Abstract
Raltegravir is a recently, Food and Drug Administration-approved, small-molecule drug that inhibits retroviral integrase, thereby preventing HIV DNA from inserting itself into the human genome. We report here that the activity profile of raltegravir on the replication of murine leukemia virus is similar to that for HIV, and that the drug specifically affects autoimmune disease in mice, in which endogenous retroelements are suspected to play a role. While NZW and BALB/c mice, which do not succumb to autoimmune disease, are not affected by raltegravir, lupus-prone (NZBxNZW) F1 mice die of glomerulonephritis more than a month earlier than untreated mice. Raltegravir-treated NZB mice, which share the H-2 haplotype with BALB/c mice, but which are predisposed to autoimmune hemolytic anemia, develop auto-antibodies to their red blood cells >3 months earlier than untreated mice of the same strain. Because nonautoimmune mice are not affected by raltegravir, we consider off-target effects unlikely and attribute the exacerbation of autoimmunity to the inhibition of retroviral integrase.
Keywords: 2-LTR circles, hemolytic anemia, lupus disease, muring leukemia virus, Trex1
Although endogenous retrovirus seems to play a small (known) role in human disease, 40% of our genome consists of retroelement sequences. That these elements may contribute to autoimmune disease is indicated by the fact that human patients with a deficiency in Trex1, an enzyme that degrades retroelement cDNA (1), may suffer from chilblain lupus (2, 3), systemic lupus erythematosus (4), or Aicardi-Goutières syndrome (2, 3, 5). Both systemic lupus erythematosus and Aicardi-Goutières syndrome are also characterized by type I IFN in the serum (6), a cytokine that is synthesized in response to cytoplasmic DNA. In Trex1-deficient mice, which suffer from myocarditis (7), there apparently is a causal relationship between accumulation of retroelement DNA and autoimmune disease; the accumulating DNA triggers the type I IFN stimulatory DNA response (1).
Approximately 10% of the mouse genome consists of retroviral sequences—many of them similar to murine leukemia virus. Most of the sequences do not produce functional particles, but it has been noted that all mice with spontaneous autoimmune disease, including NZB (a standard model for hemolytic anemia) and (NZBxNZW) F1 mice (abbreviated B/W; a standard model for lupus disease), produce large amounts of retrovirus, and the discovery of these viruses was immediately linked to the autoimmunity (8–12). Although many genetic loci are associated with it, the disease phenotype of B/W mice can be fully recovered in a normal genome when recombined with three loci (13). While the exact identity of these loci is unknown, there is a strong link to endogenous retroviral expression in B/W mice: Mouse serum gp70 (produced by the liver) is very similar to MLV gp70 and is present in virtually every mouse strain, but only lupus-prone strains produce auto-antibodies to (retroviral) gp70 (14). In the production of anti-gp70 auto-antibodies, the Toll-like receptor 7 (TLR7) plays a critical role (15). Furthermore, mice deficient in TLR7, but not mice deficient in TLR9, have ameliorated lupus disease (16). Duplication of the TLR7 gene accelerates autoimmunity (17, 18). The TLRs trigger innate immunity; because TLR7 binds viral RNA, it may be viral RNA that exacerbates the autoimmune response. Furthermore, because pathogen-free mice also develop lupus (19), the RNA may be produced by endogenous retrovirus.
Because there is conflicting evidence as to how the absence of Trex1 contributes to autoimmunity (20), we confirmed that Trex1 indeed degrades retroelement cDNA. We then probed a potential retroelement contribution to autoimmune disease in B/W and NZB mice, using the highly specific drug raltegravir. Raltegravir is a small-molecule drug that inhibits retroviral integrase of HIV (21, 22). All integrases have a characteristic catalytic core domain, the D,D35E motif (23–26), and we found that the integration of MLV is also inhibited by raltegravir. Because integrase inhibitors are known to cause accumulation of retroviral preintegration DNA (27, 28), they may exacerbate autoimmune disease, analogous to the effect in Trex1-deficient mice. Indeed, the activity profile of raltegravir for inhibition of MLV integration was similar to that for HIV, which led us to feed raltegravir to B/W and NZB mice, along with NZW and BALB/c mice as controls. This study shows that interfering with retroelement replication modulates autoimmune disease in B/W and NZB mice.
Results
Trex1 Is Coresponsible for the Deletions at the Junctions of Circular DNA.
The integration of retroelements is mediated by the preintegration complex, which is derived from the core of an infecting virion, or from an endogenous retroelement. But a fraction of the linear retroelement DNA molecules is also covalently circularized by several host factors—reactions that prevent subsequent integration of the retroelement DNA. Or, viewed from a functional perspective, preintegration cDNA that for some reason cannot integrate is circularized (Fig. 1A). Before the ends of the circles are closed, however, they are often digested by (unknown) cellular exonucleases; it is also possible that aberrant cleavage by RNase H generates insertions of retroviral polypurine tract and primer-binding site sequences, which prevent integration of the cDNA. Although in contention (20), Trex1 is a 3′ DNA exonuclease that is induced by intracellular DNA and is thought to digest reverse-transcribed endogenous retroelement DNA (1). If so, we thought that this enzyme may be, at least in part, responsible for the deletions at the circle junctions, which would be an indication of previous degradation of cDNA ends.
