Abstract
Reprogramming of DNA methylation patterns during mammalian preimplantation development involves the concurrent maintenance of methylation on differentially methylated domains (DMDs) of imprinted genes and a marked reduction of global (non-DMD) genomic methylation. In the developing mammalian embryo, one allele of a DMD is unmethylated, and the opposite parental allele is methylated, having inherited this methylation from the parental gamete. The maintenance of DMDs is important for monoallelic imprinted gene expression and normal development of the embryo. Because the DNMT1 cytosine methyltransferase governs maintenance methylation in mammals, rearrangements of non-DMD, but not DMD methylation in preimplantation embryos suggest that the preimplantation DNMT1-dependent maintenance mechanism specifically targets DMD sequences. We explored this possibility using an engineered mouse ES cell line to screen for mutant DNMT1 proteins that protect against the loss of DMD and/or global (non-DMD) methylation in the absence of the wild-type endogenous DNMT1 methyltransferase. We identified DNMT1 mutants that were defective in maintenance of either DMD and/or non-DMD methylation. Among these, one mutant maintained non-DMD methylation but not imprinted DMD methylation and another mutant maintained just DMD methylation. The mutated amino acids of these mutants reside in a mammal-specific, disordered region near the amino terminus of DNMT1. These findings suggest that DNMT1 participates in epigenetic reprogramming through its ability to distinguish different categories of methylated sequences.
Keywords: epigenetic, imprinting, methylase, methylation, reprogramming
Genomic imprinting is a mammalian epigenetic process that distinguishes maternal and paternal alleles to ensure parent-specific (monoallelic) expression of ≈80 imprinted genes (1). The molecular basis of this process is de novo methylation during gametogenesis and maintenance methylation during embryogenesis; this sequence of activities leads to the generation of imprinted DMDs (2). Methylation is placed on DMD sequences in the maternal and paternal germ lines by the DNMT3a cytosine methyltransferase (3, 4). Following fertilization DMD methylation is maintained (inherited) during preimplantation development by the combined action of different isoforms of the DNMT1 cytosine methyltransferase (5–8). The Mr 175,000 DNMT1o form is synthesized in the oocyte and maintains methylation during preimplantation development (5, 9), whereas the Mr 190,000 DNMT1s form is synthesized both in the oocyte and in the early embryo and this form also functions in preimplantation embryos (6). Along with this maintenance of DMD methylation is a significant reduction in the level of global (non-DMD) methylation (10, 11). The concurrent inheritance of DMD methylation and reduction of global methylation rearranges the genome's methylation patterns just before the formation of pluripotent embryo stem cells (12).
Maintenance of DMD methylation in the presence of a reduction in the average level of genomic methylation during preimplantation development could be explained by selective maintenance of DMD methylation (13). Because DNMT1 is the only known maintenance methyltransferase in preimplantation embryos, then DNMT1 itself may selectively maintain DMD methylation. How this would occur is not known, although we can speculate that preimplantation DNMT1 may bind to hemimethylated DMD sequences, but not to hemimethylated non-DMD sequences. Alternatively, preimplantation DNMT1 may bind all hemimethylated genomic sequences, yet function catalytically to convert only hemimethylated DMDs to fully methylated DMDs. Hemimethylated non-DMD sequences whose methylation is not actively maintained during a cell cycle would undergo replication during the next cell cycle, generating unmethylated DNA (5).
Oocyte-derived DNMT1o and embryo-derived DNMT1s are both present in nuclei of 8-cell blastomeres, yet only the oocyte-derived protein can maintain imprinted DMD methylation at this stage (5, 6). This maternal-effect DNMT1o function can be restored by oocyte-derived DNMT1s protein, but not restored by embryo-derived DNMT1o expressed from a modified endogenous Dnmt1 allele (6, 14). These findings are further support for sequence-specific DNMT1 activity during preimplantation development, and suggest that the developmental source of the protein (oocyte versus embryo) determines this specificity. A possible mechanism for sequence-specific maintenance methylation would be oocyte- or embryo-specific posttranslational modifications (PTMs) of DNMT1 proteins. For example, an oocyte-specific PTM of DNMT1o may be required for it to maintain DMD methylation at the 8-cell stage of embryogenesis; in the absence of this PTM, DNMT1o cannot maintain DMD methylation. Alternatively, sequence-specific activity of DNMT1 proteins could be mediated through proteins that interact with DNMT1. Although a number of different proteins are known to interact with DNMT1 proteins (15–17), there is no evidence that such interactions lead to sequence-specific maintenance methyltransferase activity. Because of the likely roles of sequence-specific DNA methylation in general, and DNMT1 proteins specifically in epigenetic reprogramming and the inheritance of genomic imprints, we screened mutant DNMT1 proteins for activity in maintaining methylation on different DNA sequences.
