polyclonal rabbit anti-FLAG, polyclonal rabbit anti-HA Ab, cisplatin, doxorubicin and dithiothreitol (DTT) were from Sigma-Aldrich, St. Louis, MO. Phorbol-12-myristate-13-acetate (TPA) was from Cell Signaling, Danvers, MA. HRP conjugated-goat anti-mouse secondary Ab and chemiluminescent detection kits were from Pierce Biotechnology, Rockford, IL. bis-indolylmaleimide (BIM) was from LC Laboratories, Woburn, MA, and MG132 was from Calbiochem, San Diego, CA. Optitran BA-S 85 nitrocellulose membrane was from Whatman Schleicher and Schuell, Dassel, Germany.
Constructs
CerS1 was cloned in frame with a C-terminal 3x-FLAG tag (CerS1-FLAG); an N-terminal 3x-FLAG tag (FLAG-CerS1); both an N-terminal 3x-FLAG, C-terminal HA dual tag (FLAG-CerS1-HA); or a C-terminal HA-tag (CerS1-HA) as BamHI-EcoRI fragments into pCMV3tag-1A, pCMV3tag-3A or pcDNA3-HA. C-terminal deletions encompassing the last 5, 13, and 19 C-terminal amino acids of the mouse CerS1 were generated by amplifying CerS1 with a forward primer from the N-terminal end of the molecule (and which included a BamHI sequence) and the reverse primers, containing sequences upstream from the desired deletion and a terminal EcoRI site. All deletions were cloned into pCMV3tag-3A to generate C-terminal FLAG-tagged proteins.
Cell culture, transfection and generation of HEK 293 cell lines stably expressing CerS1-tagged proteins
HEK 293 cells were cultured in Dulbecco’s modified Eagle’s medium (MEM) supplemented with 10% FBS, 2 mM L-glutamine, non-essential amino acids, 100 IU/ml penicillin and 100 μg/ml streptomycin in a 5% CO2 incubator at 37 C. Human embryonic kidney 293 (HEK 293) cell lines stably expressing the various CerS1 constructs were generated by transfecting cells in 6 well plates with 4 μg purified plasmid DNA as above. After 48 h the cells were transferred to a 75 cm2 culture flask and were selected in the presence of G418 sulfate, gradually increasing from 100 to 500 μg/ml [18]. G418 resistant cell lines were assayed for CerS1 expression by immunoblotting using anti-FLAG mAb, directed against the FLAG epitope. The cell lines were tested for ceramide synthase activity, using sphinganine and C18 acyl CoA-the preferred substrate for CerS1–using the in vitro assay described below. The cell lines were stored frozen in FBS containing 10% DMSO under liquid nitrogen, and freshly thawed cultures were used for experiments.
Drug/stress treatment
Equal numbers of HEK 293 cell lines stably expressing the CerS1 protein, at 90% confluence, were treated with the indicated amounts of drugs for the indicated times. Cisplatin and doxorubicin were prepared in Pt-buffer (3 mM NaCl/1 mM Na-PO4, pH 6.4), and DTT in water. Solvent alone was added to control untreated wells. For UV treatment, cells were exposed to 254 nm UV light (germicical lamp ITT G15T8; 1sec = 1.6 J/m2) for the indicated time, followed by 3 h incubation, prior to staining. The level of incident fluence was measured with a Black Ray meter (UVP, San Grabriel, CA). In the case of drug (inhibitor/activator) co-treatment, cells were pre-treated with indicated amounts of the inhibitor/activator for 1 h prior to the application of stress. The stresses were used at the lowest dose for which it is possible to see a cellular response in a reasonable amount of time and which is consistent with many published reports (see [22–24]). These cytotoxic agents all function in a dose- and time-dependent manner.
Immunoblotting
At the end of drug/stress treatments, cells were quickly rinsed in ice-cold PBS and lysed by adding HEPES/digitonin lysis buffer (20 mM HEPES buffer, pH 7.4, 25 mM KCl, 250 mM sucrose, 1:100 protease inhibitor cocktail (2.5 mM phenylmethylsulphonyl fluoride, 0.01 μg/ml pepstatin A, 0.001 μg/ml leupeptin in methanol), 1% digitonin) directly to the dish. The lysates were scraped from the plates, collected in 1.5 ml tubes and centrifuged at 12,000 rpm in a Sorvall desktop centrifuge for 1 min to remove cell debris. Total protein in the supernatant was quantified using the Pierce BCA protein assay reagent according to the manufacturer’s instructions. For immunoblotting, equal amounts of protein per lane in 2X urea sample buffer (50 mM Tris-HCl pH 6.8, 1.6% SDS, 7% glycerol, 8 M urea, 4% β-mercaptoethanol, 0.016% bromophenol blue) were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 15% gels and transferred onto nitrocellulose membranes by standard techniques. CerS1 was detected with anti-FLAG mAb in 0.5X casein blocking buffer, and HRP-conjugated goat anti-mouse IgG secondary Ab. The blots were developed with a chemiluminescent detection kit. All immunoblotting experiments were repeated (biological replicates) and similar results were obtained.
Immunostaining and microscopy
Circular 12 mm borosilicate cover slips were dipped in absolute alcohol and flame sterilized, and were placed inside the wells of a 24-well culture plate. The cover slips were treated for 20 min at 37° C in 1 ml poly-L-lysine (to facilitate cell adhesion), rinsed several times with autoclaved distilled water and finally rinsed with DMEM. HEK 293 cells stably expressing various CerS1 constructs were then seeded onto the cover slips and allowed to adhere overnight. After drug or stress treatment, the medium was removed from the wells, and the cover slips were rinsed once with PBS. Cells were fixed for 30 min with 4% paraformaldehyde in PBS. After rinsing 3x with PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS for 20 min. This was followed by preincubation with 0.1% BSA for 1 h and blocking with goat blocking buffer (GBB; 5% BSA, 5% normal goat serum, 0.1% Micro-O-protect in PBS) for 2 h. To detect CerS1 co-localization with the Golgi apparatus cells were stained overnight with primary antibody (rabbit polyclonal anti-FLAG, or rabbit polyclonal anti-HA (1:600 in 10% GBB)), together with anti-Golgi GM130 mAb (1:500), followed by secondary antibody (goat anti-rabbit Alexa Fluor 488 or goat anti-mouse Alexa Fluor 568 (1:600 in 10% GBB)). The nuclei were then stained for 20 min with 10 μM To-Pro 3-iodide. After the final washes, the cover slips were mounted on slides in Prolong gold anti-fade reagent. After overnight incubation in 4° C, the edges of the cover slips were sealed with nail polish. Radiance 2000 confocal system (Bio-rad Cell Sciences–Carl Zeiss Microimaging Inc., Germany) coupled to an Olympus 1X70 inverted microscope (Olympus America, Inc., Melville, NY) was used for confocal imaging by methods described elsewhere [18]. All experiments were performed multiple times.
Ceramide synthase assay
In vitro ceramide synthase 1 assays were performed as described [8, 18]. Briefly, pellets were resuspended in 1 ml homogenization buffer, homogenized as described above, and the protein concentrations were determined using the Bradford reagent. Homogenates (50 μg protein in 0.25 ml final volume) were incubated with 0.25 μCi of [4,5-3H] sphinganine/15 μM sphinganine/20 μM defatted BSA, and 50 μM C18 fatty acyl CoA for 20 min at 37° C. Lipids were extracted, and the level of dihydroceramide synthesis was analyzed by TLC, using chloroform/methanol/2 M ammonium hydroxide (40:10:1; v/v/v) as the developing solvent, and using N-stearoyl-sphingosine as an authentic standard. Lipids were visualized using a phosphorimager (Fuji), recovered from the TLC plates by scraping the silica directly into scintillation vials, and quantified by liquid scintillation counting. Specific enzyme activity is expressed as picomoles of dihydroceramide synthesized per min per mg protein. The assays were done multiple times and standard deviations were calculated using the statistical tools in Microsoft Excel.
