Abstract
The complex behavioral symptoms and neuroanatomical abnormalities observed in autistic individuals strongly suggest a multi-factorial basis for this perplexing disease. Although not the perfect model, we believe the Engrailed genes provide an invaluable “window” into the elusive etiology of Autism Spectrum Disorder. The Engrailed-2 gene has been associated with autism in genetic linkage studies. The En2 knock-out mouse harbors cerebellar abnormalities that are similar to those found in autistic individuals and, as we report here, has a distinct anterior shift in the position of the amygdala in the cerebral cortex. Our initial analysis of background effects in the En1 mouse knock-out provides insight as to possible molecular mechanisms and gender differences associated with autism. These findings further the connection between Engrailed and autism and provide new avenues to explore in the ongoing study of the biological basis of this multifaceted disease.
Keywords: autism, Engrailed, amygdala, cerebellum, mouse model, brain development
Introduction
Autism Spectrum Disorder (ASD) is a major mental health disorder afflicting up to 13 out of every 10,000 individuals [29]. Features that typify the broad range of autistic behavior include language impairments (including deficits in verbal and non-verbal communication), restricted patterns of interests and activities, abnormal responses to sensory stimuli, poor eye contact, an insistence on sameness, an unusual capacity for rote memorization, and often repetitive actions [38]. The complexity of the presentation of this malady, however, was perhaps best described by Bachevalier [5] as: “the disruption of a basic characteristic of the human species: the sophistication to generate complex displays of emotion and the ability to respond to the expressive behaviors of other individuals”. Determining the biological basis for these behaviors and neurological impairments has remained both elusive and perplexing. Unlike the situation in Parkinson’s or Alzheimer’s disease, a well defined collection of major disease genes is not yet available. No clear biomarker or tightly linked change in neurochemistry is known. Finally, the hunts for both neuroimaging signals and neuropathological changes associated with autism have produced highly variable findings with no agreed upon structural alteration emerging. Although the neuroanatomical basis of autism is still somewhat unclear, certain brain regions appear to be regularly altered in individuals with ASD. These include areas in the neocortex, cerebellum, amygdala, hippocampus and brain stem [10,11,19].
One view of ASD that fits with much of the current data is that a timely environmental insult transforms a latent genetic susceptibility into structural abnormalities during the development of the brain. Although the exact nature of the possible environmental contributions remains quite speculative, there is a large body of evidence that indicates that there is a strong genetic component to ASD. For unknown reasons, it affects males four times more frequently than females [30] and a number of reports show significant concordance rates (up to 82%) in monozygotic twins [6,28,61]. Although it is believed that there are a half-dozen or more genes remaining to be discovered, three genes that are emerging as plausible players in the etiology of ASD are reelin (RELN), the serotonin transporter gene (5HTT), and Engrailed-2 (EN2) [9].
For some time we have been intrigued with the prospect that the Engrailed mouse mutants are potentially important tools in the investigation of the biology of autism. It should come as no surprise that virtually any human disease with a complex spectrum of behavioral and biological traits such as autism would be difficult to model in any one animal system. As such, the Engrailed genes are not likely to be the sole causal elements in ASD. We prefer to think of the mouse Engrailed genes as a “window” into the neurobiological basis of autistic symptoms rather than a direct phenocopy, or even a formal “model” of them. Seen in this light, Engrailed seems to capture many aspects of the autistic brain and has the potential to unveil many more. To date, three genetic studies have linked the human En2 gene to autism [13,32,51]. These studies concur with reports that describe the broad similarities between the neuropathology of the En2−/−mouse and a subset of features commonly found in autism. For example, in the En2−/− mouse there is a modest yet reproducible disruption in the anterior/posterior pattern of cerebellar foliation and transgene expression, particularly in posterior vermis [37,43]. The folial abnormalities are similar to the distortions seen in autistic individuals as reported by Courchesne and colleagues [23]. There is also a significant decrease in the number of Purkinje, granule, deep nuclear and inferior olive cells in the En2 mutant [40]. This again is reminiscent of previously reported neuroanatomical abnormalities observed in autistic individuals [10,11].
Outward phenotypic effects such as these are likely to be the manifestation of a greater core problem. In this regard, we agree with Herbert et al. [35] who proposed that the perturbation of the balance of various brain regions lies at the root of the behavioral symptoms in ASD. One likely location for the fulcrum of this balance is the midbrain/hindbrain boundary of the early neural tube. The Engrailed genes are well-described players in the establishment of this area of the nervous system. En1 is first expressed in a strong transverse band at the junction between the mesencephalon and metencephalon at the one somite developmental stage. A null mutation of En1 results in the absence of most of the cerebellum and midbrain, and in lethality shortly after birth [66]. En2 expression is also found at the presumptive midbrain-hindbrain region but initiates later, at the 5 somite stage [25,26]. Unlike En1, null alleles of Engrailed-2 have only a subtle neurological phenotype. There is a developmental change in the banded organization of spinal cord mossy fiber afferents [65] and in the transient pattern of banded expression of En1, En2, Pax2, and Wnt7b seen during late gestation [42]. Other markers of cerebellar compartmentation are also disrupted in both the vermis and hemispheres of the En2hd null mutant. These include Zebrin II, Ppath and the expression of a L7lacZ fusion gene [40].
The Engrailed genes are commonly known for their pattern formation activity in the midbrain and hindbrain regions (the only CNS regions where they are expressed), yet both the symptomatology and the structural changes in autism point to significant involvement of telencephalic structures. Some of these include neocortical malformations such as irregular laminar patterns, thickened cortices, abnormally oriented pyramidal cells, and an increase in the number of layer one neurons [7]. In specific cortical areas, Casonanova et al. [17] have reported the presence of smaller, more compact, and more numerous minicolumns. Regions in the forebrain that show reduced neuronal size and increased cell packing density include the hippocampus, subiculum, entorhinal cortex, mammilary bodies, anterior cingulated gyrus, septum and amygdala ( [38] and see [12,27,47] for recent reviews on autism neuropathology).
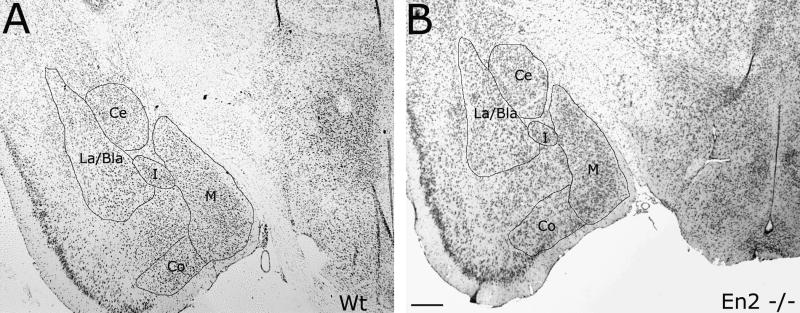
Perhaps due to the more posterior location of Engrailed expression, none of the telencephalic structures affected in autism have been analyzed in either Engrailed mutant. One particularly intriguing location to examine would be the amygdala, as it is an important constituent of the complex neural network of interconnected structures that give rise to certain aspects of social behavior. The amygdala is located at the medial edge of the temporal lobe, just rostral to the hippocampus, and consists of defined subgroups with unique cytology (Figure 1) and specific efferent and afferent connections that are indicative of its function. The lateral nucleus (La), for example, is bounded by the forked-shaped portion of the external capsule. It receives input from regions of the temporal, insular and parietal cortex, which process gustatory, visceral, somatosensory, auditory or visual information. The main outputs from the lateral nucleus include the prefrontal cortex, the medial temporal lobe and the nucleus accumbens. Adjacent to the lateral nucleus is the basal nucleus (Bla). This nucleus receives light input from the hypothalamus, olfactory system and striatum, and substantial input from the frontal cortex, hippocampal formation and thalamus. It provides an abundance of projections to the contralateral amygdala and to external target areas such as the hippocampal formation, thalamus, olfactory tubercle, nucleus accumbens and caudate putamen. The central nucleus is medial to the La/Bla region and receives substantial input from the hippocampus, hypothalamus, midbrain, pons, medula and spinal cord. The extra-amygdaloid outputs of the central nucleus include reciprocal connections from the hypothalamus, midbrain, pons and medulla as well as the thalamus, hypothalmus and bed nucleus of the stria terminalis. The medial nucleus can be found ventral to the central nucleus in the coronal aspect. It receives a large projection from the hypothalamic nuclei and the infralimbic cortex and sends moderate projections to the hippocampal formation, thalamus and hypothalamus (see [54] for review).