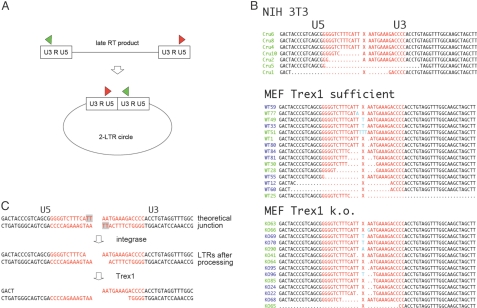
Fig. 1.
Trex1 degrades retroviral cDNA. (A) Schematic of circularization of retroelement. As a nonfunctional by-product, retroelement cDNA is circularized by the cell's DNA repair system. The circles are covalently closed. Red and green arrows represent PCR primers used to amplify the part of circular DNA that contains the site where the two LTR ends of the linear sequence are joined to form a circle. (B) Sequences of 2-LTR amplified from NIH/3T3 cells and mouse embryonic fibroblasts (MEF) that are Trex1 sufficient or deficient (Trex1 k.o.) 24 h after infection with MoMLV. Sequences are from the 0.5-kb band that contains the 2-LTR junction (X): (red) inverted repeat sequences, (green) Cru sequences from NIH/3T3 cells. Cru6 and Cru8 have the prototype sequence with perfect joint; dots indicate nucleotides missing, as compared to prototype sequence. “MEF Trex1-sufficient” sequences are from MEF Trex1 KO cells transduced with Trex1 using a vector based on the mouse stem-cell virus. Numbers in green are from cells not treated with raltegravir; in blue, with raltegravir. From Trex1-sufficient cells, the 14 sequences with insertions/mutations/deletions (i.e., nonprototype) plus one prototype are displayed, of 46 total; from Trex1 KO cells, the 13 nonprototype sequences plus one prototype, of 45 total. Nucleotides in light blue are insertions/point mutations. (C) Proposed schematic of 3′ end-processing by integrase, followed by Trex1 degradation. The processed ends identify the DNA as generated by reverse transcriptase.
Using the alkaline lysis method, we isolated circular DNA from cells that were transduced with Moloney MLV (MoMLV), and we amplified a 0.5-kb piece that ought to contain the site where the two LTRs that flank the linear cDNA are joined (in the so-called “2-LTR circles”) (see Fig. 1A). The sequences at the junctions often are “flush” LTR dimers and represent the prototypic joint, marked by an “X,” of the inverted repeats (sequences marked in red of the U5 and U3 segments of the LTR in Fig. 1B). However, in 20% of the cases in HIV 2-LTR circles, for example, one finds large deletions (28); the number of such deletions is increased in circles derived from MoMLV with a defective nucleocapsid protein (29). Of the seven sequences of MoMLV LTRs from NIH/3T3 cells shown in Fig. 1B, three have perfect junctions (prototype), and in four other sequences a cellular DNA exonuclease has nibbled nucleotides before the DNA repair system closed the circle.
Large deletions similar to the ones in NIH/3T3 were also present in mouse embryonic fibroblasts (MEF) from Trex1 KO mice reconstituted with Trex1 (Trex1 sufficient) (see Fig. 1B and Fig. S1), regardless of whether or not the cells were treated with raltegravir (see below) at the time of transduction with MoMLV vector. However, the number of large deletions was greatly reduced in MEF without Trex1 (see “Trex1 k.o.” in Fig. 1B). The two larger deletions that we did find may have been introduced by a number of other faulty processing steps, including failure to complete reverse transcription. The smaller deletions of one or two nucleotides on either side of the junction are probably caused by the integrase, which takes off dinucleotides from the 3′ ends (Fig. 1C). This “processing” reaction takes place in the cytoplasm when reverse transcriptase has finished the late double-stranded cDNA product of retrovirus, and it is required for integration into genomic DNA (27) (see Fig. 1C). Taking the integrase processing into account, the probability that the reduced number of deletions (i.e., deletions of more than two nucleotides, on either or both sides of the circle junction) in Trex1-deficient cells is a result of chance is P = 0.049. This is the probability, when all 45 sequences of Trex1 KO and 46 sequences of Trex1-sufficient cells, respectively, are considered in Fisher's exact test. Considering only the sequences with deletions, P = 0.019.
Because a nonfunctional mutant integrase greatly reduces the number and extent of deletions (28), we assume that the exonuclease that causes them attacks the processed 3′ ends, although, clearly there are other interpretations possible. The processed ends would identify the DNA as generated by reverse transcriptase and distinguish it from external plasmid DNA, which is not degraded by Trex1 (1). Although it is active on single-stranded and double-stranded DNA, Trex1 is most active on double-stranded DNA with unpaired 3′ termini (30–33). Recessed ends, as provided by the integrase processing, may thus be an in vivo target for Trex1. However, the conclusion that Trex1 degrades retroelements has the caveat that Trex1 was overexpressed in the reconstituted cells, as compared to the (uninduced) wild-type cells (see Fig. S1). It is possible that the higher concentration makes Trex1 less discriminatory; on the other hand, Trex1 expression is robustly increased by IFN stimulatory DNA (1).
Raltegravir Inhibits MLV Integration and Causes Accumulation of 2-LTR Sequences from Exogenous MLV.