Results
CerS1 translocation is a general response to stress
We previously demonstrated that two chemotherapeutic drugs, cisplatin (which forms bulky adducts on DNA by crosslinking adjacent purines) and doxorubicin (which intercalates DNA), induce a robust and specific translocation of C-terminal HA-tagged CerS1 (CerS1-HA), but not CerS4 or CerS5, from its normal ER location to the Golgi apparatus [18]. To determine the generality of this stress-response of CerS1, we now tested the effect of two other very different stresses, UV-light (which causes pyrimidine dimer formation) or DTT (which induces the unfolded protein response).
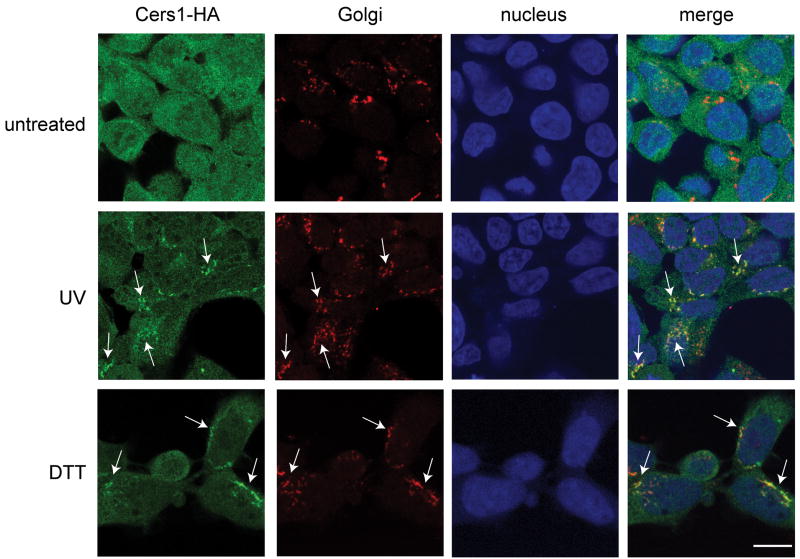
Figure 1 shows that treating HEK 293 cells, stably expressing CerS1-HA, with DTT resulted in translocation of CerS1-HA to the Golgi apparatus. The green channel shows an intense concentration of HA-tagged CerS1 after DTT treatment (indicated by arrows) in the peri-nuclear region (nuclei shown in blue channel), which co-localizes with the Golgi apparatus shown in the red channel. The merged images show this co-localization clearly as yellow areas (indicated by arrows). Similarly, UV-light treatment of the cells results in the translocation of CerS1-HA from the ER to Golgi apparatus. Again there is an intense concentration of HA-tagged CerS1 shown in the green channel (indicated by arrows) to the peri-nuclear region which co-localizes with the Golgi apparatus shown in the red channel, and this shows as yellow in the merged images (indicated by arrows). Parallel control untreated cells do not show translocation and concentration of CerS1-HA from the ER to the Golgi apparatus. In distinct contrast to the results with CerS1-HA, neither UV light nor DTT caused translocation of C-terminally tagged CerS4-HA or CerS5-HA in HEK cells expressing these genes (Supplemental Figures 1 and 2). Overall, the results extend those previously reported with cisplatin and doxorubicin [18]. These experiments show that the ER to Golgi translocation is specific to CerS1 and is not a limited response to chemotherapeutic drugs, but that a wide variety of stresses result in the specific translocation of CerS1 to the Golgi apparatus. It should be noted that even without exogenous stress, some low basal level of ER to Golgi translocation is observed, and that stress-induced translocation is 75–80% under these stress conditions (see quantitiation in Figures 6 and 7).
Figure 1. UV light and DTT induce translocation of the CerS1 protein to the Golgi apparatus.
HEK 293 cells expressing CerS1 C-terminally tagged with HA (CerS1-HA) were stressed with either 5 mM DTT for 30 minutes or UV light for 90 seconds (UV intensity, 1 sec = 1.6 J/m2), and analyzed after 30 minutes for the localization of CerS1 protein. Cells were stained with rabbit anti-HA polyclonal antibody (green) and anti-Golgi GM130 mAb (red). Translocation of CerS1-HA is indicated with arrows and co-localization with the Golgi is shown in the merged images (yellow). Size bar–20 μm.
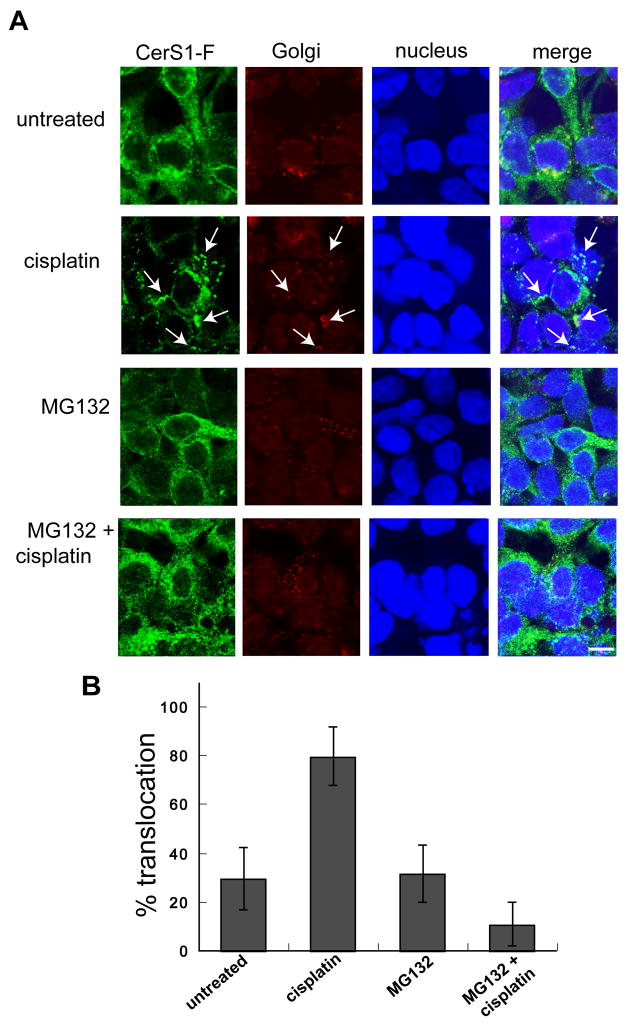
Figure 6. The proteasome-inhibitor MG132 prevents stress induced ER to Golgi translocation of CerS1-FLAG.
A. HEK 293 cells stably expressing C-terminally tagged CerS1-FLAG were treated with 50 μM cisplatin for 3 h in the presence or absence of 1 h pre-treatment with 20 μM proteasome-inhibitor MG132, and were examined for CerS1-FLAG translocation (see arrows) using rabbit anti-FLAG polyclonal Ab (green) and anti-Golgi GM130 mAb (red). Cisplatin-induced translocation of CerS1 (arrows) was inhibited by the presence of MG132, indicating that the proteasomal processing event is involved in CerS1 translocation (Size bar–20 μm). B. A quantitative representation of the above experiment. Twenty fields of approximately 20 cells were scored, and the values are the percent of cells that show stress-induced CerS1 translocation from the ER to the Golgi apparatus.