Results
Lack of Interference with Endogenous DNMT1 Function.
We approached the issue of selective maintenance methylation by DNMT1 by first determining whether DNMT1s mutants expressed in ES cells interfere with wild-type DNMT1s function and consequently alter genomic methylation patterns. The 5′ region of the Dnmt1s transcript encodes an amino acid domain of ≈160 aa (amino acids 190–350) found only in eutherian mammals (Fig. 1 A and B). Because of the possibility that this region would function in a mammal-specific, methylation-dependent process such as genomic imprinting, we generated a series of mutations in this region and expressed them in wild-type R1 ES cells (Fig. 1C). The mutations encoded for DNMT1s proteins that were deleted for contiguous stretches of amino acids. Because the mammal-specific region is N-terminal of a known region required for homodimer formation (18), it is possible that heterodimers between endogenous DNMT1s and exogenous expressed mutant DNMT1s would interfere with function of the endogenous wild-type protein. Alternatively, expression of a mutant DNMT1s protein could interfere in other ways with function of wild-type DNMT1s.
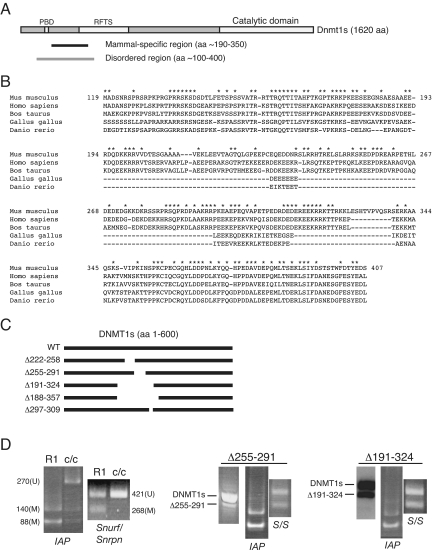
Fig. 1.
Mammal-specific region of DNMT1s and effect of expression of mutant DNMT1s proteins in ES cells. (A) Schematic of DNMT1s protein showing location of overlapping disordered and mammal-specific regions and other landmarks. Near the amino terminus, the PCNA binding domain (PBD) and targeting sequences (RFTS), which have been implicated in targeting DNMT1 to replication foci, are highlighted (23, 30, 31). (B) Comparison of amino acid sequences of the N-terminal region of DNMT1. Region shown is approximately that defined as “part A” of DNMT1o (22). Mouse PBD is TRQTTITAHFTKG (amino acids 158–170). Every amino acid that is identical in the three mammalian species is marked by an asterisk (*). (C) Schematic of mutations within amino terminus of DNMT1s. The analyzed deletion (Δ) mutants and the positions of their deleted amino acids are shown. (D) Effects of expression of two DNMT1s deletion mutants in R1 ES cells. The level of expression of the Δ255–291 mutant transcript compared to endogenous Dnmt1s transcript was measured by RT-PCR followed by electrophoretic separation of wild-type and mutant products. Forward primer for PCR: GAG TCG GAA GAG GGG AAC TC; reverse primer for PCR: CAT GAA TTG CTT TGG CAC AC. The mutant product is 700 bp and the wild-type product is 810 bp. The level of expression of Δ191–324 compared to endogenous DNMT1s was assessed on an immunoblot using the anti-DNMT1s UPT82 antibody. Non-DMD methylation was measured by an IAP COBRA assay and DMD methylation was measured by an Snurp/Snrpn COBRA assay (29, 30). S/S, Snurf/Snrpn. Numbers are sizes in nucleotides of methylated (M) or unmethylated (U) COBRA assay digestion products.
Deletion of a small portion (Δ255–291) of the mammal-specific region did not alter non-DMD [as assessed by a COBRA assay for methylation of the repetitive intracisternal A particle (IAP) retroelement] or DMD (assessed by a COBRA assay for Snurf/Snrpn DMD) methylation in the presence of wild-type DNMT1s (Fig. 1D). For comparison, COBRA assays for IAP and Snurf/Snrpn sequences in wild-type R1 and Dnmt1-null (Dnmt1c/c) ES cells are shown. Deletion of the majority of the mammal-specific region (Δ191–324) also did not alter non-DMD or DMD methylation (Fig. 1D). We conclude from this analysis that there are no measurable effects on genomic DNA methylation that would suggest that these mutants interfere with endogenous wild-type protein function.