We found that raltegravir inhibits the integrase of MLV. Fig. 2A shows that increased concentrations of raltegravir decreased the number of proviruses in NIH/3T3 cells transduced with a MoMLV-based retroviral vector containing GFP. In fact, we saw an effect even at 10 nM, and retroviral integration was almost completely blocked at 300 nM (see Fig. 2A). The EC95 was <100 nM, which is in the same order of magnitude as that reported for HIV (31 ± 20 nM).
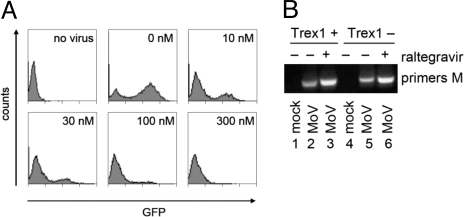
Fig. 2.
Raltegravir activity on MLV. (A) Inhibition of MLV integration into host DNA by raltegravir. Flow cytometry graphs displaying GFP intensity: y axis, cell number; x axis, fluorescence intensity on a logarithmic scale. A MoMLV-based vector encoding GFP was added to NIH/3T3 cells with 0-, 10-, 30-, 100-, or 300-nM raltegravir. (B) Amplification of circular DNA isolated from MEF cells infected with MoMLV, done in duplicates; primers used covered sequences of MoMLV (primers M). MoV was transduced with MoMLV in the presence (+) or absence (−) of raltegravir in Trex1-sufficient (Trex +) or -deficient (Trex −) cells.
Sequencing across the circle junctions of MoMLV-infected cells allowed us to assess whether the activity profile of raltegravir in MLV replication is similar to the one for HIV. While diketo acid-derivative drugs inhibit the DNA strand transfer into the genome, they do not inhibit the 3′ processing reaction of integrase. These drugs also do not inhibit reverse transcription of retroviral RNA. We found this to be the case for MoMLV as well, as there was no difference in the 2-LTR junctions with or without raltegravir treatment (see Fig. 1B, green versus blue sequence numbers). It is also well established that integrase inhibitors of the diketo acid-derivative family, of which raltegravir is a member, increase the concentration of the preintegration (circular) cDNA up to 10-fold without affecting total viral cDNA synthesis (27, 28). Similarly, in the presence of raltegravir, more MoMLV 2-LTR circles accumulated (Fig. 2B). These circles are long-lived (34); in the experiment shown in Fig. S2, the circular DNA was recovered after the cells had been transduced with the replication-defective GFP vector and cultured for 2 weeks.
From these experiments, we conclude that, in MLV replication: (i) Raltegravir inhibits the DNA strand transfer (27) but not the 3′ processing; (ii) Because it increases rather than decreases 2-LTR circle concentration, reverse transcriptase activity and DNA repair are not inhibited either; (iii) Trex1 degrades the 3′ ends of retroviral DNA and also is not inhibited by raltegravir; and (iv) Because insertions at the circle junctions are generated by defective RNase H activity (35), but raltegravir treatment does not seem to increase their number (see Fig. 1B), RNase H probably is also unaffected. Raltegravir thus exhibits an activity profile in MLV replication similar to the one for HIV: that is, it specifically targets integrase.
Accumulation of 2-LTR Sequences from Endogenous MLV by Raltegravir in B/W Cells.
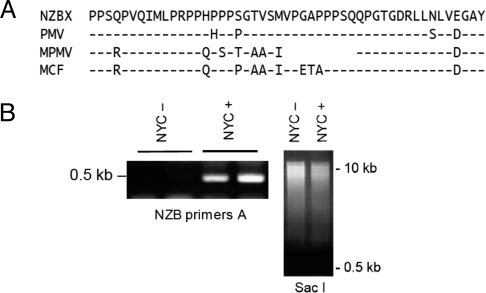
In the presence of raltegravir, not only the exogenous MLV, but some of the endogenous retroelements also accumulate; this depends on exogenous virus, raltegravir or Trex1 (see Fig. S2). Therefore, we investigated the effect of the drug in B lymphocytes of B/W mice, where retroelements are suspected to play a role in autoimmunity. B/W mice are known to produce a host of endogenous viruses. Although one of the parent strains, NZB, is thought to produce only nonecotropic virus, ecotropic MLV that is derived from the NZW strain (36) can be isolated from the B/W mouse (10, 37). When injected with anti-CD4 antibody (38) or with CTLA4Ig (39), and thus prevented from succumbing to lupus disease, B/W mice develop B-cell leukemia. We adapted leukemia cells to culture (designated NYC cells) and found that they produce large amounts of retrovirus (40) (Fig. S3A). To get an overview of what kind of MLV strains are present in the B/W mice, we typed the proline-rich region of the gp70 envelope sequence. In this sequence, the ecotropic MLV differs greatly from all of the nonecotropic MLVs, which differ less between themselves. The sequence is flanked by identical segments, which we used for primer design (41). Fig. 3A shows the amino acid sequence of that diagnostic segment of all types of nonecotropic viruses in the two different mice, B/W and NZB. Besides the xenotropic NZB virus, B/W mice contain the mink cell focus-forming virus, which is the classic de novo oncogenic virus contributing to mouse leukemia and the modified polytropic MLV, with its ORF for gp70. Similar results were reported for B/W mice by Izui and colleagues (42). To show that raltegravir also inhibits the endogenous retroelements in B/W mice, we isolated circular DNA from NYC cells and amplified the 0.5-kb segment that contains the breakpoint with primers NZB2. While there was only a faint band in cells without raltegravir [NYC − (difficult to see in Fig. 3B); loading control, SacI digest of circular DNA], there was a strong band in cells with raltegravir (NYC +) (see Fig. 3B); apparently, inhibition of integrase caused an accumulation of endogenous retrovirus. Similar to the 0.5-kb band from MoMLV, the band contained the ligated inverted repeats of the U5 and U3 regions; that is, they were derived from (various) bona fide retroviral preintegration circular DNA (Fig. S3B). We also isolated circular DNA of at least four different viruses from NZB mice that were or were not fed raltegravir (three of them are shown in Fig. S3B).