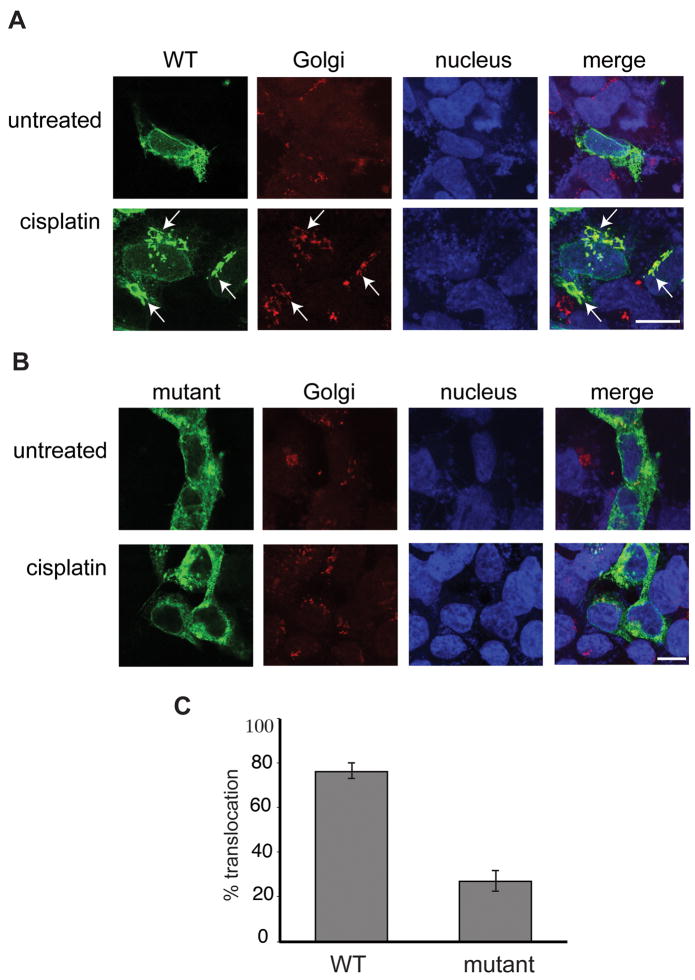
Figure 7. The catalytically inactive CerS1 mutant shows decreased Golgi translocation in response to stress.
HEK 293 cells stably expressing either CerS1-HA or the catalytically inactive CerS1 (H182A, H183A)-HA were treated with 50 μM cisplatin for 3 h and stained with rabbit anti-HA polyclonal antibody (green) and mouse anti-Golgi GM130 antibody (red). A. Cisplatin treatment of cells expressing catalytically active CerS1-HA shows translocation to the Golgi as indicated by co-localization of CerS1-HA with the Golgi marker GM130 (see arrows). B. Cells expressing the catalytically inactive mutant CerS1 (H182A, H183A)-HA shows decreased CerS1 translocation to the Golgi apparatus following treatment with cisplatin when compared to normal control CerS1. C. Percent translocation observed in cells expressing either catalytically inactive or control CerS1. Twenty fields of 20 cells were scored in each experiment. Size bar–20 μm.
Stress-induced translocation of CerS1 can be detected with C-terminal but not N-terminal tagged protein
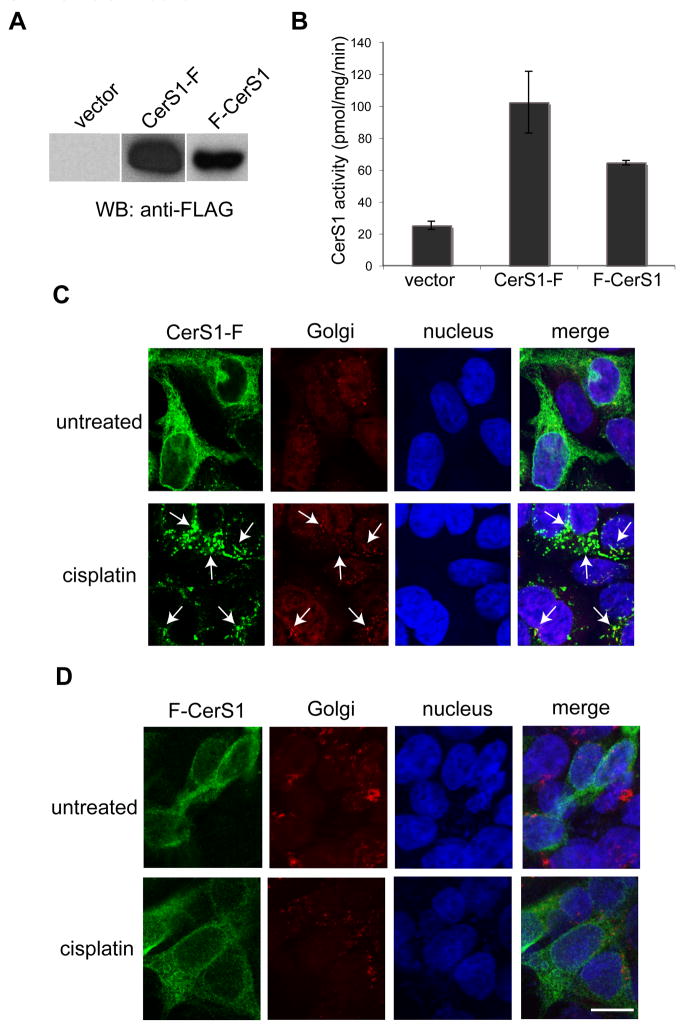
Figure 2 shows that cells stably expressing C-terminal or N-terminal FLAG-tagged CerS1 express the full length protein (37 kDa; Figure 2A), and that the expressed protein is enzymatically active with C18-ceramide as substrate (Figure 2B). The C-terminally FLAG–tagged CerS1 (CerS1-FLAG) translocates and concentrates in the Golgi apparatus following stress with cisplatin (Figure 2C), in precisely the same manner as we previously demonstrated for the CerS1-HA protein (see Figure 1 and reference [18]), indicating that the specific type of tag (HA or FLAG) does not affect the result. Surprisingly however, despite many attempts we did not observe any stress-induced translocation with an N-terminal FLAG-tagged version of CerS1 (FLAG-CerS1) when probed with the same anti-FLAG antibodies (Figure 2D).
Figure 2. Stress-induced translocation of CerS1 can be detected with C-terminal but not N-terminal tagged protein.
Western blot analysis (A) - 15 μg extracts of HEK 293 cells, stably expressing C-terminally FLAG-tagged CerS1 (CerS1-FLAG), N-terminally FLAG-tagged CerS1 (FLAG-CerS1) and vector control were detected with anti-FLAG Ab. Both constructs express protein of the expected size (37 kDa). Enzyme activity (B) - Ceramide synthase in vitro enzyme activity of FLAG-CerS1 and CerS1-FLAG using C18 as a substrate as described in the Materials and Methods section. Immunofluorescence analysis of CerS1 localization - HEK 293 cells expressing CerS1-FLAG (panel C) or FLAG-CerS1 (panel D) were treated with 50 μM cisplatin for 3 h and stained for CerS1 using anti-FLAG polyclonal Ab (green) and anti-GM-130 mAb (red). Translocation of C-terminal FLAG tagged CerS1 in (C), is indicated with arrows. No ER to Golgi translocation was detected with the N-terminal FLAG tagged CerS1 (D). Size bar–20 μM.