Transient Absence of DNMT1 Expression Leads to Loss of Imprinted Methylation.
We next considered the possibility that mutant DNMT1s proteins with mutations in the mammal-specific region may be innately defective in maintaining DMD methylation. To address this we developed a genetic screen to identify DNMT1 mutants that are defective in maintaining DNA methylation. The foundation of this approach is an ES cell line in which endogenous DNMT1s expression is extinguished by the addition of 2 μg/mL doxycycline (TET-OFF regulation). These Dnmt1tet/tet ES cells were generated using targeted mutagenesis to introduce tet regulatory cassettes into exon 1s sequences of both Dnmt1 alleles of R1 ES cells (Fig. 2A). The placement of the cassettes results in expression of the tTA activator gene from the endogenous Dnmt1s promoter. tTA protein binds to the tet operator sequences located 5′ of the initiation codon in the modified exon 1s to promote transcription of a novel Dnmt1s transcript in trans. The binding of doxycycline to tTA prevents this transactivation.
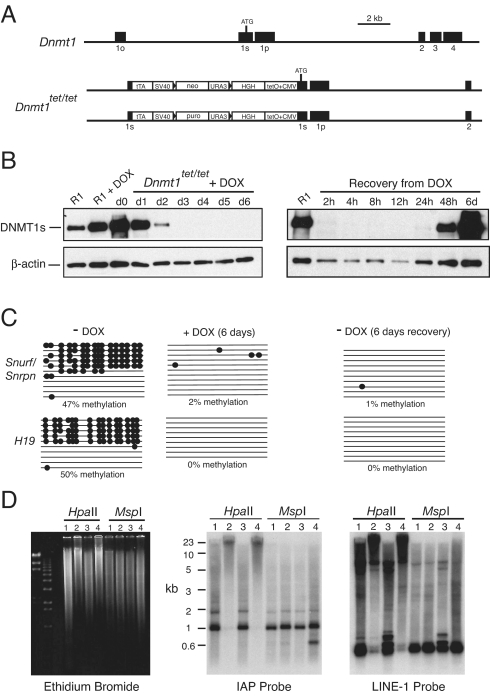
Fig. 2.
Responses of Dnmt1tet/tet ES cells to addition and removal of doxycycline (DOX). (A) Description of mutant alleles generated by insertion of TET-OFF cassettes into exon 1s of both Dnmt1 alleles in R1 ES cells. tTA, tet activator; TetO+CMV, an element containing multiple copies of the tet operator followed by a minimal promoter from cytomegalovirus; neo, the neomycin resistance gene; and puro, the puromycin resistance gene. (B) Addition of 2 μg/mL DOX to the culture medium results in loss of DNMT1s protein in Dnmt1tet/tet ES cells. DNMT1s protein was detected on immunoblots using the UPT82 antibody. β-actin protein levels were measures of protein loading. R1, wild-type R1 ES cells; d0, start of doxycycline exposure. After 3 days of treatment with DOX, DNMT1s was not detected. Upon removal of doxycycline from the culture medium, DNMT1s protein is again expressed. (C) Snurf/Snrpn DMD methylation is lost when DNMT1s expression is extinguished in Dnmt1tet/tet cells and remains low when DNMT1s is reexpressed. Each horizontal line is a Snurf/Snrpn allele whose methylation is determined by bisulfite genomic sequencing. Filled circles represented methylated CpG dinucleotides and the absence of circles indicate unmethylated CpGs. (D) Ethidium-stained gel and Southern blots showing the IAP and LINE-1 methylation in Dnmt1tet/tet cells. 1, Dnmt1c/c; 2, Dnmt1tet/tet in the absence of DOX; 3, Dnmt1tet/tet after 6 days of exposure to DOX; 4, Dnmt1tet/tet after 6 days of recovery from DOX exposure.