Fig. 3.
Accumulation of preintegration circular DNA of endogenous retrovirus in B/W leukemia cells cultured with raltegravir. (A) One-letter amino acid sequence of part of the proline-rich region of the gp70 envelope protein of various MLVs found in B/W leukemia. NZBX, NZB xenotropic virus NZB-9–1; PMV, polytropic murine virus; MPMV, modified polytropic murine virus; MCF, mink cell focus-forming virus. Protein sequences were deduced from the sequence of RT-PCR-amplified mRNAs isolated from peripheral blood lymphocytes. Dashes indicate identity to xenotropic NZB-9–1. (B) Amplification, with NZB primers (NZB primers A), of circular DNA isolated from NYC cells without (NYC−) or with (NYC+) raltegravir, all done in duplicates for each sample of circular DNA isolation triplicates. SacI, SacI digest of circular DNA from NYC− and NYC+ cells, to show similar input; SacI linearizes the 16.3-kb mitochondrial DNA and digests circular DNA of various sizes.
Raltegravir Exacerbates Kidney Disease in Lupus-Prone B/W Mice.
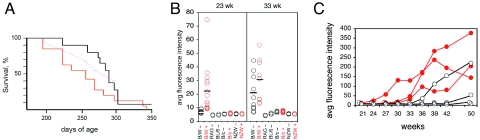
Because raltegravir caused the accumulation of retroviral DNA in B/W cells, which in turn could trigger the innate type I IFN stimulatory immune response, we investigated the effect of the drug on B/W mice. We fed a cohort of 11 B/W mice with regular drinking water and fed another cohort of 14 B/W mice from conception on with 40 mg/kg per day raltegravir; this amounted to 1-mg per mouse per day, which resulted in a fourfold exposure above that of the recommended human dose (800 mg/day, or 10 mg/kg per day). The mice were observed daily, the number of dead mice was recorded, and the results are displayed in a Kaplan–Meier survival curve for untreated and raltegravir-treated mice (Fig. 4A). Comparing the cohorts, raltegravir-treated B/W mice died >35 days (at 50%) and 55 days (at 70%) earlier than untreated mice. It is likely that the mice died from glomerulonephritis: we determined kidney dysfunction, as assessed by proteinuria (essentially, albumin excretion in urine), using the semiquantitative method of dipsticks.
Fig. 4.
Effect of raltegravir on B/W and NZB mice. (A) Kaplan–Meier survival curve of B/W mice fed raltegravir. Cohorts of 14 mice fed (red curve; exact 95% confidence limits in magenta) and 11 mice not fed (black curve) with raltegravir. y axis, percent survival; x axis, age of mice, in days. (B) Mean fluorescence intensities in flow cytometry of RBC from individual mice (B/W mice from the cohorts, as shown in A) reacted with anti-IgG antibodies as a function of age of mice, in weeks (23 and 33 weeks): (red circles) individual mice fed with raltegravir (B/W+, 14 mice; B/c+, 10 mice; and NZW+, 4 mice); (black circles) without raltegravir (B/W−, 11 mice; RAG−, 3 mice; BL/6−, 3 mice; B/c−, 5 mice; and NZW−, 4 mice); (bar) mean fluorescence intensities of RBC, averaged over all mice in the cohort. B/W, female B/W mice; RAG, RAG-2-deficient mice on C57BL/6 background; BL/6, C57BL/6 mice; B/c, BALB/c mice; NZW, NZW mice. y axis, relative fluorescence intensity. (C) Development of anti-RBC reactivity over time in individual NZB mothers fed raltegravir (red curves) or not (black curves). y axis, mean fluorescence intensities in flow cytometry of RBC reacted with anti-IgG; x axis, age of mice, in weeks. Data taken from Table S1.
It was possible that the effect of raltegravir is not specific; that is, raltegravir is not a result of its effect on retroviral integrase, but that it causes or exacerbates disease for other reasons. For example, raltegravir might act as a hapten that, by modifying a self-epitope, elicits an immune response. In that case, all mouse strains might be affected by the drug, whether or not they are prone to autoimmunity. To address this possibility, we also followed the four fathers (NZW) and the three mothers (NZB; one mouse died of unknown causes at the birth of her babies) of the B/W cohorts. While half of the raltegravir-treated B/W mice were dead after 8 months, the raltegravir-treated NZB mice died of hemolytic anemia mice after 16 months; the untreated NZB mice and all treated- and untreated-NZW mice were alive at that age.