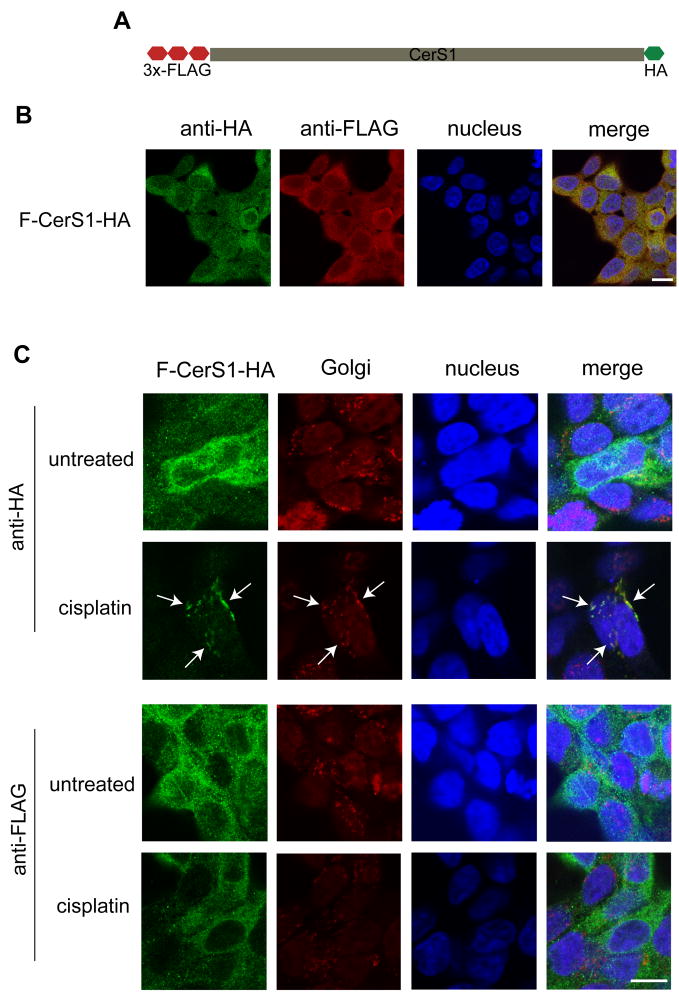
To explore the possibility that the N-terminal tag inhibits translocation, we generated cells expressing a dual-tagged CerS1 protein with an N-terminal FLAG tag, and a C-terminal HA tag (FLAG-CerS1-HA; Figure 3A). As expected, Figure 3B shows precise co-localization of the FLAG (red channel) and the HA (green channel) tags, indicating that the full length, dual-tagged protein is produced in these cells. Following stress, translocation of the FLAG-CerS1-HA from the ER to the Golgi apparatus could be detected when probing for the HA-tagged C-terminus (green), using anti-HA antibody, consistent with the previous results (Figure 3C upper 2 panels, see arrows). However, no ER to Golgi apparatus translocation could be detected with the anti-FLAG antibody (green) directed against the N-terminus of the molecule (Figure 3C lower 2 panels). These results exclude the possibility that the presence of a tag on the N-terminus inhibited translocation, and suggested the possibility that the translocated CerS1 protein underwent a proteolytic cleavage following stress, which resulted in the loss of the N-terminal tag, thus making it appear that translocation is not occurring when probing with the anti-FLAG antibody.
Figure 3. With a dual tagged CerS1, only the C-terminal tag is observed to translocate to the Golgi apparatus after stress.
A. Schematic representation of the dual-tagged (N-terminal FLAG and C-terminal HA) construct (FLAG-CerS1-HA). B. FLAG-CerS1-HA stably expressing cells were stained with mouse anti-HA mAb (green) and rabbit anti-FLAG polyclonal antibody (red). The figure shows co-localization in the ER of both tags (yellow in merged images), indicating that the full, dual tag protein has been expressed. C. HEK 293 cells, stably expressing FLAG-CerS1-HA were treated with 50 μM cisplatin for 3 h and stained with rabbit anti-HA or rabbit anti-FLAG polyclonal antibody (both green) along with mouse anti-Golgi GM130 antibody (red). When treated with cisplatin, the CerS1 C-terminal tag but not the N-terminal tag shows translocation (see arrows) and co-localization with the Golgi apparatus. Size bar–20 μm.
Stress-induced formation of a C-terminal CerS1 fragment
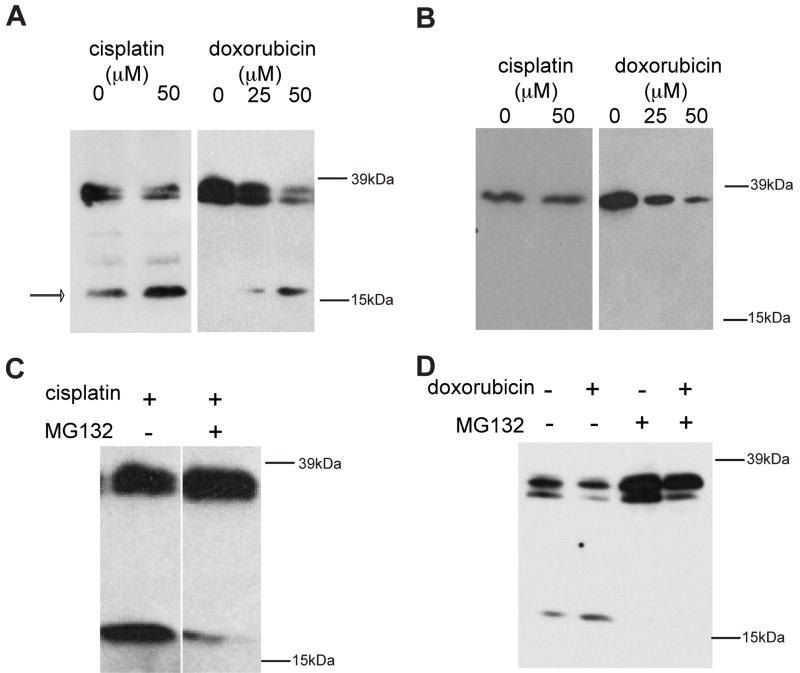
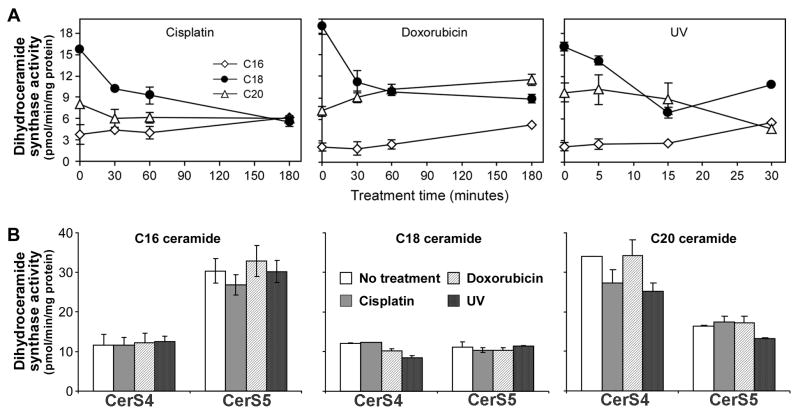
To directly test the hypothesis that CerS1 undergoes stress-induced processing which removes the N-terminal end, cells stably expressing either C- or N-terminally FLAG-tagged CerS1 were stressed, and were compared to equal amounts of the untreated controls by SDS-PAGE and Western blotting (Figure 4). (Note, that FLAG was chosen for these experiments because of consistent difficulty in visualizing HA-tagged CerS1 on blots). Blots of the untreated C-terminal FLAG-tagged CerS1 expressing cells showed the major band of full length CerS1-FLAG protein at 37 kDa, and second band (C-terminal tagged) of weaker intensity at approximately 17 kDa. Stressing the cells with either cisplatin or doxorubicin induced an increase in the amount of the 17 kDa band and some decrease in the full length 37 kDa band indicating that the lower molecular weight band is processed from the full length CerS1 protein (Figure 4A). In contrast, when N-terminal-tagged FLAG-CerS1 was examined following stress with either cisplatin or doxorubicin (Figure 4B), there was a decrease in the amount of full length 37 kDa protein, but the smaller 17 kDa fragment was never detected (because it was not C-terminally tagged), suggesting that the N-terminal part of the molecule is subject to cleavage and therefore the loss of the N-terminal tag. Figure 5 shows that there is a concomitant loss of CerS1 (but not CerS4 or CerS5) enzyme activity following stress with UV light, doxorubicin or cisplatin.