The response of Dnmt1tet/tet cells to the addition and removal of doxycycline is shown in Fig. 2B. In the absence of doxycycline, there was a significantly higher level of DNMT1s expression in Dnmt1tet/tet cells, presumably due to the strong tTA transactivation. This level of increase in DNMT1s expression is not likely to influence genomic methylation (14, 19). Following 3 days of exposure of Dnmt1tet/tet cells to doxycycline, the level of DNMT1s protein fell to undetectable levels. As a consequence of the doxycycline-induced loss of DNMT1s, there was a substantial loss of Snurf/Snrpn DMD, H19 DMD, IAP, and LINE-1 (L1) methylation (Fig. 2 C and D). Following removal of doxycycline from the culture medium, DNMT1s protein levels returned to the starting level (Fig. 2B). Coincident with the restoration of DNMT1s protein expression in Dnmt1tet/tet cells, IAP and L1 methylation levels returned to normal, yet the Snurf/Snrpn DMD and H19 DMD methylation levels remained very low (Fig. 2 C and D). We conclude from this characterization that DNMT1s maintenance methyltransferase protein can be effectively regulated in an ON-OFF fashion in Dnmt1tet/tet ES cells and that profound changes in genomic methylation accompany changes in DNMT1s protein levels.
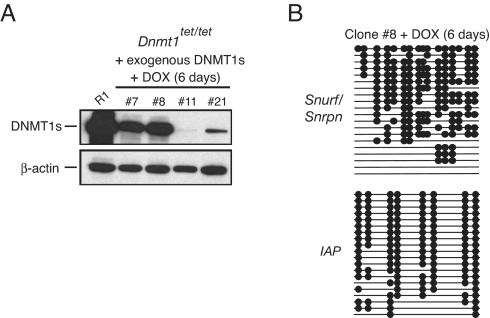
We next addressed whether full-length DNMT1s expressed from a stably integrated expression plasmid in Dnmt1tet/tet ES cells can prevent the doxycycline-induced loss of DNA methylation. In this experiment, a Pgk-hygromycinR plasmid was transfected (electroporated) along with the Dnmt1s expression plasmid, and a number of individual hygromycin-resistant colonies collected and expanded. Following 6 days of exposure to doxycycline, which extinguished endogenous DNMT1s expression, as expected there was clonal variability in the expression of exogenous DNMT1s (Fig. 3A). Some clones (#11 and #21) expressed very low amounts of DNMT1s, whereas other clones (#7 and #8) showed much higher levels of DNMT1s, although less than in R1 cells. Importantly, the level of Snurf/Snrpn DMD and IAP methylation were unaffected in clones #7 and #8 in the presence of doxycycline for 6 days, an exposure that results in loss of methylation on these sequences in Dnmt1tet/tet cells (data for clone #8 shown in Fig. 3B). The return of IAP and L1 methylation to normal or near normal levels following the removal of doxycycline (Figs. 2D and 3B) is analogous to the increase in global methylation when DNMT1s is expressed from an exogenous plasmid construct in Dnmt1-null (Dnmt1c/c) cells (20). Overall, these findings indicate that the Dnmt1tet/tet ES cells can enable identification of regions in DNMT1 regulating the maintenance of genomic imprints.
Fig. 3.
Expression of exogenous DNMT1s from a stably integrated expression plasmid prevents DOX-induced loss of methylation in Dnmt1tet/tet cells. (A) Relative levels of exogenous DNMT1s in four ES cell clones. (B) Snurf/Snrpn DMD and IAP methylation in ES cell clone #8 expressing wild-type DNMT1s protein in the presence of DOX. Methylation was determined by bisulfite genomic sequencing.
DNMT1 Mutants Show Defects in Maintenance Methylation.
To identify DNMT1 mutants that are capable of maintaining non-DMD but not DMD methylation, we focused on the amino terminus of the protein (Fig. 1 A and B). Amino acids 1–118 of DNMT1s define a region that interacts with the DMAP1 protein (21), and this region has been shown to be dispensable for viability and fertility in mice (14). The region defined by amino acids 119–410 is located just C terminal to the DMAP1-interaction domain, and is approximately the same as part A of the tripartite structure of DNMT1o proposed by Margot et al. (22). Although the precise functions of this region are unknown, there is experimental evidence supporting its role in nuclear localization, replication-associated maintenance methylation, and functional interactions with other regions of DNMT1 (22–24). Much of this region is present and evolutionarily conserved only in mammals (Fig. 1B) and, when this region is deleted, DNMT1 exhibits a partial loss of DNA methyltransferase activity in vitro (22). Although this in vitro evidence might indicate that the region is dispensable for maintaining DNA methylation in cells, we postulate that this region is instead required for mammal-specific regulatory functions, and therefore studied the maintenance methylation function of DNMT1 proteins containing mutations in this region.