Raltegravir Exacerbates the Anti-RBC Antibody Response in B/W Mice.
We also investigated the effect of raltegravir on hemolytic anemia in the B/W mice. The B/W mice die of glomerulonephritis, but in addition to producing antibodies to nucleic acids and nucleoprotein complexes, they also make antibodies to RBC. In this they are similar to their parent-strain NZB, which is a prominent mouse model for hemolytic anemia. To determine the amount of autoantibody on RBC by flow cytometry, we took RBC from nonautoimmune and autoimmune mice and reacted them with a fluorescein-coupled antibody to IgG. RAG-deficient mice, which make no antibody, served as controls for nonautoimmune mice. There was no difference between RAG-deficient mice and normal C57BL/6 mice (Table S1), but at 23 weeks, untreated B/W mice (Fig. 4B) had slightly higher levels than the other mouse strains tested. The raltegravir-treated B/W mice, however, clearly had more auto-antibodies attached to the surface of their RBC (see Fig. 4B). The means (± SD) for treated vs. untreated mice were 22.6 ± 13.3 and 7.41 ± 1.19, respectively; P = 0.009. At 33 weeks, the untreated B/W mice had increased their anti-RBC antibody concentration to levels almost as high as those of the raltegravir-treated cohort (20.20 ± 13.70 vs. 29.90 ± 15.8, respectively; P = 0.17) (see Fig. 4B). At that time, 5 of the 14 raltegravir-treated B/W mice and 1 of the 11 control mice were dead.
Raltegravir Exacerbates the Anti-RBC Antibody Response in NZB Mice, but Has no Effect on NZW and BALB/c Mice.
With raltegravir exacerbating hemolytic anemia in the B/W mice, how would the parent mice, NZB and NZW mice be affected? The NZB mice also make anti-DNA antibodies and circulating immune complexes, causing glomerulonephritis to some extent. However, unlike the B/W mice, NZB mice eventually die of anemia, while NZW mice are healthy. At 36 weeks of age, one of the four untreated NZB mice had higher than the initial level of anti-RBC antibodies (at age 21 weeks), and at 50 weeks, two of the four mice had higher than that level (see Table S1). However, as early as 21 weeks of age, the three treated NZB mice clearly produced more auto-antibodies to RBC than did the four untreated mice (10.7 ± 1.20 vs. 7.90 ± 0.57, respectively; P = 0.009 according to Student's t test) (see Table S1). By 50 weeks, the raltegravir-treated mice had generated high concentrations of auto-antibodies, while the (untreated) C57BL/6 mice and the (untreated and raltegravir-treated) NZW mice were still at base levels (see Table S1; an excerpt of Table S1 is shown in Fig. 4C). From these data, it appears that raltegravir exacerbates the autoimmune disease phenotypes; that is, in NZB mice, raltegravir exacerbates autoimmunity to RBC, while in B/W mice it exacerbates kidney disease and also hastens autoimmunity to RBC—the mice die before developing hemolytic anemia.
We attribute the restriction of the raltegravir effect on B/W and NZB mice to the retroviruses they synthesize. More complicated explanations for the exacerbation of autoimmune disease are, of course, possible. For example, the difference between the NZB and NZW mice could be caused by a modified RBC epitope that is presented by the H-2d haplotype of NZB but not by the H-2z haplotype of NZW. Although they differ in the TL and Qa1 antigens, NZB mice share the classical H-2d haplotype with BALB/c mice (43); therefore, we also treated BALB/c mice with raltegravir and followed their anti-RBC response by flow cytometry. At 42 weeks of age, both male and female raltegravir-treated BALB/c mice had only background levels of anti-RBC antibodies (see Table S1), which is consistent with the view that raltegravir does not modify RBC epitopes. In line with this, the serum of autoimmune mice stained the RBC from untreated RAG-minus mice, whereas untreated mice producing no auto-antibodies did not (Fig. S4).
Discussion
Many genes are known or suspected to predispose to spontaneous lupus disease, and well over 50 manipulated genes in transgenic mice are responsible for enhanced activation of B or T cells, enhanced antigen presentation, reduced apoptosis, proautoimmune cytokine milieu, and decreased clearance of apoptotic debris or immune complexes (reviewed in ref. 44). Some of the genes may break tolerance via dysregulated receptor-editing (45–47); and somatic mutation (46) of nuclear antigen-specific B cell receptors can lead to self-reactivity. In the present study, we examined the potential contribution of endogenous retroelements.
The work of Stetson et al. (1) has shown that the accumulation of retroelement DNA is likely a proximate cause for autoimmunity in Trex1-deficient mice. Because there is conflicting evidence as to the role of Trex1 deficiency in autoimmunity (20), we set out to confirm that Trex1 indeed degrades retroelement cDNA. We demonstrated that the 3′ exonuclease activity of Trex1 is responsible for the large deletions in 2-LTR retroelement circles. Because it does not affect exogenous plasmid DNA (1), retroelement degradation by Trex1 apparently has some degree of specificity. We explain this by suggesting that Trex1 digests 3′ ends only when previously processed by integrase, which specifically targets retroelement cDNA. Confirmation of the function of Trex1 gives a clear precedence for the long-suspected involvement of retrovirus in spontaneous autoimmune disease in mice.