Figure 1. Amygdalar subdivisions in En2−/− and control sections.
Cresyl violet-stained, 10 μm representative sections in the coronal aspect from control (A) and En2 mutant (B) brains from 3 month old male mice indicate the marked similarity in amygdalar subdivisions: central, medial, and lateral/basolateral nuclei. The intermediate nucleus and the cortical nucleus are also quite conspicuous and serve as a reference for slide alignment. Ce, central nucleus; Co, cortical nucleus; M, medial nucleus; La/Bla, lateral and basolateral nucleus; I, intermediate nucleus. Scale bar equals 0.2mm.
The complex circuitry of the amygdala and its interconnection with other limbic system structures as well as with other brain regions strengthens its association to autism. The heavily reciprocal pathways provide the amygdala with visual cues of faces and facial expression, body postures and motions, and cues of specific sounds and intonations [5]. The amygdala is responsible for the initial, rapid assignment of meaning or an “emotional value” to the cues it receives and then the rapid transmittance of this information to higher order areas of the brain. In individuals with autism, three fMRI studies have shown the amygdala to be hypoactive during face perceptual tasks [8,24,53]. Studies where the amygdala was lesioned in adult and infant monkeys has led Amaral et al. to propose that damage to the amygdala may contribute to the increased anxiety that is often a feature of autism [3]. In a recent study by Pandey et al. [48], the authors concluded that decreased BDNF function in the central and medial amygdala (but not in the basolateral division) produced anxiety-like effects in rats.
Because of both the observed and intuitive relationship of the amygdala to autism, as well as the high fidelity of the En2−/− cerebellar and hindbrain structural changes to the changes found in the autistic brain, we decided to explore the effects of the En2−/− mutation on the size and structure of the mouse amygdala. Also, as we have postulated that the Engrailed system is a likely component of the strong genetic basis of ASD, we have chosen to begin an analysis of the modifier loci that alter the phenotype of the Engrailed-1 gene. What follows is a progress report on our search for the links between the biology of autism and the biology of the Engrailed pattern formation genes.
Materials and Methods
The En2 mouse strain
Mice carrying the En2hd allele [37] were transferred to a 129/S1 inbred background from a 129/Sv inbred background. Generally, homozygotes either do not breed or have low birth rates. Therefore, the colony was maintained primarily by breeding heterozygotes. To obtain Engrailed-2 homozygous mutants, +/En-2 mice were mated to En-2/En-2 mutants. Wildtype controls were generated by crossing +/+ mice to each other, and therefore were not littermates to the En-2 homozygotes used in this study. Offspring were genotyped via PCR with the following primers: 5′-AGCAAAGCCT-TCAAGCCTTC-3′ and 5′-GGGACTGTTTAGGGTTTCAG-3′ which amplify a 500 bp band in wild-type animals and 5′-TACTTTCTCGGCAGGAGCAAG-3′ and 5′-TGGATTGGCACGC-AGGTTCTC-3′ which amplify a 300 bp band in Engrailed-2 mutants. All animals were housed in microisolators in the American Association for the Accreditation of Laboratory Animal Care-accredited Case University animal facility, where they were maintained on a 14:10 hour light/dark cycle. Food and water were available ad libitum.
The En1 mouse strain
En1 mice were maintained and genotyped as described previously [14]. For the mapping experiments, two types of crosses were performed. Both began with the mating of a C57BL/6•En1−/+ heterozygote with a wild-type 129/S1 mate. An intercross mating scheme was conducted by mating two of the resulting F1 heterozygotes to each other to produce an F2 generation. Timed matings were used to produce these F2 animals, all of which were taken during late embryogenesis in order to insure no loss of mutant progeny. The backcross mating scheme began with the same F1 heterozygotes but crossed them instead to heterozygotes of the parental strain, C57BL/6•En1−/+. We adopted this second approach when we determined that the number of rescues in the intercross matings was too small to meaningfully use for mapping. All embryos were taken at embryonic day 17 (E17). Samples of the viscera were taken for DNA extraction; brains were fixed in 4% formaldehyde overnight and bisected on the midline. Cerebellar phenotype was scored by examining the midsagittal cut under a dissecting scope.
Histology
Mice were anesthetized with Avertin (0.025ml/gm body weight) and transcardially perfused with phosphate buffered saline (PBS) for 2 minutes, followed by 10 minutes of 4% paraformaldehyde in 0.1M phosphate buffer. Dissected brains were placed in fixative overnight (O/N) and rinsed with PBS prior to being placed in a solution of 30% sucrose in PBS and stored at 4°C. Brains were sectioned coronally at 10μm using a cryostat. Slides were air dried for a minimum of 1 day and then stained for Nissl substance with cresyl violet. For a working solution of cresyl violet, a stock solution of cresyl violet acetate (2gm/100ml distilled water) was diluted by a factor of 1:20 followed by the addition of 15 drops of 10% glacial acetic acid per 100ml of solution. The slides were rinsed in distilled water two times for 2 minutes each, placed in the cresyl stain for 2 minutes, rinsed in water twice, then dehydrated for 2 minutes each in 50%, 75%, 95% and 100% ethanol. They were then placed in 50% ethanol: 50% Histoclear (National Diagnostics) for 2 minutes and 100% Histoclear prior to coverslipping with Permount (Fisher Scientific).
In Situ Hybridization
Mice were sacrificed by cervical dislocation and the brains were removed immediately. Brains were stored in 30% sucrose and PBS O/N at 4°C, and then were bisected coronally before embedding for cryostat sectioning (10μm). After drying slides for 1 hour, they were stored at −70°C until ready for use. All probes were generated by reverse transcription PCR using RNA from the amygdaloid region of a 4-week old 129/Sv mouse brain. A 512 bp fragment was amplified for the Lamb3 probe, using the following primers: 5′-AAGGAAAAAAGCGGCCGCTAGGTGCCCGGAAAGATATG-3′ and 5′-CCCAAGCTTGGTGGCATTACGGAGACTGT-3′. For the PGPx probe, a 511 bp fragment was amplified using the following primers: 5′-AAGGAAAAAAGCGGCCGCTCTACGAGTATGGAGCCCTCA-3′and 5′-CCCAAGCTTTCCATCTTGACGTTGCTGAC-3′. The Gad65 probe was generated using the following primers to amplify a 531 bp fragment: 5′-AAGGAAAAAAGCGGCCGCCAGCCTT-AGGGATTGGAACA-3′ and 5′-CCCAAGCTTACCCAGTAGTCCCCTTTGCT-3′. Fragments were subcloned into pBluescript (using NotI and HindIII restriction enzyme sites), and were then linearized for T7 transcription (RNAMaxx High Yield Transcription Kit, La Jolla, CA) during which probes were labeled with digoxygenin-UTP from Roche (Indianapolis, IN). After transcription, probes were incubated with DNAse I for 15 minutes at 37°C, and reactions were stopped with 0.5M EDTA, followed by precipitation with tRNA, ammonium acetate, and 100% ethanol O/N at −20°C. Pellets were rinsed with 70% ethanol, resuspended in ddH20, and stored at −20°C.
Slides were fixed in 4% paraformaldehyde and PB for 10 minutes at room temperature (RT), rinsed three times with PBS, acetylated (1.32% Triethanolamine, 0.8% HCl, 0.25% Acetic Anhydride for 10 minutes at RT), and then rinsed again three times with PBS. Hybridization solution (50% Deionized Formamide, 5X SSC, 5X Denhardts, 0.25 mg/ml yeast RNA, 0.5 mg/ml Salmon Sperm DNA) was placed on the slides for 2 hours at RT. Digoxygenin-labeled probes were diluted in fresh hybridization solution (after being previously tested to find optimal dilution), heated to 80°C for 5 minutes, and then placed on ice for 2 minutes. Probe solution was then placed on slides, followed by coverslips and rubber cement to seal them. Slides were incubated at 72°C O/N. The following day, the rubber cement and coverslips were removed, and slides were incubated in 0.2X SSC for 1–3 hours at 72°C. Slides were rinsed for 5 minutes with 0.2X SSC, and then with solution B1 (0.1M Tris pH 7.5, 0.15M NaCl). Blocking was done with B1 and 10% heat-inactivated goat serum for 1 hour at RT. The solution was replaced with fresh blocking solution containing anti-digoxygenin antibody (Roche) at a 1:2500 dilution, and slides were incubated at 4°C O/N. After three rinses with B1 solution, slides were equilibrated with B2 (0.1M Tris pH 9.5, 0.1M NaCl, 50mM MgCl2) for 5 minutes at RT. Finally, slides were developed in B2 solution containing 6.8μl/ml nitroblue-tetrazolium-chloride, 3.5μl/ml 5-bromo-4-chloro-indolyl-phosphate, and 0.48 mg/ml levamisole for 2 hours to O/N at RT. Reaction was stopped by submerging slides in ddH20, and slides were coverslipped using PBS:glycerol (1:1).