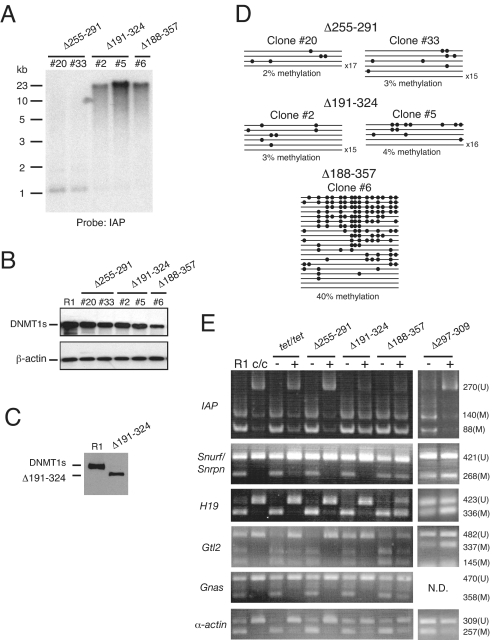
Deletion mutations of Dnmt1 were generated by site-directed mutagenesis. Each DNMT1s mutant was stably expressed in Dnmt1tet/tet cells using the same expression vector used to express wild-type DNMT1s. The level of mutant protein expression relative to the expression of β-actin was determined in a number of different hygromycin-resistant clones, and only those clones expressing at least 50% of the DNMT1s levels in wild-type (R1) ES cells were studied further. After 6 days of exposure of these clones to doxycycline, endogenous DNMT1s protein expression had been extinguished, and only the exogenous mutant DNMT1s proteins were expressed. At this point the level of methylation of different sequences was determined. As shown in Fig. 4A, IAP sequences in cells expressing a form of DNMT1s missing amino acids 188–357 (Δ188–357), a region that roughly defines the mammal-specific portion of DNMT1, were methylated. Because IAP retrotransposons are an abundant class of repetitive genomic sequences, IAP methylation reflects non-DMD genomic methylation (5). In addition, methylation was maintained on the imprinted DMDs of the Snurf/Snrpn, H19, Gtl2, and Gnas genes, as well as maintained on the promoter of the single-copy α-actin gene (Fig. 4E). This normal DMD and non-DMD methylation was seen in four out of four Δ188–357 clones examined. These observations are consistent with previous observation that DNMT1 mutant proteins devoid of these sequences retain maintenance methyltransferase activity in vitro (22) and consistent with the finding that human imprinted DMD sequences retain their methylation in a chick–human hybrid cell line expressing the chicken (Gallus gallus) DNMT1 (25).
Fig. 4.
Effects of mutant DNMT1s proteins on maintenance of genomic methylation. (A) Southern hybridization of HpaII-digested genomic DNA samples with an IAP LTR probe. DNA was isolated from individual clones after 6 days of exposure to DOX. Individual ES cell clone numbers are indicated at the top. Clones #20 and #33 (Δ255–291), #2 and #5 (Δ191–324), and #6 (Δ188–357). (B) Immunoblots showing wild-type (R1 ES cells) and mutant DNMT1 proteins in same ES clones depicted in panel A. (C) Immunoblot comparing relative sizes of wild-type DNMT1s protein expressed in R1 ES cells and the Δ191–324 DNMT1s mutant expressed in Dnmt1tet/tet ES cells following 6 days of exposure to 2 μg/mL DOX. (D) Snurf/Snrpn DMD methylation in DNA samples from panel A. Methylation was determined by bisulfite genomic sequencing, and symbols defined as in Fig. 1C. (E) COBRA assays. Gnas PCR products were digested with HpyCH4IV and other PCR products digested as described previously (29, 30). −, no DOX; +, 6 days of exposure to 2 μg/mL DOX. Fragments representing methylated (M) or unmethylated (U) DNA and their sizes in nucleotides are indicted. R1, wild-type R1 ES cells; c/c, Dnmt1c/c ES cells; tet/tet, Dnmt1tet/tet ES cells. ND, not done.