We found that retroelement inhibitors can influence autoimmune disease where retroelements are suspected. Our work does not show that retroelements trigger autoimmune disease in these mice, but it does suggest that retroviral factors can contribute to it. To explain the fact that raltegravir exacerbates lupus disease in B/W mice and hemolytic anemia in B/W and NZB mice, we consider the following possible mechanisms. (i) We know that raltegravir binds to retroelement integrase; the bound drug molecule may represent a new T- or B-cell epitope, which, in turn, will exacerbate the autoimmune response. (ii) Raltegravir selects mutants of integrase. It is well known that in vitro and in patients treated with raltegravir alone, within 3 weeks most of the HIV is of mutant type (21, 22). These mutants may be mimetopes of self-molecules. (iii) Raltegravir increases the concentration of 2-LTR circles (and, presumably, 1-LTR circles), which may increase the IFN-β response. Thus, the effect of the drug potentially mimics the increase in concentration of preintegration cDNA in Trex1-deficient mice, with the innate immune system triggered as a consequence. Indeed, we have shown that 2-LTR circles of endogenous virus accumulate, although we have not yet probed the putative innate immune response.
The findings reported here may also have some relevance to the treatment of HIV infection. Of all classes of drugs, the integrase inhibitor raltegravir (21, 22) causes the fastest decline of HIV viremia in humans, even with monotherapy (48) and its rapid accumulation of HIV mutants. This result is puzzling, because raltegravir is no better an antiretroviral drug than efavirenz or protease inhibitors in vitro (49). Thus, accumulation of the reverse-transcribed preintegration DNA might signal beyond the time of the acquisition of resistance, triggering, for example, the type I IFN stimulatory DNA response (1). Because all integrases share a characteristic catalytic core domain (23–26), raltegravir may accumulate cDNA encoded not only by HIV, but also by other, endogenous retroelements. However, what may be a beneficial adjuvant effect in HIV therapy may exacerbate the autoimmune response, in which endogenous retroelements are suspected to play a role, including chilblain lupus, systemic lupus erythematosus, or Aicardi-Goutières syndrome. Therefore, it is conceivable that raltegravir increases the concentration of retroelement cDNA in autoimmune patients with familial or sporadic disease and, as a consequence, exacerbates their condition. Some in vitro findings, along with the limited clinical data available, point to a potential risk of raltegravir causing autoimmune and lymphoproliferative disorders in the long term (50). These findings also raise the question as to whether lupus-inducing drugs, such as procainamide, hydralazine, phenytoin, and isoniazid activate retroelements or affect processing of cDNA.
Materials and Methods
Mice.
At the time of mating, NZB females and NZW males were fed with 40 mg/kg per day raltegravir; this amounted to 1 mg per mouse per day, which resulted in a fourfold exposure above that of the recommended human dose (800 mg/day, or 10 mg/kg per day). BALB/c mice were treated in the same way. We dissolved raltegravir in drinking water, at 0.2 mg/mL, assuming that a mouse drinks an average of 5 mL of water per day. Using an in vitro transduction assay (see Fig. 1), we compared the activity of freshly made raltegravir solution with that of solutions 1 or 5 months old, and there was no difference in efficacy.
Cohorts of B/W mice and BALB/c progeny were fed from conception on with regular drinking water, and another cohort was fed with raltegravir. We assessed kidney dysfunction in B/W mice by proteinuria (essentially, albumin excretion in urine), using dipsticks (Chemstrip 2 GP; Roche), which are impregnated with a drop of urine; positive values of 1+, 2+, 3+, and 4+ correspond to 0.3, 1, 3, and 20 g/L, respectively.
The experiments were approved by the Institutional Animal Care and Use Committee of University of California, San Francisco.
In Vitro Transduction Assay to Measure MLV Integrase Inhibition.
The Phoenix ecotropic packaging cells (based on the 293T cell line) were plated at day 0 in 60-mm dishes at a concentration of 5 × 105 cells/5 mL. The next day, cells were transfected with the pCru5-EGFP vector (51) using a lipid-based transfection reagent according to the manufacturer's protocol (Lipofectamine from Invitrogen). Forty-eight hours later, supernatant from the Phoenix cells was harvested and filtered through 0.45-μm syringe filter units (Millex-HV Millipore), and 5 × 104/mL per well NIH/3T3 cells (plated the evening before in 24-well plates) were transduced in the presence of 4 μg/mL polybrene. At the same time, raltegravir was added in various concentrations (10 to 900 nM). Two to 3 days later, the NIH/3T3 cells were evaluated for GFP expression by flow cytometry.
Retroviral Envelope Sequences.
Total RNA was extracted from peripheral blood lymphocytes using the RNeasy Mini Kit (Qiagen). All RNA samples were treated with rDNase 1 (Ambion) before reverse transcription. RNA (100 ng) from each sample was reverse transcribed with oligo(dT)12–18 using the SuperScript First-Strand Synthesis System (Invitrogen). PCR primers were as follows: gp70 forward 5′ CCGCGTCCCCATTGGGCCTAATCC 3′; gp70 reverse 5′ GGACGGCAACCCCTTCGTAGTAGG 3′. The 50-μl PCR mixture contained 1 μL of cDNA, 0.2 μM each primer, 200 μM dNTP, 1.5 mM MgCl2, 1 U of recombinant TaqDNA polymerase in 1x PCR buffer (final concentration). The cycling conditions were as follows: 94 °C 5“; 60 °C 1′, 72 °C 1′ for 35 cycles and 72 °C for 5′.