Volumetric Assessment
Images of the amygdaloid complex on the left side of three male corteces were taken at approximately 80μm intervals. The images were incorporated into the NIH imaging program, Image J, and the scale was set to 370 pixels/mm (as determined with the linear scale on a hemocytometer). The lateral and basolateral, central, and medial nuclei were identified with the aid of The Mouse Brain in Steriotaxic Coordinates [50]. Each region was outlined and area measurements were obtained using Image J. For each animal, the anterior-most section in which the anterior commissure crossed the midline was used as the “zero-point” of reference. The distance of a given section from this point was used in the following equation to determine volume: Volume = the sum of (Cn+Cn+1)/2 × (Dn+1−Dn) where C = the measured area and D = the distance from where the anterior commissure traverses the midline. Statistical analysis to calculate standard deviations and P(T<=t)two-tail values (as reported in Table 1) was done with the Microsoft Excel analysis Toolpak module.
Table 1.
Volume of amygdalar nuclei (mm3)
| Animal | Nuclei | ||
|---|---|---|---|
| La/Bla | Central | Medial | |
| Wt1 | 0.649 | 0.371 | 0.29 |
| Wt2 | 0.669 | 0.44 | 0.371 |
| Wt3 | 0.605 | 0.379 | 0.317 |
| Wt Avg.(std dev.) | 0.641(.033) | 0.397(.038) | 0.326(.041) |
| En4 | 0.633 | 0.355 | 0.358 |
| En5 | 0.613 | 0.344 | 0.324 |
| En6 | 0.53 | 0.263 | 0.277 |
| En Avg. (std dev.) | 0.592(.055) | 0.321(.500) | 0.32(.041) |
| En as %Wt | 92% | 81% | 98% |
| P(T<=t) two-tail | 0.105 | 0.131 | 0.888 |
To determine the values for the percent of total volume attained in Figures 3 and 4 (Y-axis), we first multiplied the area of each section by the distance from the previous measurement to calculate the volumes of a series of three-dimensional slices. The volumes of these slices were then summed to produce a total volume. The volume of each individual slice was divided by this total to estimate the proportion of the volume contained within each slice. Cumulative volume attained, as plotted in Figures 3 and 4, was calculated by sequentially adding the proportional volume of each slice to the sum total of all of the proportional volumes of the previous slices. To compare the average of the two groups for each sample, we imputed a set of extrapolated sections that were spaced every ten micrometers and assigned these sections area measurements. Using these sections, we created uniformly spaced three-dimensional slices using the same method described above. Positionally identical slice volumes from the subnuclei of the three En2−/− and the three wild type animals were averaged and plotted as shown in Figures 3 and 4.
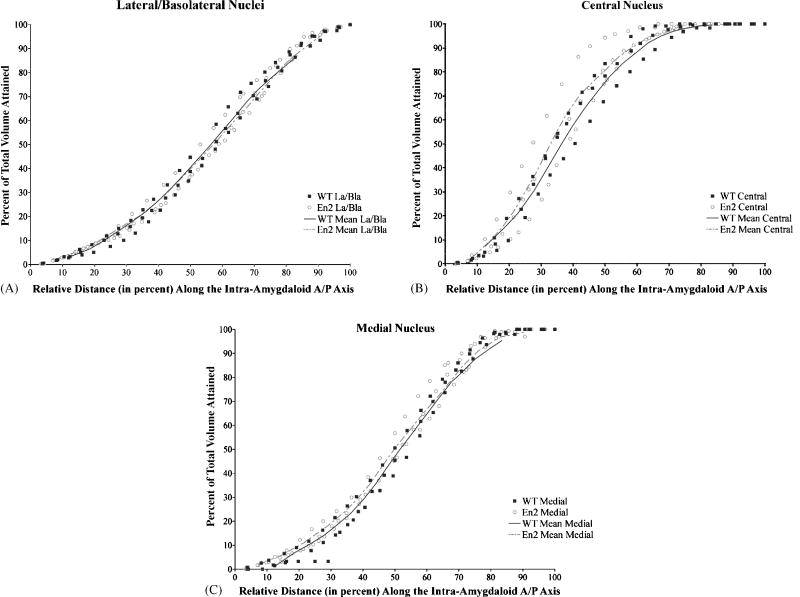
Figure 3. Distribution of intra-amygdaloid volume in En2−/− and controls along the A/P axis.
Area measurements were taken (using Image J software) of the amygdalar subdivisions on each slide of about 25 representative slides bearing coronal histological sections from 3 mutant and 3 control animals. The percentage of the total amygdalar volume attained is plotted as a function of the distance from the anterior most section of each nucleus. Solid squares represent measurements converted to a percentage of total volume at a given distance from control animals, while open circles represent the corresponding measurements from En2−/− animals (see methods section for a detailed description of conversion calculations). Each line (dashed in the case of the En2−/− group of three animals studied) connects the mean value points of all three animals within each genotype. For example, compare the mean value lines at ½ the way through the lateral/basolateral nuclei (or at a 50% distance from the anterior most section). Here about 35% of the volume of this subdivision is accounted for in both mutants and controls. In each of the nuclei examined, the distribution of the En2−/− volume tracks closely to controls, indicating a normal distribution of volume and shape of the En2−/− amygdala when compared to controls.
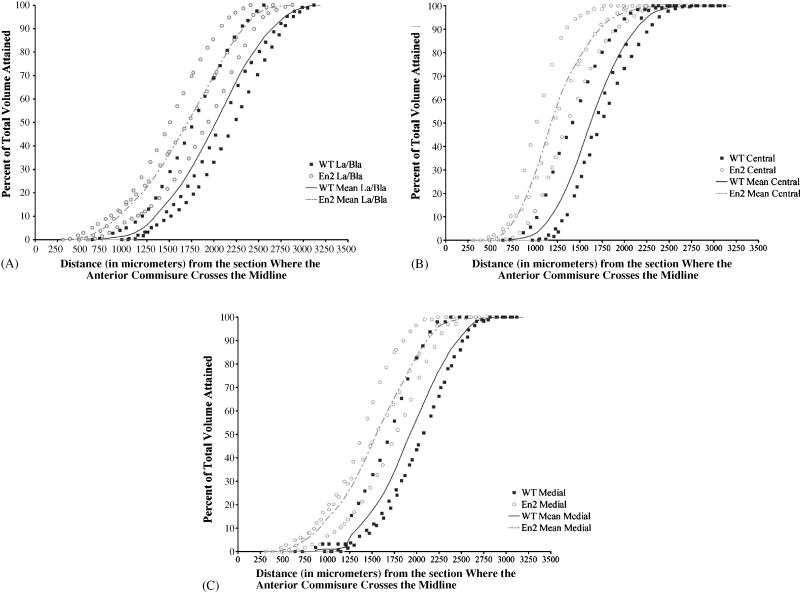
Figure 4. The amygdala is shifted to a more anterior location in the cortex of En2−/− mice.
The percentage of the total amygdalar volume attained is plotted as a function of the distance (along the anterior/posterior axis) from an arbitrary reference point: the first section where the anterior commisure traverses the midline in the coronal aspect (see figures 29 and 30 of The Mouse Brain Atlas [50]). As in Figure 3 above, solid squares represent converted measurements from control animals and open circles represent the converted measurements from En2−/− animals. Each “S” shaped line (dashed in the case of the En2−/− group) connects the mean value points of the three animals within each genotype. Comparing the mean value lines in the three En2−/− amygdalar nuclei we have studied here (panels A, B, C), we find that all lie in the region that starts about 250 μm from our arbitrary reference point and ends about 2625 μm later, thus spanning a total distance of about 2.375 mm. In the controls, each subdivision lies within the region delimited by about 750 and 3125 μm from our cortical reference point. The control amygdalar nuclei span an equivalent distance (2.375mm), but begin and end about 500 μm more posterior than the En2−/− amygdalar nuclei. Thus, the lateral/basolateral nuclei (A), central nucleus (B), and medial nucleus (C) in the En2−/− mice are shifted to a location in the cortex that is approximately 500 μm more anterior than in controls.