Figure 4. Stress increases CerS1 proteolysis.
HEK 293 cells stably expressing CerS1-FLAG (A) or FLAG-CerS1 (B) were treated with cisplatin (50 μM), or doxorubicin (25–50 μM). After 6 h the control and treated cells were lysed with HEPES/digitonin lysis buffer on the plates and harvested. The lysates were assayed for protein concentration (BCA protein assays), and equal amounts of protein for each sample were separated by SDS-PAGE and subjected to Western analysis, using anti-FLAG mAb. All treatments caused an increase in the level of the C-terminal tagged 17 kDa fragment (A), but a fragment was not detected with the N-terminally FLAG tagged CerS1 (B). HEK 293 cell expressing CerS1-FLAG was treated with cisplatin (C) or doxorubicin (D), in the presence or absence of 1 h pre-treatment with the proteasome inhibitor MG132 and subjected to Western blotting analysis as described above. MG132 co-treatment inhibited the formation of the 17 kDa CerS1 proteolytic product.
Figure 5. Stress reduces the level of ceramide synthase activity of CerS1 but not CerS4 or CerS5.
Cells stably expressing CerS1-HA, CerS4-HA or CerS5-HA were treated with cisplatin, doxorubicin or UV light. Cells were solubilized and assayed for dihydroceramide synthase activity as described in Material and Methods. A. Synthesis of C16, C18 and C20-dihydroceramide by extracts of CerS1 expressing cells after treatment. 50μM Cisplatin; 25 μM Doxorubicin; 1.6J/m2 UV-light. B. Synthesis of C16-, C18- and C20-dihydroceramides by extracts of CerS4 and CerS5 expressing cells following treatment with cisplatin (3 h; 50μM), doxorubicin (3 h; 25 μM) or UV-light (30 min; after 90 seconds of 1.6 J/m2). Each assay was repeated three times in three separate extracts. ± s.d.
Translocation of CerS1 is dependent on proteasomal processing
CerS1 is subject to ubiquitination and proteasomal turnover and several different proteasome inhibitors blocked this effect [19]. We now show that the generation of the CerS1 17 kDa C-terminally tagged fragment after stress is dependent on proteasomal function. Indeed, treatment with the proteasomal inhibitor MG132 greatly reduced the CerS1 17 kDa product, in CerS1-FLAG expressing cells treated with either cisplatin or doxorubicin (Figure 4C and D). Taken together, these results indicate that CerS1 undergoes a proteasomal dependent processing to generate a C-terminal 17 kDa fragment, in response to stress.
The preceding data suggested that proteasomal processing was a prerequisite for the stress-induced ER to Golgi translocation of CerS1, and that blocking the proteasome should interfere with CerS1 translocation. To test this hypothesis, cells stably expressing C-terminally tagged CerS1-FLAG were treated with the proteasome-inhibitor MG132 followed by stress with cisplatin, and were examined for CerS1-FLAG ER to Golgi apparatus translocation. As shown in Figure 6A treatment of cells with MG132 alone has no effect on the CerS1 localization in the ER, and itself did not induce translocation to the Golgi apparatus. However, MG132 clearly inhibits the stress-induced CerS1-FLAG translocation to the Golgi apparatus. Figure 6B shows a histogram of the percent of cells which show translocation in each condition. These results strongly indicate that CerS1 translocation is dependent on proteasomal processing.
CerS1 stress-induced ER to Golgi translocation requires CerS1 catalytic activity
Ectopic expression of sphingosine kinase 1 has been shown to block stress induced translocation of CerS1, indicating that the balance between ceramide and S-1-P is involved in regulating CerS1 translocation [18]. This in turn, implied that the activity of CerS1 should be necessary for translocation. Mutation of the 182 and 183 histidines in the conserved Lag1p motif of CerS1 has been reported to significantly affect CerS1 activity [12]. This molecular genetic approach is more specific than using relatively non-specific inhibitors of ceramide synthesis such as fumonisin or myriocin which inhibit either all ceramide synthases or the upstream serine palmitoyl transferase, respectively. The change of two amino acids probably does not alter the folding or membrane topology of CerS1 as previously shown [25].
Thus, to determine if CerS1 translocation is dependent upon its activity, we generated, cloned, and tested a C-terminally tagged mutant CerS1 with both histidines mutated to alanines (H182A and H183A). The mutant enzyme was confirmed to have less than 10% of the wild-type level of catalytic activity when assayed with its preferred substrate, C18 acyl CoA [19]. When stressed with cisplatin, cells stably expressing the inactive mutant CerS1 showed a significant decrease in ER to Golgi apparatus translocation when compared to cells expressing the wild-type protein (Figure 7). These results strongly suggest that CerS1 enzyme activity is required for its stress-induced ER to Golgi translocation, and complements the earlier results on cell expressing sphingosine kinase [18].