Although the mammal-specific region of DNMT1s between amino acids 187 and 358 is not absolutely required for maintaining DMD and non-DMD methylation in Dnmt1tet/tet ES cells, we were interested in knowing whether this region, when present in the DNMT1s protein, regulates the maintenance of DMD and/or non-DMD methylation. To address this issue, we first screened the same set of DNMT1s deletion mutants previously screened in R1 ES cells in Dnmt1tet/tet ES cells. The positions of these deletions are shown schematically in Fig. 1C. The two mutants (Δ222–258 and Δ255–291) containing 36-amino-acid deletions in the N-terminal portion of the mammal-specific region failed to maintain DMD and non-DMD methylation after 6 days of exposure to DOX (Fig. 4 A and E and supporting information (SI) Table S1). These findings indicate that disruptions in the normal integrity of the mammal-specific region can affect the protein's function in maintaining both DMD and non-DMD methylation in living cells.
Based on the observations that relatively small deletions in the amino-terminal portion of the mammal-specific region prevented DNMT1s from maintaining genomic methylation patterns in ES cells, but deletion of the entire mammal-specific region did not affect genomic methylation patterns, we reasoned that other deletions within the mammal-specific region would produce intermediate molecular phenotypes. We therefore constructed a DNMT1s mutant with a deletion of the N-terminal four-fifths of the mammal-specific region (Δ191–324) and evaluated its function in Dnmt1tet/tet ES cells. Interestingly, in contrast to the mutants with smaller deletions in the amino-terminal portion of the mammal-specific region (Δ222–258 and Δ255–291), this mutant maintained non-DMD methylation but not DMD methylation (Fig. 4 A and E). This specific maintenance of non-DMD methylation was seen in six out of six Δ191–324 clones examined. We conclude from this analysis that mutant DNMT1s proteins can be identified that are selectively defective in maintaining methylation on imprinted DMD sequences. This result is consistent with a role of the mammal-specific region in selectively regulating the maintenance of different types of genomic methylation patterns.
We also determined the function of a number of DNMT1s mutants with small deletions (12 aa) within the mammal-specific region (Table S1). Two of these mutants (Δ288–300 and Δ305–317) were unable to maintain DMD and non-DMD methylation, whereas the Δ322–334 mutant maintained both DMD and non-DMD methylation in Dnmt1tet/tet cells in the absence of wild-type DNMT1s protein. None of these mutant proteins therefore exhibited any evidence of selective maintenance methylation. In contrast to these, the Δ297–309 mutant maintained DMD methylation but not non-DMD methylation (Fig. 4E). This specific maintenance of DMD methylation was seen in six out of six Δ297–309 clones examined. The abnormal maintenance methylation phenotype of Δ297–309 is thus opposite that of the Δ191–324 mutant, indicating that different regions of the mammal-specific DNMT1s region regulate DMD and non-DMD methylation. As expected, DNMT1s mutants with partial or complete maintenance methyltransferase activity localize to the nucleus (Fig. S1).
Discussion
The analyzed DNMT1s proteins contained mutations within a region of DNMT1s found only in mammalian species (amino acids 188–357). Because the entirety of this mammal-specific region is embedded in a predicted disordered region [amino acids 100–400; Disopred2 disorder prediction (26)], it appears that a unique and uncharacterized feature of the DNMT1s protein is involved in a mammal-specific function(s) of DNMT1s. We obtained a number of different molecular phenotypes by analyzing DNMT1s mutants for their ability to maintain DMD and non-DMD methylation in the absence of wild-type DNMT1s protein. Observed mutant phenotypes ranged from a failure to maintain both DMD and non-DMD methylation to a wild-type phenotype in which both DMD and non-DMD methylation were maintained. Between these two extreme phenotypes, we observe partial phenotypes in which either just DMD methylation or just non-DMD methylation was maintained. Our findings indicate that one function of this mammal-specific, disordered region is to distinguish DMD from non-DMD sequences.
During normal preimplantation development non-DMD methylation is not maintained while DMD methylation is maintained; a failure to maintain DMD methylation leads to a loss of imprinted monoallelic expression and fetal death (5, 13). For these reasons, distinguishing these two main categories of genomic sequences is likely to be a crucial aspect of the molecular mechanism underlying genomic imprinting. In principle, distinguishing among categories of genomic sequences could be through a direct interaction of the disordered region with hemimethylated sequences or through interactions with unknown accessory proteins that, in turn, bind to different hemimethylated sequences. Regardless of whether accessory proteins are involved in maintaining methylation on different genomic sequences, the mammal-specific region of DNMT1s most likely plays a role in the maintenance of all genomic methylation.