Results
Volumetric assessment in the amygdala at 90 days
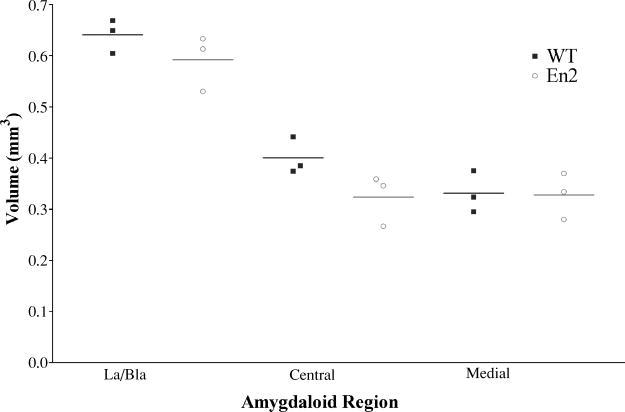
The anatomy and connectivity of the amygdala has been well studied (see [54,57] for review). In the adult rat, the amygdala is composed of roughly 600,000 neurons parceled into thirteen nuclei and cortical regions, some of which are further subdivided. Each of these regions has a unique cytoarchitecture, cheomoarchitecture and connectivity. For volumetric analysis, we examined four nuclei: lateral (La), basolateral (Bla), central (Ce) and medial (M) because of their defined cytoarchitecture in cresyl violet stained, coronal sections (Figure 1). In preliminary studies, we found significant variability in volumetric measurements across different animals. As the genetic background was constant, we attributed the variability to age, laterality (either the right or left amygdala), and sex (both males and females were analyzed). As a result, we were careful to use material exclusively from males at postnatal day 90 (P90) +/−2, and examined only the left amygdala in our current analysis. Wild type controls were 129/S1 male animals of the same age that were derived from crossing +/+ animals to each other. We examined approximately 25 representative sections from each brain. The lateral and basolateral nuclei (La/Bla) generally lie together within the fork-shaped external capsule and thus were grouped together for measurement. In both control and mutant animals, the lateral and basolateral nuclei have the largest volume (ranging around 0.6mm3). The central and medial nuclei are smaller and similar in volume with average sizes between 0.3 and 0.4mm3. In these adult animals, we found no significant difference between mutants and controls with respect to the volumes of the lateral/basolateral, central and medial nuclei. We note, however, that comparison of mean volumes suggests a trend (although not statistically significant) for a volume decrease that is most apparent in the central subdivision of the En2−/− amygdala (see Table 1 and Figure 2). Further, although we did not perform actual cell counts, we did not observe obvious changes in neuronal packing or cell size in the nuclei examined.
Figure 2. Volumetric analysis of amygdaloid regions in En2−/− and control animals.
Volumes were calculated for the three subdivisions shown (La/Bla, Central and Medial) on the left side of 3 month old male mice. Solid squares and open circles represent actual volume measurements (as reported in Table 1) for the three wild type and three En2−/− mice examined. Horizontal lines indicate the average of the three volume measurements in each amygdalar nucleus of each genotype. Statistical analysis indicates no significant difference in the amygdalar volumes of the lateral/basolateral, central, and medial nuclei of En2−/− and wild type animals.
Amygdala shape and location
In order to determine if the structure of the En2−/− amygdala was analogous to controls, we compared the percent of the amygdalar volume attained as a function of the distance from the starting point of each nucleus (i.e., the histological section where the first area measurement was made). This allowed us to determine if the intra-amygdaloid areas are similarly distributed along the anterior/posterior (A/P) axis. In Figure 3, the solid line represents a plot of the average values of the measurements taken from the three wild type mice. The hatched line indicates a plot of the average measurements in the three En2−/− mice. In Figure 3A, for example, at one-half of the way through the lateral/basolateral amygdalar nuclei along the A/P axis (or a 50% distance on the X axis), about 35% of both the En2−/− and control La/Bla nucleus is accounted for. Although there is some variation, each nucleus in both the mutant and control brains generally yields a graph with a characteristic “S” shape; and the En2−/− plot closely tracks that of the controls (Figure 3A, B, C). This indicates that the intra-amygdaloid volume is appropriately distributed throughout the En2−/− amygdala when compared to controls in the regions we have studied here. We next compared the distance from the start of each nucleus to an arbitrary reference point: the location where the anterior commisure crosses the midline in the coronal aspect (see figures 29 and 30 of The Mouse Brain Atlas [50]). This allowed us to determine if the amygdalar nuclei are properly positioned within the En2−/− cortex. In Figure 4A, B, and C, we again compared the percentage of total volume of each nuclei reached at a particular location along the A/P axis, but this time it was plotted as a function of the distance from our arbitrary reference point in the cortex. For example, comparing the solid and hatched lines that represent the mean values obtained from three mice of each genotype, we find that by 1500μm from the section where the anterior commisure traverses the midline, about 35% of the La/Bla nucleus has been accounted for in the En2 −/− brain, as compared to about 15% of the corresponding control nucleus. In the mutant, this nucleus first appears at about 250μm from the anterior commisure and disappears about 2.375 mm later (at about 2625 μm from our reference point). In the controls, the nucleus is first seen at about 750μm and last seen at about 3125 μm, yet it still spreads over an equivalent 2.375 mm of cortex. In each of the three regions we examined, the La/Bla, central and medial nuclei, we observed a shift in position measuring approximately 500μm to a more anterior location in the cortex of the En2−/− brain.
In situ hybridization of amygdala nuclei
To validate that the boundaries of the nuclei were correct in the volumetric analysis and to further analyze expression patterns in the amygdalar subdivisions, in situ analysis was performed. Previous work has identified several genes that are specific to individual amygdala subnuclei [62,63,67]. We generated RNA in situ probes from several of these genes in order to more distinctly analyze some of amygdala regions in question. Laminin β3 (Lamb3) and neuronal pentraxin receptor (Nptxr) both robustly labeled the lateral and basolateral regions; glutamate decarboxylase (Gad65) marked medial and central regions; LIM-homeodomain protein (Lhx6) showed expression in the medial, lateral, and basolateral nuclei; and plasma glutathione peroxidase (PGPx) was an excellent marker for medial and cortical amygdala. Comparison of wild-type, En2 +/−, and En2−/− mutant animals using in situ hybridization on adult cryostat sections showed no obvious difference in the expression domains of any of the markers mentioned above. Since many of the gene expression patterns overlap, an overall representation of our findings is shown in Figure 5.
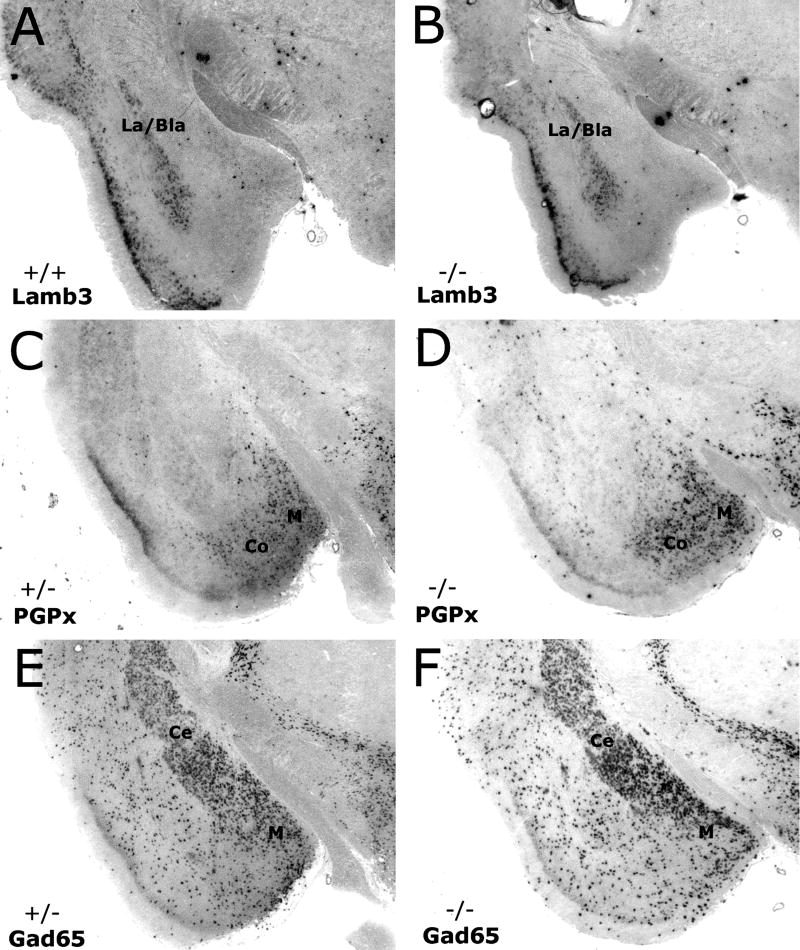
Figure 5. Expression Pattern of Amygdala markers in En2−/− and control animals.