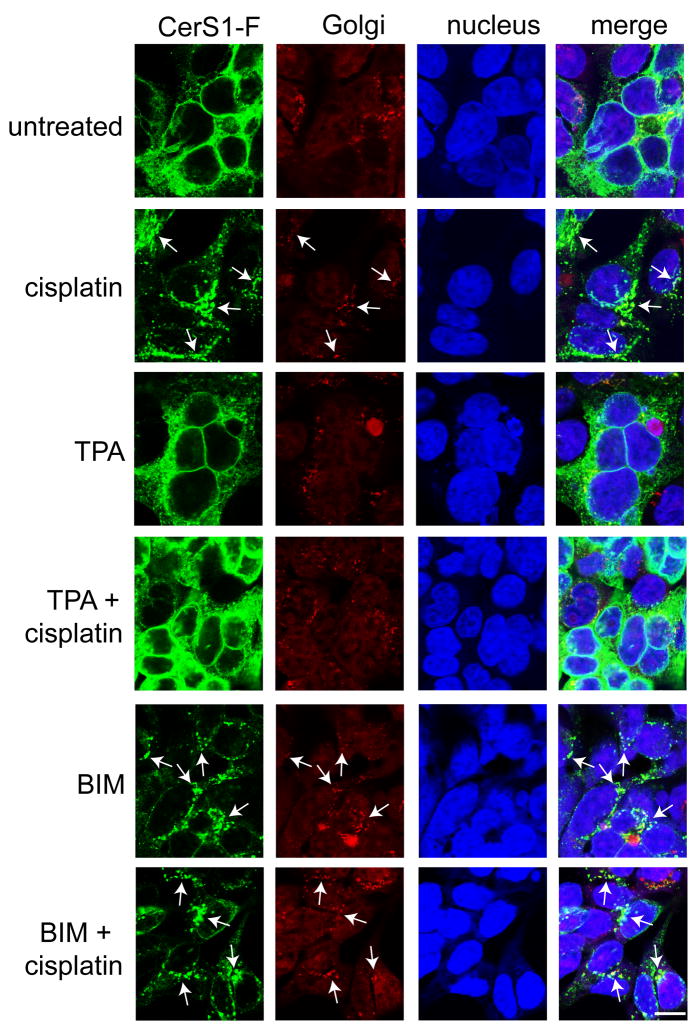
CerS1 translocation is dependent on protein kinase C (PKC)
It was previously shown that the proteasomal turnover of CerS1 is regulated by PKC, and CerS1 is phosphorylated following activation of PKC [19]. Hence, we hypothesized that modulation of PKC activity may also affect CerS1 ER to Golgi translocation. To this end, cells stably expressing C-terminal-tagged CerS1 were treated with either the PKC activator TPA (12-o-tetraphorbol-13 acetate) or the PKC inhibitor BIM (bis-indolelyl maleimide) followed by cisplatin treatment. Consistent with previous experiments, cisplatin treatment caused CerS1-FLAG translocation from the ER to the Golgi apparatus (Figure 8, arrows). Treatment with the PKC activator TPA prevented stress-induced CerS1 translocation to the Golgi apparatus in response to stress, while in contrast, the PKC inhibitor BIM caused CerS1 translocation (arrows), either in the absence or presence of cisplatin treatment. It should be noted that BIM inhibits 5 of 8 PKC isoforms and further experiments will be necessary to identify which specific isoform(s) is regulating CerS1. In summary, these studies indicate that PKC is intimately involved in regulating CerS1 translocation.
Figure 8. Protein kinase C (PKC) regulates CerS1 translocation in response to stress.
HEK 293 cells stably expressing CerS1-FLAG were treated with 50 μM cisplatin for 3 h in the presence or absence of 1 h pre-treatment with 200 nM PKC activator 12-o-tetradecanoyl-phorbol-13 acetate (TPA) or 5 μM PKC inhibitor bis-indolelyl maleimide (BIM). Cells were stained with rabbit anti-FLAG polyclonal antibody (green) and mouse anti-Golgi GM130 antibody (red) and analyzed for CerS1 translocation and co-localization with the Golgi apparatus (see arrows). Cisplatin-induced translocation of CerS1 was blocked by TPA, while enhanced translocation (see arrows) was seen in the presence of BIM (Size bar–20 μm).
Analysis of the C-terminus of CerS1 with respect to ER to Golgi translocation
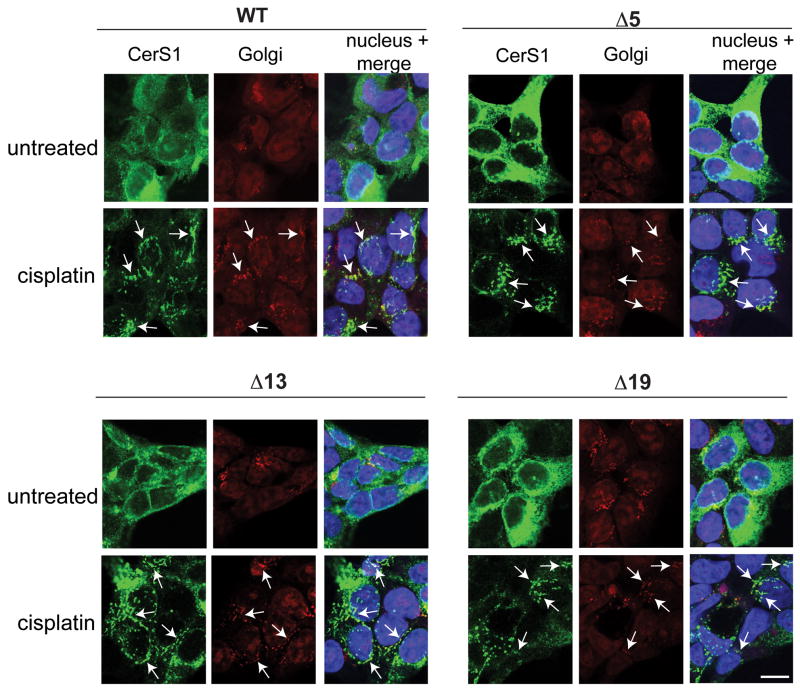
The apparent absence of translocation of the N-terminal part of the CerS1 molecule suggested that the C-terminal end of CerS1 functions in regulating the stress-induced translocation. This idea was consistent with the presence of two KxKxx-like domains in the C-terminus of CerS1, but not in the other CerS enzymes, and the fact that this domain has been reported to function in regulating ER to Golgi apparatus protein movement [26, 27]. In an attempt to define the region that is necessary for translocation, we generated and expressed a series of mutant CerS1-FLAG genes that lacked the last 5, 13, and 19 amino acids of the enzyme (see Materials and Methods). The -5 amino acid deletion represents a deletion of the C-terminal KxKxx domain. The -13 amino acid deletion mimics a naturally occurring splice variant of CerS1, which terminates 13 amino acids upstream of the commonly accepted stop codon for CerS1. The -19 amino acid deletion is missing a second KxxKxx domain of CerS1 penultimate to the C-terminus of the splice variant. Mutants missing the C-terminal 5, 13 and 19 amino acids were as active as the parent construct in the ceramide synthase in vitro assay using C18 acyl CoA (data not shown). The mutant proteins missing the last 5, 13 and 19 amino acids translocated from the ER to the Golgi apparatus when the cells stably expressing these mutant proteins were stressed with cisplatin (Figure 9), indicating that there is no domain between -19 to the end of the molecule that is indispensable for translocation. It is important to emphasize that these mutant CerS1 molecules did not constitutively accumulate in the Golgi apparatus (i.e., in the absence of stress) indicating that the stress-induced translocation is not merely the result of preventing retrieval to the ER by blocking the KxKxx motif with the C-terminal tag.
Figure 9. CerS1 C-terminal deletion mutants lacking 5, 13, and 19 amino acids show ER to Golgi translocation.
HEK 293 cells stably expressing wild type CerS1 or the various C-terminal deletion mutants were treated with 50 μM cisplatin for 3 h and stained for stress-induced translocation using anti-FLAG polyclonal Ab (green) and anti GM-130 mAb (red). Translocation of CerS1 and co-localization with the Golgi apparatus is indicated with arrows. WT and the CerS1 deletion mutants show cisplatin-induced translocation to the Golgi apparatus (Size bar–20 μM).
Discussion
CerS1 is one of a family of enzymes responsible for the de novo synthesis of ceramide in mammalian cells. A growing number of studies indicate that CerS1 is structurally and functionally distinct from the other enzymes in the family [7]. One prime example of this is the fact that although all the CerS enzymes are ER resident, CerS1, but not CerS4 or CerS5, has been shown to translocate to the Golgi apparatus upon stress [18]. It will be interesting to examine CerS2, 3 and 6 with respect to stress-induced translocation. The CerS1 translocation is robust and independent of the type of stress, and occurs in response to different chemotherapeutic drugs, UV light and an inducer of the unfolded protein response, DTT ([18] and this study). Thus, although the diverse stresses each initiate responses through different signaling pathways, they ultimately converge to regulate CerS1.