Insights into the specific function of the mammal-specific region can be derived from the observed effects of the different DNMT1s mutants on DMD and non-DMD methylation. The maintenance of DMD and non-DMD methylation in the Δ188–357 mutant indicates that DNA methylation patterns can be maintained in the absence of the mammal-specific region. This finding is consistent with the observation that human imprinted DMD sequences retain their methylation in chick–human hybrid cells expressing the chicken (G. gallus) DNMT1, which is missing the mammal-specific region (Fig. 1B and ref. 25). However, because mutations within this mammal-specific region (Δ191–324, Δ222–258 and Δ255–291, Δ297–309) affect DMD and/or non-DMD methylation, the mammal-specific region, when present, regulates genomic methylation. The main region of overlap between the Δ187–357 and Δ191–324 mutants (amino acids 325–357) is likely to be involved in regulating the maintenance of DMD methylation, and mammal-specific sequences other than the amino acids deleted in the Δ297–309 mutant are likely to be involved in regulating the maintenance of non-DMD methylation. However, this straightforward interpretation may be inaccurate because it does not account for the possible effects of amino acid deletions on the function of nearly amino acids. Therefore, the accurate assignment of amino acids to different maintenance methylation functions will require a much more extensive and detailed mutational analysis of this region. In vivo studies of DNMT1 mutants will be required to determine whether this region has a role in developmental changes in DNA methylation.
The identification of DNMT1s mutants with specific maintenance methyltransferase activity in ES cells suggests that the wild-type DNMT1s protein can fail to maintain all DNA methylation patterns. On the basis of our analysis of mutations within the mammal-specific region, this would occur by activating an inhibitory function of the region, such that DMD and/or non-DMD methylation is not maintained. One such means by which an inhibitory function might be acquired is through stage-specific posttranslational modifications or interactions with other proteins. Interestingly, there are two stages in normal development where such effects may occur. First, during proliferation of primordial germ cells (PGCs) and their migration to the genital ridges, both DMD and non-DMD methylation are lost, despite the presence of DNMT1s protein in the nuclei of the PGCs. We speculate that during this period of development, DNMT1s protein inhibits the maintenance of both DMD and non-DMD methylation. Second, during preimplantation development of the blastocyst, DMD methylation is maintained, whereas a significant portion of non-DMD methylation is not maintained (10, 11). We speculate that during this developmental window, as observed with the Δ297–309 mutant, DNMT1s protein selectively inhibits the maintenance of non-DMD methylation.
Materials and Methods
Mutant DNMT1s Expression.
Deletions were generated using a wild-type mouse Dnmt1s cDNA as template. XbaI restriction sites were introduced into the cDNA at defined positions by site-directed mutagenesis (QuikChange kit,Stratagene; Table S2). Site-directed mutagenesis products were transformed into Escherichia coli, mutant plasmids were digested with XbaI, gel purified, ligated, and again transformed into E. coli. Mutant DNMT1s proteins were expressed in R1 and Dnmt1tet/tet ES cells by coelectroporation with a plasmid expressing a mutant Dnmt1 cDNA from the mouse Pgk-1 promoter, and a Pgk-1-hygromycinr plasmid. Following selection with hygromycin, ES clones expressing exogenous DNMT1s were identified after doxycycline (DOX) treatment by Western blotting using the UPT82 antibody (27). These cells maintained undifferentiated colony morphologies throughout treatment with antibiotics (hygromycin and doxycycline) and during their proliferation. Mutant Dnmt1s transcripts were detected using RT-PCR assays as previously described (27), incorporating restriction enzyme digestion of RT-PCR products in some cases to distinguish wild-type and mutant products.
Dnmt1tet/tet ES cells.
The two alleles of the endogenous Dnmt1 locus of R1 ES cells were sequentially modified using two similar targeting vectors to generate the DOX-OFF Dnmt1tet/tet cell line. The isolated 5′ portion of the mouse Dnmt1 gene used for targeted mutagenesis (14) was first modified by site-directed mutagenesis to introduce a unique MluI site 30 nucleotides 5′ to the Dnmt1s initiation codon. Each TET-OFF cassette, containing either a neor or a puror gene, was then cloned into the MluI site (28).
Immunoblot Assay.
Detection of DNMT1s proteins was performed using an immunoblot assay as previously described (6). Affinity-purified rabbit anti-DNMT1 antibody UPT82 was used at a 1:1,000 dilution. The anti-rabbit secondary antibody was used at a 1:50,000 dilution. The enhanced chemiluminescence detection kit was from Amersham Pharmacia.