In situ Hybridization with RNA probes that are specific to the lateral & basolateral nuclei (Lamb3; A, B), medial & cortical nuclei (PGPx; C, D), and central & medial nuclei (Gad65; E, F), indicate there is no obvious difference in the pattern of expression in these regions when comparing adult control (A, C, E) and En2 mutants (B, D, F).
Lateral and basolateral nuclei were examined using Lamb3 expression, which clearly marks these amygdala territories in both wild-type (Fig 5A) and mutant (Fig 5B) animals. While the overall pattern differed slightly depending on the location along the A/P axis, it never varied between control and mutants of comparable sections. The same results were seen in the medial and cortical nuclei when labeled with PGPx. Although the expression domain in the cortical region decreased in more posterior sections while remaining apparent in the medial nucleus, there was no obvious difference between controls (Fig 5C) and mutants (Fig 5D) when comparing similar A/P sections. Likewise, when examining the central and medial nucleus using the Gad65 probe (Figs 5E, F), the area of expression in controls and En2−/− brains was nearly identical. Although there is some scattered expression in regions of the amygdala outside of these nuclei, the central and medial divisions are obvious by the dense expression in these areas. The pattern seen with the Gad65 probe remained consistent throughout the A/P axis. Again, although we did not perform cell counts for any of these markers, the overall expression pattern showed no obvious difference in the En2 mutants.
Genetic modifiers of Engrailed
In addition to our structural studies of the En2 amygdala, we have been searching for genes whose expression can modify the phenotype of the midbrain/hindbrain region through interaction with either of the two Engrailed genes. While the phenotype of the Engrailed-2 mutant is mostly resistant to changes in its expressed phenotype on different genetic backgrounds the Engrailed-1 null mutation is exquisitely sensitive. Depending on the background, the same Engrailed-1 mutation can result in phenotypes ranging from perinatal death and a complete absence of the cerebellum and caudal midbrain (on a 129/S1 genetic background, [66]) to a nearly normal lifespan and a nearly normal sized cerebellum with a full complement of constituent cells (on a C57Bl/6 genetic background, [14]). Most interestingly, this apparent rescue of the En1−/− cerebellum on the C57Bl/6 background is completely ablated by the deletion of just one copy of En2, suggesting that the rescue mechanism likely functions through En2. With this in mind, we have been conducting a genome wide screen for genes that can “modify” the En1 null phenotype (Figure 6). Genetic modifiers identified in this screen should be relevant not only to the En1 gene and normal cerebellar development, but also to En2 and Autism. For a review or our approach, please see [46].
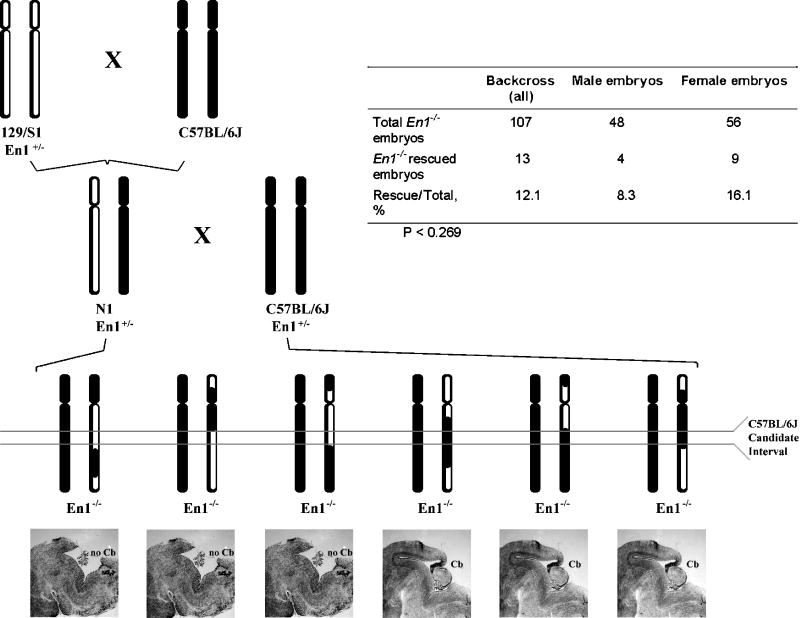
Figure 6. Backcross mapping strategy and sex bias in the rescue embryos.
C57BL/6 mice (solid chromosomes) were crossed to 129/S1• En1+/− mice (hollow chromosomes). The resultant En1+/− F1 animals were then backcrossed to C57BL/6• En1+/− mice to produce En1−/− N2 embryos. DNA collected from the En1−/− N2 embryos was genotyped at loci spanning the genome (A). The presence or absence of a cerebellum in these animals was assessed visually (B). The ability to amplify Y-specific markers or the identification of heterozygous loci on the X chromosome allowed for sexual assignment. Though not statistically significant, female embryos were twice as likely to rescue as male embryos (C). Note: No Y-specific markers amplified for three embryos but all X chromosome markers assayed for these embryos were homozygous, preventing definitive determination of sex.
While we have yet to complete this study, several principles have already been established. First, only a small number of genes accounts for the rescue. This is apparent because we can find structurally intact cerebella among the En1−/− offspring resulting from both the intercross and backcross mapping schemes (although the number of such rescues is very low in the intercross). Second, although we had determined previously that the Engrailed-1 rescue seen on the C57BL/6 background must occur through the actions of the parologous Engrailed-2 gene, our preliminary genetic analysis has failed to localize any of the potential modifiers at a genomic position at or near the Engrailed-2 locus. This suggests that the genes we will identify in our screen must function through modulation of En2 expression rather than through a differential interaction with a modified En2 gene, message or protein. Given the association between En2 and the autism spectrum disorders, we thought it prudent to assay our samples for any sex bias that might exist. Both male and female embryos were recovered in roughly equal numbers among the total mutant progeny. However, mutant female embryos were twice as likely to rescue as male embryos (Figure 6C). Unfortunately, our sample size was not sufficient to produce a statistically significant P-value. Despite this, these results further encourage us in our proposal that using the Engrailed system as an animal model for autism will provide many useful insights into the genetic basis of ASD.
Discussion
The Cerebellum and Autism
A role for the cerebellum in ASD is in agreement with a number of studies indicating that the cerebellum is involved in more than just motor functioning. It has been suggested that the cerebellum plays an important role in sensory tracking, prediction, association, and anticipatory learning [15,41,49,64]. Studies involving spatial attention tasks have shown that autistic individuals as well as individuals with cerebellar lesions took significantly longer to orient their attention than controls [64]. In fMRI studies, abnormalities in cerebellar activation were observed during both motor and non-motor tasks. In autism, cerebellar activation was found to be abnormally low during a task of selective attention [1] and abnormally high during a simple motor task [2]. Courchesne et al. [20] have proposed a model by which the cerebellum functions to anticipate and prepare internal systems for future events. In this model, the cerebellum constantly tracks sensory, cognitive and motor information to formulate and relay predictions to other brain systems thereby generating appropriate and timely attentional responses. This model suggests the cerebellum interacts with a number of brain systems involved in both motor and nonmotor function.
We hypothesize that a disruption in the balance between major brain regions is an integral part of autism. Further, we believe the fulcrum of this balance to be the mid/hindbrain region. The cerebellum, and the development and maintenance of its normal functional relationship with other brain regions, is central to our model. As the cerebellum lies just caudal to our proposed fulcrum, we believe that it is anatomically well positioned to reflect changes in this balance. Indeed, structural changes such as a decreased number of cerebellar Purkinje cells and smaller sized neurons in the deep cerebellar nuclei have been reported in individuals with autism [7,39,56]. Cellular differences such as these may translate into some of the functional differences found in autism. The reduction in Purkinje cells in specific regions of the autistic cerebellum, for example, could release specific neurons in the deep cerebellar nuclei from inhibition. This could result in an abnormally strong physical connectivity along the cerebello-thalamocortical circuit. This altered pattern of excitation could be responsible for autistic features such as abnormalities in cortical maps of motor function and face processing, as well as abnormal overgrowth in frontal lobes [16,45,53].