We present data that strongly support the idea that proteasomal processing is required for stress induced ER to Golgi apparatus translocation. This was discovered because stress-induced translocation could be detected only with C-terminally tagged protein. Thus, we hypothesized that CerS1 might be processed such that the N-terminus is removed prior to ER to Golgi translocation, and therefore an N-terminal-tagged CerS1 protein would not be observed to translocate. Indeed, in this study we have shown that CerS1 undergoes processing which generates a 17 kDa C-terminal polypeptide, and this fragment is more abundant when cells are stressed. Moreover, both the generation of the 17 kDa CerS1 C-terminal fragment and the stress-induced ER to Golgi apparatus translocation are dependent on proteasome function. The data present a consistent parallel to earlier work on the ubiquitination/proteasomal regulation of CerS1 [19]. The current study suggests that in response to a variety of diverse stresses, there is a proteasomal dependent process that results in the removal of the N-terminal half of CerS1, leaving a 17 kDa fragment. Proteasomal processing down to a large polypeptide fragment has been shown in the processing of NF-κB [28–30].
We have not been able to demonstrate translocation with endogenous protein because the cell lines that we have found which express CerS1 do so at a low level and the available antibody does not detect the protein by immunofluorescence. Clearly, studies with ectopically expressed protein have limitations due to increased levels of protein. However, this approach is extremely useful in facilitating biochemical and cell biological studies. In the case of CerS1, ectopic expression does not cause altered cellular localization of the enzyme, and translocation to the Golgi apparatus is observed only after stress. The specificity for CerS1 argues that stress-induced translocation is an important element in the regulation of this enzyme and thus in ceramide homeostasis. Indeed, both sphingosine kinase 1 and sphingosine kinase 2 have been reported to change intracellular location in response to stress, suggesting that cellular trafficking of sphingolipid metabolizing enzymes is a common form of regulation [20, 21]. Other reported examples of translocation in response to stress include the ATF6 protein, which is part of the unfolded protein response, and translocates from the ER to the Golgi in response to a variety of metabolic, chemical or genetic stresses [31], and the sterol-regulatory element-binding protein (SREB), which undergoes ER to Golgi translocation in response to sterol depletion [32]. In both cases, the translocation of the proteins involves proteolytic processing.
The data with the inactive H182A, H183A mutant suggest that enzymatic activity is necessary for translocation. The data do not completely exclude the possibility of cleavage by an ER resident protease that acts at the site of the tandem histidines that are mutated in the inactive CerS1 mutant used in these studies, and that MG132 may inhibit such an enzyme. Further in vitro studies could directly test for such a protease. In this regard, we are attempting to isolate the C-terminal fragment in sufficient quantity that will allow the identification of the cleavage site. Only the 17 kD fragment appears to be transported to the Golgi, which is consistent with the observation that neither the N-terminally tagged CerS1 nor the N-terminal tag of the dual tagged protein, are observed to translocate to the Golgi apparatus. In addition, early attempts to biochemically fractionate ER and Golgi indicate that the 17 kDa fragment only co-fractionates with the Golgi apparatus following stress. At present we do not know the function of the translocation and this will require ectopic expression of the C-terminal fragment to determine if it is constitutively localized to the Golgi apparatus, and whether there is any resultant phenotype.
PKC activity appears to prevent stress-induced translocation. Earlier work showed that CerS1 is phosphorylated and that activation of PKC increases the phosphorylation [19]. This suggests that de-phosphorylation is involved in translocation. Thus, it will be important determine the specific PKC isoforms that are involved, and identify which of 5 putative serine and threonine PKC phoshorylation sites are important in this process by mutation analysis and the analysis of phosphatases that act on PKCs. These studies will help to determine if PKC is acting directly on CerS1 or via an intermediate.
The deletion analysis of the C-terminal end revealed two important points. 1) The C-terminal tag is not blocking a C-terminal domain that is required for ER retention or retrieval (such as the KxKxx domain) because removing up to 19 amino acids does not result in constitutive translocation, and 2) there is no obvious domain within the last 19 amino acids that is absolutely required for translocation.
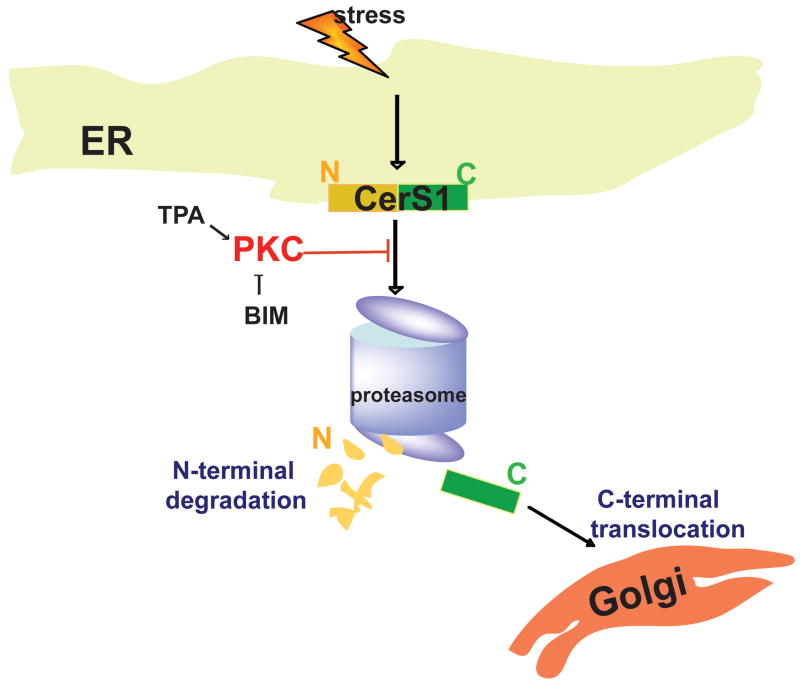
The CerS enzymes are important regulators of ceramide homeostasis, and CerS1 is central to regulating C18 ceramide, which has been shown to be important in cancer and the response to chemotherapeutic drugs [1, 15, 16, 18]. CerS1 responds uniquely to stress via proteasome processing to a C-terminal 17 kDa polypeptide that appears to be translocated from the ER to the Golgi apparatus. CerS1 translocation in response to stress is robust and not observed with CerS4 or CerS5. A schematic model of the results in depicted in Figure 10. Overall, the data provide an increasingly detailed understanding of the regulation of this important enzyme in normal and stressed cells and support the idea that it is uniquely regulated with respect to the other CerS enzymes.
Figure 10. Proposed model for stress-mediated ER to Golgi translocation of CerS1.
Stresses activate proteasome-mediated CerS1 processing, which is regulated by PKC. CerS1 proteolysis leads to translocation of the C-terminal portion of CerS1 to the Golgi apparatus, while the N-terminal fragment gets degraded.
Supplementary Material
Supplemental Figure 1. UV light and DTT do not induce translocation of the CerS4 protein to the Golgi apparatus.
HEK 293 cells expressing CerS4 C-terminally tagged with HA (CerS1-HA) were stressed with either 5 mM DTT for 30 minutes or UV light for 90 seconds (UV intensity, 1 sec = 1.6 J/m2), and analyzed after 30 minutes for the localization of CerS4 protein. Cells were stained with rabbit anti-HA polyclonal antibody (green) and anti-Golgi GM130 mAb (red). There is no translocation of CerS4-HA (green) with the Golgi (red). Size bar–20 μm.