Isolation of Circular DNA.
Isolation of circular DNA was performed as described (52, 53).
Amplification of Retroviral 2-LTR Circle Sequences.
The following primers were used for the plasmid containing Moloney circular DNA: 5′-GAG TGA GGG GTT GTG GGC TCT-3′ (U3 reverse) and 5′-ATC CGA CTT GTG GTC TCG CTG-3′ (U5 forward). For the NZB virus, “NZB primers A”: 5′-CAG TGT GGA GTT CTT ACC CTT-3′ (U3 reverse) and 5′-ATC CGA ATC GTG GTC TCG CTG-3′ (U5 forward); and “NZB primers B”: 5′-TAC CCT TTT TAT AGG GCT GGG G-3′ (U3 reverse) and 5′-GCG CCA GTC CTC CGA TAG ACT G-3′ (R“c” forward). The 50-μl PCR mixture contained 2 μL of circular DNA, 0.2 μM each primer, 200 μM dNTP, 1.5 mM MgCl2, 1 U of recombinant TaqDNA polymerase (Invitrogen) in 1x PCR buffer (final concentration). The cycle conditions were as follows: 94 °C 5“; 60 °C 1′, 72 °C 1′ for 35 cycles; and 72 °C for 5′. PCR-amplified products were gel purified using a Qiaquick gel extraction kit (Qiagen), cloned into pCR2.1 TOPO vector using a TOPO TA cloning kit (Invitrogen) and sequenced with M13 sequencing primers.
Supplementary Material
Acknowledgments.
We thank Jay Levy, John Coffin, Finn Pedersen, Dan Stetson, Mehrdad Matloubian, Alisa Oberschelp, Jay Lalezary, and Leonard Evans for numerous discussions; Martin Weigert for encouragement; Leonard Evans for monoclonal antibody 83A25; Abul Abbas and Jay Levy for comments on the manuscript; and Mary McKenney for editing the manuscript. Dan Stetson generously provided the Trex1 KO and reconstituted cells and Fig. S1. This work was supported by National Institutes of Health Grant R01 AI041570; a grant from the Lupus Research Institute; and in part by a research grant from the Investigator-Initiated Studies Program of Merck and Co., Inc. (to M.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908074106/DCSupplemental.
References
- 1.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee-Kirsch MA, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 3.Rice G, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutières syndrome. Am J Hum Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 5.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 6.Preble OT, Black RJ, Friedman RM, Klippel JH, Vilcek J. Systemic lupus erythematosus: presence in human serum of an unusual acid-labile leukocyte interferon. Science. 1982;216:429–431. doi: 10.1126/science.6176024. [DOI] [PubMed] [Google Scholar]
- 7.Morita M, et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′–>5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mellors RC, Huang CY. Immunopathology of NZB/BL mice. VI. Virus separable from spleen and pathogenic for Swiss mice. J Exp Med. 1967;126:53–62. doi: 10.1084/jem.126.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshiki T, Mellors RC, Strand M, August JT. The viral envelope glycoprotein of murine leukemia virus and the pathogenesis of immune complex glomerulonephritis of New Zealand mice. J Exp Med. 1974;140:1011–1027. doi: 10.1084/jem.140.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy JA. Autoimmunity and neoplasia. The possible role of C-type viruses. Am J Clin Pathol. 1974;62:258–280. doi: 10.1093/ajcp/62.2.258. [DOI] [PubMed] [Google Scholar]
- 11.Levy JA. Xenotropism: the elusive viral receptor finally uncovered. Proc Natl Acad Sci USA. 1999;96:802–804. doi: 10.1073/pnas.96.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bielschowsky F, Bielschowsky M. Carcinogenesis in the pituitary dwarf mouse. The response to methylcholanthrene injected subcutaneously. Br J Cancer. 1959;13:302–305. doi: 10.1038/bjc.1959.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morel L, et al. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci USA. 2000;97:6670–6675. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izui S, et al. Retroviral gp70 immune complexes in NZB × NZW F2 mice with murine lupus nephritis. J Exp Med. 1981;154:517–528. doi: 10.1084/jem.154.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshinobu K, et al. Selective up-regulation of intact, but not defective env RNAs of endogenous modified polytropic retrovirus by the Sgp3 locus of lupus-prone mice. J Immunol. 2009;182:8094–8103. doi: 10.4049/jimmunol.0900263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen SR, et al. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 18.Fairhurst AM, et al. Yaa autoimmune phenotypes are conferred by overexpression of TLR7. Eur J Immunol. 2008;38:1971–1978. doi: 10.1002/eji.200838138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogelweid CM, Wright DC, Johnson JC, Hewett JE, Walker SE. Evaluation of memory, learning ability, and clinical neurologic function in pathogen-free mice with systemic lupus erythematosus. Arthritis Rheum. 1994;37:889–897. doi: 10.1002/art.1780370617. [DOI] [PubMed] [Google Scholar]
- 20.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Hazuda DJ, et al. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 22. http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm?fuseaction=Search.DrugDetails.