Immunostaining.
Dnmt1v/v primary mouse embryonic fibroblasts were grown on glass cover slips and transfected with circular plasmids expressing wild-type and mutant Dnmt1s cDNAs from the human EF-1α promoter (pEF1/myc, Invitrogen) using Lipofectamine 2000 (Invitrogen). Twenty-four hours later cells were fixed, immunostained, and imaged as previously described (6).
CpG Methylation Assays.
IAP, α-actin, Snurf/Snrpn, H19, Gnas, and Gtl2 DMD methylation levels were determined using combined bisulfite restriction analysis (COBRA) assays (29, 30) Table S3), bisulfite genomic sequencing (6) and Southern hybridizations (27).
Supplementary Material
Acknowledgments.
The authors thank Dr. Andrew VanDemark for many helpful discussions, and Dr. En Li for the gift of Dnmt1c/c ES cells. This work was supported by a grant to J.R.C. from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905668106/DCSupplemental.
References
- 1.Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart B, Chaillet JR. Genomic imprinting: Cis-acting sequences and regional control. Int Rev Cytol. 2005;243:173–213. doi: 10.1016/S0074-7696(05)43003-X. [DOI] [PubMed] [Google Scholar]
- 3.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 4.Kaneda M, et al. Essential role for de novo methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 5.Howell CY, et al. Genomic imprinting disrupted by a maternal-effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 6.Cirio MC, et al. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008;8:9. doi: 10.1186/1471-213X-8-9. Jan 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirasawa R, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurihara Y, et al. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev Biol. 2008;313:335–346. doi: 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 9.Mertineit C, et al. Sex-specific exons control DNA methyltransferase activity in mammalian germ cells. Development. 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- 10.Chaillet JR, Vogt TF, Beier DR, Leder P. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell. 1991;66:77–83. doi: 10.1016/0092-8674(91)90140-t. [DOI] [PubMed] [Google Scholar]
- 11.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 12.Branco MR, Oda M, Reik W. Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev. 2008;22:1567–1571. doi: 10.1101/gad.1690508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhart B, Paoloni-Giacobino A, Chaillet JR. Specific differentially methylated domain sequences direct the maintenance of methylation at imprinted genes. Mol Cell Biol. 2006;26:8347–8356. doi: 10.1128/MCB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding F, Chaillet JR. In vivo stabilization of the Dnmt1 (cytosine-5)- methyltransferase protein. Proc Natl Acad Sci USA. 2002;99:14861–14866. doi: 10.1073/pnas.232565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 16.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 17.Sajedi E, et al. DNMT1 interacts with the developmental transcriptional repressor HESX1. Biochim Biophys Acta. 2008;1783:131–143. doi: 10.1016/j.bbamcr.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellinger K, Rothbauer U, Felle M, Längst G, Leonhardt H. Dimerization of DNA methyltransferase 1 is mediated by its regulatory domain. J Cell Biochem. 2009;106:521–528. doi: 10.1002/jcb.22071. [DOI] [PubMed] [Google Scholar]
- 19.Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)- methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damelin M, Bestor TH. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol Cell Biol. 2007;27:3891–3899. doi: 10.1128/MCB.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 22.Margot JB, et al. Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J Mol Biol. 2000;297:293–300. doi: 10.1006/jmbi.2000.3588. [DOI] [PubMed] [Google Scholar]
- 23.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 24.Margot JB, Ehrenhofer-Murray AE, Leonhardt H. Interactions within the mammalian DNA methyltransferase family. BMC Mol Biol. 2003;30(4):7. doi: 10.1186/1471-2199-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horike S, et al. Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum Mol Genet. 2000;9:2075–2083. doi: 10.1093/hmg/9.14.2075. [DOI] [PubMed] [Google Scholar]
- 26.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Ratnam S, et al. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev Biol. 2002;245:304–314. doi: 10.1006/dbio.2002.0628. [DOI] [PubMed] [Google Scholar]
- 28.Bond CT, et al. Respiration and parturition affected by conditional overexpression of the Ca2+-activated K+ channel subunit, SK3. Science. 2000;289:1942–1946. doi: 10.1126/science.289.5486.1942. [DOI] [PubMed] [Google Scholar]
- 29.Schermellah L, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J-Y, Lees-Murdock DJ, Xu G-L, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84:952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Chuang LS, et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.