Engrailed, the cerebellum and autism
Upon cursory examination, the mouse En2−/− cerebellum is about one-third smaller than its wild type counterpart and harbors subtle abnormalities in its folial pattern. Cell counts reveal that all the major cell types of the olivocerebellar circuit (Purkinje, granule, inferior olive and deep nuclear) are reduced by 30–40% in the En2 mutant [40]. In the sagittal plane, there is a misalignment of posterior lobules 8 and 9 at the midline, and a fusion of the Crus II and pml lobules in the hemispheres [37]. Although the structural and cellular changes in the En2−/− cerebellum are not completely congruent with those reported in the autistic brain, they are still quite analogous. At a functional level, Pierce and Courchesne [52] provide insight as to how cerebellar abnormalities may be linked with autistic behavior. In a study of 14 children with autism, they found that abnormal exploratory behavior was significantly associated with the magnitude of the hypoplasia of cerebellar vermal lobules VI and VII. However, a study by Gerlai et al. [31] has indicated no difference in exploratory behavior between En2−/− mice and controls. In this same study, Engrailed-2 mutants performed normally in several tests of motor function, but both hetero- and homozygotes performed below the level of wild type mice in a motor learning paradigm, indicative of a possible learning disability. In this paradigm, mice had to learn to master a progressively difficult motor task: an increase in rotorod rotation speed, in accordance with a set learning criterion. These studies, both in the human and the mouse, suggest that subtle changes in cerebellar architecture can have read outs in non-motor activities such as behavior and learning. Clearly, comprehensive behavioral studies will be of great value in further illuminating the relationship between Engrailed and autism.
It is highly likely that genetic background plays a significant role in the etiology of autism spectrum disorder. This supposition makes our findings of background effects of the Engrailed-1 gene all the more intriguing. The En1 null, which results in a complete loss of the cerebellum and in perinatal lethality on a 129/S1 background, is essentially rescued on a C57BL/6 background. The rescue is blocked, however, when even a single copy of the remaining two En2 genes is missing [14]. Therefore, it is likely that the rescue mechanism includes genetic modifiers that function through the En2 gene. These early studies on their own are consistent with three alternative possibilities. The first is that the C57BL/6 En2 allele contains a coding sequence change that alters its interactions with its binding partners. The second is that there is an alteration in the regulatory regions surrounding the En2 locus that change its response to the developmental patterning biochemistry. It is a third alternative – one that we considered unlikely as we began the study – that is most consistent with our data. It would appear that there are coordinated changes in a few trans-acting regulatory factors, and these changes work in concert to alter the expression pattern of an unchanged Engrailed-2 locus.
Though still incomplete, our mapping studies offer insights into how the Engrailed-1 rescue is brought about at the molecular level. As discussed above, the analyses suggest that a relatively small number of recessive modifiers from the C57BL/6 background are responsible for this shift in En1−/− phenotype. This need not have been the case as the known genetic differences between the two strains are large in number. Further, in our attempts to identify these genes, we have become aware of distinct sex effects. For example, the En1−/− rescued embryos obtained from our analysis of the backcross were twice as likely to be females (Figure 6). If this finding holds up with additional animals, it offers a possible model in which to study the sexually dimorphic pattern of presentation of ASD. The implications of this observation could be important. Though it is far more severe in its outcome, consider for a moment that the Engrailed-1 mouse is the analog of autism in humans. Our findings are clear that if this were the case, then independently segregating factors in the genetic background can suppress the defects (and, by analogy, cure autism). It may be a semantic point, but the fact that this genetic ‘cure’ is more effective in females than in males means that rather than males being more sensitive, females are more resistant to the condition. Where these semantics become important is in the design of molecular models to explain the gene to phenotype transition.
The Amygdala, Engrailed and Autism
MRI assessments in autistic individuals have shown variable changes in the volume of the amygdala. These changes have been alternately reported as increases, decreases and no change [4,34,36,59,60] (and see [18] for review). The discrepancies in these findings may be attributed to variations in the studies with regard to diagnoses, age range of participants, imaging techniques and the different procedures for defining and parceling the amygdala for analysis. The heterogeneity of the disorder itself is also a potential conflicting feature. A combination of contributors such as etilology, genetic susceptibility and environmental factors may vary between subjects and lead to unique differences in brain structure, even in individuals who show similar behavioral symptoms. Changes in amygdalar volume may be representative of a certain category of change within the broad range of ASD abnormalities.
It is also worth considering the oft reported transient nature of certain brain anomalies in ASD. Schumann et al., [59] found a significant enlargement of amygdala volume in younger children with autism, but not in adolescents. At birth, brain size, as determined by head circumference, was found to be normal in children who later developed autism [21]. However, by 2–3 years of age, using quantitative MRI analysis, a 10% increase in brain volume was reported, indicative of an acceleration in brain growth [22]. Another more recent study reported the abnormal enlargement of both cerebral white and grey matter in 2 year old autistic children [33]. Findings from this study suggest that the increased brain growth in autism may begin in the latter part of the first year of life. Interestingly, an analysis of 12 MRI studies showed that by adolescence, the difference in brain volume between autistic individuals and controls was only 1 to 2 % [55]. Volumetric changes such as these likely provoked Schumann and Amaral [58] to quantitatively measure and compare the number of amygdalar neurons in post mortem autistic and control brains. Using stereological methods, they found a significant reduction in neuronal number in both the total amygdala and in the lateral nucleus of autistic individuals. Interestingly, they reported no overall change in total amygdalar volumes. As 17 of the 19 male individuals in this study were age 14 or older at the time of their death, this finding is consistent with their earlier study (mentioned above, [59]) where they reported no change in amygdalar volume in individuals that were 12.75–18.5 years of age. In this regard, our finding of no significant change in amygdalar volume in the 3 month old En2−/− mouse may well be consistent with the transient nature of amygdalar and brain volume enlargement seen in autistic individuals. This suggests that it may be informative to repeat this analysis in younger En2−/− and control mice.
In addition to having essentially normal volumes, amygdaloid regions in En2 mutant mice exhibited characteristically well defined shapes and typical expression patterns of certain molecular markers. However, in the En2 mutant, we found that the lateral, basolateral, central and medial amygdalar nuclei reside in a more anterior position in the cortex when compared to controls. This parallel anterior shift of all four of the nuclei we examined indicates that a noteworthy amygdaloid defect is present in the En2−/− mouse. Interestingly, Miyazaki et al. [44] have reported a more caudal (posterior) shift in the location of 5-HT immunostained cells in the dorsal raphe nucleus of post-natal day 50 rats that were exposed to thalidomide or valoproic acid during early embryogenesis. Animals exposed to these teratogens prenataly generally exhibit abnormalities similar to those found in autism and as such, can be useful in modeling the disease. The authors surmise that the locational shift of the 5-HT cells is most likely the result of aberrant neuronal migration early in development. We believe that something similar could be happening in the Engrailed-2 mutant.
It is initially somewhat counterintuitive that a gene known to be expressed exclusively in the mid/hindbrain region of the CNS would have an effect on a forebrain structure such as the amygdala. One plausible explanation for this phenotypic outcome brings us back to the notion of balance. From many lines of inquiry, it seems apparent that the organization of the CNS places a high premium on developing a proper balance among its many components. Our hypothesis is that perturbations that influence the locations of structures at the midbrain/hindbrain fulcrum during CNS patterning could account for a shift in structures that reside far beyond the fulcrum itself. We further hypothesize that altering the locations of neurons relative to their efferent and afferent partners can have negative effects on cognitive function. One such abnormality, suggested by our results, is the anterior shift of the amygdaloid complex within the temporal lobe. If the incoming fibers do not appropriately correct for the abnormal position, then misassignment of the meaning or emotional value to a variety of stimuli, as well as the aberrant transmittance of this information to other brain regions, might be the result. It will be of interest in our ongoing studies to determine if other structures in the limbic system, such as the hippocampus, exhibit a similar shift in position in the En2−/− mouse.