- 23.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 25.Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz RA, Skalka AM. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 27.Hazuda DJ, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 28.Svarovskaia ES, et al. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J Virol. 2004;78:3210–3222. doi: 10.1128/JVI.78.7.3210-3222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorelick RJ, et al. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from moloney murine leukemia virus. J Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′–>5′ exonucleases. J Biol Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 31.Hoss M, et al. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′–>5′ exonucleases. Characterization of the recombinant proteins. J Biol Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 33.Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J Biol Chem. 2008;283:31649–31656. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cara A, Klotman ME. Retroviral E-DNA: persistence and gene expression in nondividing immune cells. J Leukoc Biol. 2006;80:1013–1017. doi: 10.1189/jlb.0306151. [DOI] [PubMed] [Google Scholar]
- 35.Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc Natl Acad Sci USA. 2002;99:9515–9520. doi: 10.1073/pnas.142123199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chattopadhyay SK, Lander MR, Rands E, Lowy DR. Structure of endogenous murine leukemia virus DNA in mouse genomes. Proc Natl Acad Sci USA. 1980;77:5774–5778. doi: 10.1073/pnas.77.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner MB, et al. Type C virus expression and host response in diet-cured NZB/W mice. Nature. 1977;268:341–344. doi: 10.1038/268341a0. [DOI] [PubMed] [Google Scholar]
- 38.Wofsy D, Seaman WE. Reversal of advanced murine lupus in NZB/NZW F1 mice by treatment with monoclonal antibody to L3T4. J Immunol. 1987;138:3247–3253. [PubMed] [Google Scholar]
- 39.Finck BK, Linsley PS, Wofsy D. Treatment of murine lupus with CTLA4Ig. Science. 1994;265:1225–1227. doi: 10.1126/science.7520604. [DOI] [PubMed] [Google Scholar]
- 40.Jack HM, Beck-Engeser G, Lee G, Wofsy D, Wabl M. Tumorigenesis mediated by an antigen receptor. Proc Natl Acad Sci USA. 1992;89:8482–8486. doi: 10.1073/pnas.89.18.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Neill RR, Buckler CE, Theodore TS, Martin MA, Repaske R. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J Virol. 1985;53:100–106. doi: 10.1128/jvi.53.1.100-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baudino L, et al. Dissection of genetic mechanisms governing the expression of serum retroviral gp70 implicated in murine lupus nephritis. J Immunol. 2008;181:2846–2854. doi: 10.4049/jimmunol.181.4.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer Lindahl K. On naming H2 haplotypes: functional significance of MHC class Ib alleles. Immunogenetics. 1997;46:53–62. doi: 10.1007/s002510050242. [DOI] [PubMed] [Google Scholar]
- 44.Kono DH, Theofilopoulos AN. Genetics of SLE in mice. Springer Semin Immunopathol. 2006;28:83–96. doi: 10.1007/s00281-006-0030-7. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, et al. Lupus susceptibility genes may breach tolerance to DNA by impairing receptor editing of nuclear antigen-reactive B cells. J Immunol. 2007;179:1340–1352. doi: 10.4049/jimmunol.179.2.1340. [DOI] [PubMed] [Google Scholar]
- 46.Brard F, Shannon M, Prak EL, Litwin S, Weigert M. Somatic mutation and light chain rearrangement generate autoimmunity in anti-single-stranded DNA transgenic MRL/lpr mice. J Exp Med. 1999;190:691–704. doi: 10.1084/jem.190.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yachimovich-Cohen N, Fischel R, Bachar N, Yarkoni Y, Eilat D. Autoimmune NZB/NZW F1 mice utilize B cell receptor editing for generating high-affinity anti-dsDNA autoantibodies from low-affinity precursors. Eur J Immunol. 2003;33:2469–2478. doi: 10.1002/eji.200324025. [DOI] [PubMed] [Google Scholar]
- 48.Murray JM, et al. Antiretroviral therapy with the integrase inhibitor raltegravir alters decay kinetics of HIV, significantly reducing the second phase. Aids. 2007;21:2315–2321. doi: 10.1097/QAD.0b013e3282f12377. [DOI] [PubMed] [Google Scholar]
- 49.Shen L, et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med. 2008;14:762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anonymous. Raltegravir: New drug. Alternative to enfuvirtide/darunavir in multidrug-resistant HIV. Prescrire Int. 2008;17:135–137. [PubMed] [Google Scholar]
- 51.Lorens JB, Jang Y, Rossi AB, Payan DG, Bogenberger JM. Optimization of regulated LTR-mediated expression. Virology. 2000;272:7–15. doi: 10.1006/viro.2000.0353. [DOI] [PubMed] [Google Scholar]
- 52.Carroll SM, et al. Characterization of an episome produced in hamster cells that amplify a transfected CAD gene at high frequency: functional evidence for a mammalian replication origin. Mol Cell Biol. 1987;7:1740–1750. doi: 10.1128/mcb.7.5.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Schwedler U, Jack HM, Wabl M. Circular DNA is a product of the immunoglobulin class switch rearrangement. Nature. 1990;345:452–456. doi: 10.1038/345452a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.