Supplemental Figure 2. UV light and DTT do not induce translocation of the CerS5 protein to the Golgi apparatus.
HEK 293 cells expressing CerS1 C-terminally tagged with HA (CerS1-HA) were stressed with either 5 mM DTT for 30 minutes or UV light for 90 seconds (UV intensity, 1 sec = 1.6 J/m2), and analyzed after 30 minutes for the localization of CerS5 protein. Cells were stained with rabbit anti-HA polyclonal antibody (green) and anti-Golgi GM130 mAb (red). There is no translocation of CerS5-HA (green) with the Golgi (red). Size bar–20 μm.
Acknowledgments
This work was supported by NIH grant GM53929 (SA/HA), University of Missouri Research Board Grant 09-01 (SA), a U.S.-Israel Binational Science Foundation Grant (SA/AHF), and NIH Grant AT003899 (MH). SA acknowledges the support of a Morris Belkin Visiting Professorship at the Weizmann Institute of Science, and AHF is the Joseph Meyerhoff Professor of Biochemistry at the Weizmann Institute of Science. This work partially fulfills the requirements for the Ph.D. degree at the University of Missouri (PS). We thank the members of the Alexander and Futerman labs for support and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zeidan YH, Hannun YA. Translational aspects of sphingolipid metabolism. Trends Mol Med. 2007;13:327–36. doi: 10.1016/j.molmed.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Hannun YA, Obeid LM. The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J Biol Chem. 2002;277:25847–50. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 3.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 4.Spiegel S, Milstien S. Sphingosine 1-phosphate, a key cell signaling molecule. J Biol Chem. 2002;277:25851–25854. doi: 10.1074/jbc.R200007200. [DOI] [PubMed] [Google Scholar]
- 5.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–16. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 6.Spiegel S, Milstien S. Functions of the multifaceted family of sphingosine kinases and some close relatives. J Biol Chem. 2007;282:2125–9. doi: 10.1074/jbc.R600028200. [DOI] [PubMed] [Google Scholar]
- 7.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–5. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- 8.Lahiri S, Lee H, Mesicek J, Fuks Z, Haimovitz-Friedman A, Kolesnick RN, Futerman AH. Kinetic characterization of mammalian ceramide synthases: determination of K(m) values towards sphinganine. FEBS Lett. 2007;581:5289–94. doi: 10.1016/j.febslet.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH, Jr, Futerman AH. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem. 2008;283:5677–84. doi: 10.1074/jbc.M707386200. [DOI] [PubMed] [Google Scholar]
- 10.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–71. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riebeling C, Allegood JC, Wang E, Merrill AH, Jr, Futerman AH. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J Biol Chem. 2003;278:43452–9. doi: 10.1074/jbc.M307104200. [DOI] [PubMed] [Google Scholar]
- 12.Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, Hannun YA, Obeid LM. Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J Biol Chem. 2006 doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- 13.Mizutani Y, Kihara A, Igarashi Y. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem J. 2006;398:531–8. doi: 10.1042/BJ20060379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH, Jr, Futerman AH. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J Biol Chem. 2002;277:35642–9. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 15.Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, Ogretmen B. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol Cancer Ther. 2007;6:712–22. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- 16.Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, Ogretmen B. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. J Biol Chem. 2007;282:10922–34. doi: 10.1074/jbc.M610157200. [DOI] [PubMed] [Google Scholar]
- 17.Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, Day TA, Jiang JC, Jazwinski SM, Hannun YA, Obeid LM, Ogretmen B. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J Biol Chem. 2004;279:44311–9. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- 18.Min J, Mesika A, Sivaguru M, Van Veldhoven PP, Alexander H, Futerman AH, Alexander S. (Dihydro)ceramide Synthase 1 Regulated Sensitivity to Cisplatin Is Associated with the Activation of p38 Mitogen-Activated Protein Kinase and Is Abrogated by Sphingosine Kinase 1. Mol Cancer Res. 2007;5:801–12. doi: 10.1158/1541-7786.MCR-07-0100. [DOI] [PubMed] [Google Scholar]
- 19.Sridevi P, Alexander H, Laviad EL, Pewzner-Jung Y, Hannink M, Futerman AH, Alexander S. Ceramide synthase 1 is regulated by proteasomal mediated turnover. Biochim Biophys Acta. 2009;1793:1218–1227. doi: 10.1016/j.bbamcr.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maceyka M, Sankala H, Hait NC, Le Stunff H, Liu H, Toman R, Collier C, Zhang M, Satin LS, Merrill AH, Jr, Milstien S, Spiegel S. SphK1 and SphK2, sphingosine kinase isoenzymes with opposing functions in sphingolipid metabolism. J Biol Chem. 2005;280:37118–29. doi: 10.1074/jbc.M502207200. [DOI] [PubMed] [Google Scholar]
- 21.Wattenberg BW, Pitson SM, Raben DM. The sphingosine and diacylglycerol kinase superfamily of signaling kinases: localization as a key to signaling function. J Lipid Res. 2006;47:1128–39. doi: 10.1194/jlr.R600003-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JY, Knudsen ES. RB-dependent S-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–63. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyriakis J, Liu H, Chadee D. Actication od SAPKs/JNK and p38s in vitro. In: Seger R, editor. MAP kinase signaling protocols. Humana Press; Totowa, NJ: 2004. [Google Scholar]
- 24.Wang X, Martindale JL, Holbrook NJ. Requirement for ERK activation in cisplatin-induced apoptosis. J Biol Chem. 2000;275:39435–43. doi: 10.1074/jbc.M004583200. [DOI] [PubMed] [Google Scholar]
- 25.Kageyama-Yahara N, Riezman H. Transmembrane topology of ceramide synthase in yeast. Biochem J. 2006;398:585–93. doi: 10.1042/BJ20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duden R. ER-to-Golgi transport: COP I and COP II function (Review) Mol Membr Biol. 2003;20:197–207. doi: 10.1080/0968768031000122548. [DOI] [PubMed] [Google Scholar]
- 27.Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 30.Fan CM, Maniatis T. Generation of p50 subunit of NF-kappa B by processing of p105 through an ATP-dependent pathway. Nature. 1991;354:395–8. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 31.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–8. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–40. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. UV light and DTT do not induce translocation of the CerS4 protein to the Golgi apparatus.
HEK 293 cells expressing CerS4 C-terminally tagged with HA (CerS1-HA) were stressed with either 5 mM DTT for 30 minutes or UV light for 90 seconds (UV intensity, 1 sec = 1.6 J/m2), and analyzed after 30 minutes for the localization of CerS4 protein. Cells were stained with rabbit anti-HA polyclonal antibody (green) and anti-Golgi GM130 mAb (red). There is no translocation of CerS4-HA (green) with the Golgi (red). Size bar–20 μm.
Supplemental Figure 2. UV light and DTT do not induce translocation of the CerS5 protein to the Golgi apparatus.
HEK 293 cells expressing CerS1 C-terminally tagged with HA (CerS1-HA) were stressed with either 5 mM DTT for 30 minutes or UV light for 90 seconds (UV intensity, 1 sec = 1.6 J/m2), and analyzed after 30 minutes for the localization of CerS5 protein. Cells were stained with rabbit anti-HA polyclonal antibody (green) and anti-Golgi GM130 mAb (red). There is no translocation of CerS5-HA (green) with the Golgi (red). Size bar–20 μm.