Engrailed as a window into autism
The En2 mutant mouse is unlikely to be a perfect model for autism. Rather, we believe that its phenotype is representative of a subset of autistic features. The findings from the mutant suggest that the anatomical position of the change in the adult need not reflect the true site of gene action. The irregularities in the cerebellum (Purkinje cell deficit and abnormal folia) and in the amygdala (positional shift in cortical location) in the En2 knockout mouse, as well as the gender differences in the En1 rescued embryos (greater rescue rate in females) are relevant findings in the ongoing search for the mechanisms underlying Autism Spectrum Disorder. Certainly, the Engrailed genes are not the only players in this multi-factorial illness. However, these genes can still provide many clues as to the basis of ASD and many new “leads” to follow up in future studies done in both animal models and afflicted individuals. We are hopeful that our findings, as well as forthcoming structural and behavioral studies, will further the correlation between Engrailed and autism and thus advance our understanding of this complex syndrome.
Acknowledgments
This research was supported by funds granted from The National Alliance for Autism Research, NAAR (also known as Autism Speaks, www.autismspeaks.org) and The National Institutes of Health, NS18381.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allen G, Courchesne E. Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: an fMRI study of autism. Am J Psychiatry. 2003;160:262–73. doi: 10.1176/appi.ajp.160.2.262. [DOI] [PubMed] [Google Scholar]
- 2.Allen G, Muller RA, Courchesne E. Cerebellar function in autism: functional magnetic resonance image activation during a simple motor task. Biological Psychiatry. 2004;56:269–78. doi: 10.1016/j.biopsych.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Amaral DG, Bauman ML, Schumann CM. The amygdala and autism: implications from non-human primate studies. Genes Brain Behav. 2003;2:295–302. doi: 10.1034/j.1601-183x.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 4.Aylward EH, Minshew NJ, Goldstein G, Honeycutt NA, Augustine AM, Yates KO, Barta PE, Pearlson GD. MRI volumes of amygdala and hippocampus in non-mentally retarded autistic adolescents and adults. Neurology. 1999;53:2145–2150. doi: 10.1212/wnl.53.9.2145. [DOI] [PubMed] [Google Scholar]
- 5.Bachevalier J. The amygdala, social cognition, and autism. In: Aggleton JP, editor. The Amygdala. New York: Oxford University Press; 2003. pp. 509–543. [Google Scholar]
- 6.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 7.Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M, Rutter M, Lantos P. A clinicopathological study of autism. Brain. 1998;121:889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- 8.Baron-Cohen S, Ring HA, Wheelwright S, Bullmore ET, Brammer MJ, Simmons A, Williams SC. Social intelligence in the normal and autistic brain: an fMRI study. Eur J Neurosci. 1999;11:1891–8. doi: 10.1046/j.1460-9568.1999.00621.x. [DOI] [PubMed] [Google Scholar]
- 9.Bartlett CW, Gharani N, Millonig JH, Brzustowicz LM. Three autism candidate genes: a synthesis of human genetic analysis with other disciplines. Int J Devl Neurosci. 2005;23:221–234. doi: 10.1016/j.ijdevneu.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Bauman ML. Microscopic neuroanatomic abnormalities in autism. Pediatrics. 1991;87:791–6. [PubMed] [Google Scholar]
- 11.Bauman ML, Kemper TL. Histoanatomic observations of the brain in early infantile autism. Neurology. 1985;35:866–74. doi: 10.1212/wnl.35.6.866. [DOI] [PubMed] [Google Scholar]
- 12.Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci. 2005;23:183–7. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, Kamdar S, Bruse SE, Tischfiels S, Smith B, Zimmerman RA, DiCicco-Bloom M, Brzustowicz LM, Millonig JH. Support for the homeobox transcription factor gene Engrailed-2 as an Autism Spectrum Disorder suseptibility locus. Am J Hum Genet. 2005;77:851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilovocky NA, Romito-DiGiacomo RR, Murcia CL, Maricich SM, Herrup K. Factors in the genetic background suppress the engrailed-1 cerebellar phenotype. J Neurosci. 2003;23:5105–5112. doi: 10.1523/JNEUROSCI.23-12-05105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bower JM, Kassel J. Variability in tactile projection patterns to cerebellar folia crus IIA of the Norway rat. Journal of Comparative Neurology. 1990;302:768–78. doi: 10.1002/cne.903020409. [DOI] [PubMed] [Google Scholar]
- 16.Carper RA, Courchesne E. Inverse correlation between frontal lobe and cerebellum sizes in children with autism. Brain. 2000;123 ( Pt 4):836–44. doi: 10.1093/brain/123.4.836. [DOI] [PubMed] [Google Scholar]
- 17.Casanova MF, Buxhoeveden DP, Brown C. Clinical and macroscopic correlates of minicolumnar pathology in autism. J Child Neurol. 2002;17:692–5. doi: 10.1177/088307380201700908. [DOI] [PubMed] [Google Scholar]
- 18.Cody H, Pelphrey K, Piven J. Structural and functional magnetic resonance imaging of autism. Int J Dev Neurosci. 2002;20:421–38. doi: 10.1016/s0736-5748(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 19.Courchesne E. New evidence of cerebellar and brainstem hypoplasia in autistic infants, children and adolescents: the MR imaging study by Hashimoto and colleagues. J Autism Dev Disord. 1995;25:19–22. doi: 10.1007/BF02178164. [DOI] [PubMed] [Google Scholar]
- 20.Courchesne E, Allen G. Prediction and preparation, fundamental functions of the cerebellum. Learning and Memory. 1997;4:1–35. doi: 10.1101/lm.4.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Courchesne E, Carper R, Akshoomoff N. Evidence of brain overgrowth in the first year of life in autism. JAMA. 2003;290:337–44. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- 22.Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, Chisum HJ, Moses P, Pierce K, Lord C, Lincoln AJ, Pizzo S, Schreibman L, Haas RH, Akshoomoff NA, Courchesne RY. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57:245–54. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- 23.Courchesne E, Yeung-Courchesne R, Press GA, Hesselink JR, Jernigan TL. Hypoplasia of cerebellar vermal lobules VI and VII in autism. N Engl J Med. 1988;318:1349–54. doi: 10.1056/NEJM198805263182102. [DOI] [PubMed] [Google Scholar]
- 24.Critchley HD, Daly EM, Bullmore ET, Williams SC, Van Amelsvoort T, Robertson DM, Rowe A, Phillips M, McAlonan G, Howlin P, Murphy DG. The functional neuroanatomy of social behaviour: changes in cerebral blood flow when people with autistic disorder process facial expressions. Brain. 2000;123:2203–12. doi: 10.1093/brain/123.11.2203. [DOI] [PubMed] [Google Scholar]
- 25.Davis CA, Joyner AL. Expression patterns of the homeo box-containing genes En-1 and En-2 and the proto-oncogene int-1 diverge during mouse development. Genes Dev. 1988;2:1736–44. doi: 10.1101/gad.2.12b.1736. [DOI] [PubMed] [Google Scholar]
- 26.Davis CA, Noble-Topham SE, Rossant J, Joyner AL. Expression of the homeo box-containing gene En-2 delineates a specific region of the developing mouse brain. Genes Dev. 1988;2:361–71. doi: 10.1101/gad.2.3.361. [DOI] [PubMed] [Google Scholar]
- 27.DiCicco-Bloom E, Lord C, Zwaigenbaum L, Courchesne E, Dager SR, Schmitz C, Schultz RT, Crawley J, Young LJ. The developmental neurobiology of autism spectrum disorder. J Neurosci. 2006;26:6897–6906. doi: 10.1523/JNEUROSCI.1712-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Pyschol Psychiatry Allied Disciplines. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 29.Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J Clin Pyschiatry. 2005;66:3–8. [PubMed] [Google Scholar]
- 30.Fombonne E. The epidemmiology of autism: a review. Psychol Med. 1999;29:769–786. doi: 10.1017/s0033291799008508. [DOI] [PubMed] [Google Scholar]
- 31.Gerlai R, Millen K, Herrup K, Fabien K, Joyner AL, Roder J. Impaired motor learning performance in cerebellar En-2 mutant mice. Behav Neurosci. 1996;110:126–33. doi: 10.1037//0735-7044.110.1.126. [DOI] [PubMed] [Google Scholar]
- 32.Gharani N, Benayed R, Mancuso V, Brzustowicz LM, Millonig JH. Association of the homeobox transcription factor, ENGRAILED 2, 3, with Autism Spectrum Disorder. Mol Psychiatry. 2004;9:474–484. doi: 10.1038/sj.mp.4001498. [DOI] [PubMed] [Google Scholar]
- 33.Hazlett HC, Poe M, Gerig G, Smith RG, Provenzale J, Ross A, Gilmore J, Piven J. Magnetic resonance imaging and head circumference study of brain size in autism: birth through age 2 years. Arch Gen Psychiatry. 2005;62:1366–76. doi: 10.1001/archpsyc.62.12.1366. [DOI] [PubMed] [Google Scholar]
- 34.Haznedar MM, Buchsbaum MS, Wei TC, Hof PR, Cartwright C, Bienstock CA, Hollander E. Limbic circuitry in patients with autism spectrum disorders studied with positron emission tomography and magnetic resonance imaging. Am J Psychiatry. 2000;157:1994–2001. doi: 10.1176/appi.ajp.157.12.1994. [DOI] [PubMed] [Google Scholar]
- 35.Herbert MR, Ziegler DA, Deutsch CK, O’Brien LM, Lange E, Bakardjiev A, Hodgson J, Adrien KT, Steele S, Makris N, Kennedy D, Harris GJ, Caviness VS. Dissociations of cerebral cortex, subcortical and cerebral white matter volumes in autistic boys. Brain. 2003;126:1182–92. doi: 10.1093/brain/awg110. [DOI] [PubMed] [Google Scholar]
- 36.Howard MA, Cowell PE, Boucher J, Broks P, Mayers A, Farrant A, Roberts N. Convergent neuroanatomical and behavioural evidence of an amygdala hypothesis of autism. NeuroReport. 2000;11:2931–2935. doi: 10.1097/00001756-200009110-00020. [DOI] [PubMed] [Google Scholar]
- 37.Joyner AL, Herrup K, Auerbach BA, Davis CA, Rossant J. Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science. 1991;251:1239–43. doi: 10.1126/science.1672471. [DOI] [PubMed] [Google Scholar]
- 38.Kemper TL, Bauman ML. Neuropathology of infantile autism. J Neuropathol Exp Neurol. 1998;57:645–52. doi: 10.1097/00005072-199807000-00001. [DOI] [PubMed] [Google Scholar]
- 39.Kemper TL, Bauman ML. The contribution of neuropathologic studies to the understanding of autism. Neurol Clin. 1993;11:175–187. [PubMed] [Google Scholar]
- 40.Kuemerle B, Zanjani H, Joyner A, Herrup K. Pattern deformities and cell loss in Engrailed-2 mutant mice suggest two separate patterning events during cerebellar development. J Neurosci. 1997;17:7881–9. doi: 10.1523/JNEUROSCI.17-20-07881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miall RC, Weir DJ, Wolpert DM, Stein JF. Is the Cerebellum a Smith Predictor? Journal of Motor Behavior. 1993;25:203–216. doi: 10.1080/00222895.1993.9942050. [DOI] [PubMed] [Google Scholar]
- 42.Millen KJ, Hui CC, Joyner AL. A role for En-2 and other murine homologues of Drosophila segment polarity genes in regulating positional information in the developing cerebellum. Development. 1995;121:3935–3945. doi: 10.1242/dev.121.12.3935. [DOI] [PubMed] [Google Scholar]
- 43.Millen KJ, Wurst W, Herrup K, Joyner AL. Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development. 1994;120:695–706. doi: 10.1242/dev.120.3.695. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki K, Narita N, Narita M. Maternal administration of thalidomide or valproic acid causes abnormal serotonergic neurons in the offspring: implication for pathogenesis of autism. Int J Dev Neurosci. 2005;23:287–297. doi: 10.1016/j.ijdevneu.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Muller RA, Pierce K, Ambrose JB, Allen G, Courchesne E. Atypical patterns of cerebral motor activation in autism: a functional magnetic resonance study. Biological Psychiatry. 2001;49:665–76. doi: 10.1016/s0006-3223(00)01004-0. [DOI] [PubMed] [Google Scholar]
- 46.Murcia CL, Gulden FO, Cherosky NA, Herrup K. A genetic study of the suppressors of the Engrailed-1 cerebellar phenotype. Brain Res. 2006 doi: 10.1016/j.brainres.2006.06.076. Epub. [DOI] [PubMed] [Google Scholar]
- 47.Palmen SJ, van Engeland H, Hof PR, Schmitz C. Neuropathological findings in autism. Brain. 2004;127:2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- 48.Pandey SC, Zhang H, Roy A, Misra K. Central and medial amygdaloid brain-derived neurotrophic factor signaling plays a critical role in alcohol-drinking and anxiety-like behaviors. J Neurosci. 2006;26:8320–8331. doi: 10.1523/JNEUROSCI.4988-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paulin M. The role of the cerebellum in motor control and perception. Brain, Behavior and Evolution. 1993;41:39–50. doi: 10.1159/000113822. [DOI] [PubMed] [Google Scholar]
- 50.Paxinos G, Franklin K. The Mouse Brain in Steriotactic Coordinates. New York: Academic Press; 2001. [Google Scholar]
- 51.Petit E, Herault J, Martineau J, Perrot A, Barthelemy C, Hameury L, Sauvage D, Lelord G, Muh JP. Association study with two markers of a human homeogene in infantile autism. J Med Genet. 1995;32:269–74. doi: 10.1136/jmg.32.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierce K, Courchesne E. Evidence for a cerebellar role in reduced exploration and stereotyped behavior in autism. Biological Psychiatry. 2001;49:655–64. doi: 10.1016/s0006-3223(00)01008-8. [DOI] [PubMed] [Google Scholar]
- 53.Pierce K, Muller RA, Ambrose J, Allen G, Courchesne E. Face processing occurs outside the fusiform ‘face area’ in autism: evidence from functional MRI. Brain. 2001;124:2059–73. doi: 10.1093/brain/124.10.2059. [DOI] [PubMed] [Google Scholar]
- 54.Pitkanen A. Connectivity of the rat amygdaloid complex. In: Aggleton JP, editor. The Amygdala: A functional analysis. New York: Oxford University Press; 2003. pp. 31–115. [Google Scholar]
- 55.Redcay E, Courchesne E. When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biological Psychiatry. 2005;58:1–9. doi: 10.1016/j.biopsych.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 56.Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, Guthrie D, Ritvo A. Lower Purkinje cell counts in the cerebella of four autistic subjects: initial findings of the UCLA-NSAC Autopsy Research Report. Am J Psychiatry. 1986;143:862–6. doi: 10.1176/ajp.143.7.862. [DOI] [PubMed] [Google Scholar]
- 57.Sah P, Faber ES, Lopez De Armentia M, Power J. The amygdaloid complex: anatomy and physiology. Physiol Rev. 2003;83:803–34. doi: 10.1152/physrev.00002.2003. [DOI] [PubMed] [Google Scholar]
- 58.Schumann CM, Amaral DG. Stereological Analysis of Amygdala Neuronal number in Autism. J Neuros. 2006;26:7674–7679. doi: 10.1523/JNEUROSCI.1285-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schumann CM, Hamstra J, Goodlin-Jones BL, Lotspeich LJ, Kwon H, Buonocore MH, Lammers CR, Reiss AL, Amaral DG. The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J Neurosc. 2004;24:6392–6401. doi: 10.1523/JNEUROSCI.1297-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sparks BF, Friedman SD, Shaw DW, Aylward EH, Echelard D, Artru AA, Maravilla KR, Giedd JN, Munson J, Dawson G, Dager SR. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59:184–192. doi: 10.1212/wnl.59.2.184. [DOI] [PubMed] [Google Scholar]
- 61.Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Pyschol Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 62.Stenman J, Yu RT, Evans RM, Campbell K. Tlx and Pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development. 2003;130:1113–22. doi: 10.1242/dev.00328. [DOI] [PubMed] [Google Scholar]
- 63.Swanson LW, Petrovich GD. What is the amygdala? Trends in Neurosci. 1998;21:323–31. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- 64.Townsend J, Courchesne E, Covington J, Westerfield M, Harris NS, Lyden P, Lowry TP, Press GA. Spatial attention deficits in patients with acquired or developmental cerebellar abnormality. J Neurosci. 1999;19:5632–43. doi: 10.1523/JNEUROSCI.19-13-05632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vogel MW, Ji Z, Millen K, Joyner AL. The Engrailed-2 homeobox gene and patterning of spinocerebellar mossy fiber afferents. Brain Res Dev Brain Res. 1996;96:210–8. doi: 10.1016/0165-3806(96)00122-8. [DOI] [PubMed] [Google Scholar]
- 66.Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–2075. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]
- 67.Zirlinger M, Kreiman G, Anderson D. Amygdala-enriched genes identified by microarray technology are restricted to specific amygdaloid subnuclei. Proc Natl Acad Sci U S A. 2001;98:5270–5. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]