Abstract
Acetyl-CoA carboxylases (ACCs) are crucial metabolic enzymes and are attractive targets for drug discovery. Haloxyfop and tepraloxydim belong to two distinct classes of commercial herbicides and kill sensitive plants by inhibiting the carboxyltransferase (CT) activity of ACC. Our earlier structural studies showed that haloxyfop is bound near the active site of the CT domain, at the interface of its dimer, and a large conformational change in the dimer interface is required for haloxyfop binding. We report here the crystal structure at 2.3 Å resolution of the CT domain of yeast ACC in complex with tepraloxydim. The compound has a different mechanism of inhibiting the CT activity compared to haloxyfop, as well as the mammalian ACC inhibitor CP-640186. Tepraloxydim probes a different region of the dimer interface and requires only small but important conformational changes in the enzyme, in contrast to haloxyfop. The binding mode of tepraloxydim explains the structure-activity relationship of these inhibitors, and provides a molecular basis for their distinct sensitivity to some of the resistance mutations, as compared to haloxyfop. Despite the chemical diversity between haloxyfop and tepraloxydim, the compounds do share two binding interactions to the enzyme, which may be important anchoring points for the development of ACC inhibitors.
Keywords: diabetes, fatty acid metabolism, herbicides, structure-based drug design
Acetyl-CoA carboxylases (ACCs) catalyze the production of malonyl-CoA from acetyl-CoA and CO2 in two steps (1–3). The biotin carboxylase (BC) activity catalyzes the ATP-dependent carboxylation of a biotin cofactor, and then the carboxyltransferase (CT) activity catalyzes the transfer of this activated carboxyl group to the acceptor acetyl-CoA. In bacteria, the BC and CT activities reside in separate subunits of the ACC holoenzyme. In comparison, ACCs are large, multidomain enzymes in most eukaryotes, with highly conserved BC and CT domains.
ACCs are crucial enzymes for the metabolism of fatty acids. Two isoforms of ACCs are present in mammals, and mice lacking ACC2 have elevated fatty acid oxidation and reduced body fat and body weight (4). ACCs are attractive targets for the discovery of anti-diabetes and anti-obesity agents (5, 6), and currently there is significant interest in understanding their catalysis, regulation, and mechanism of inhibition.
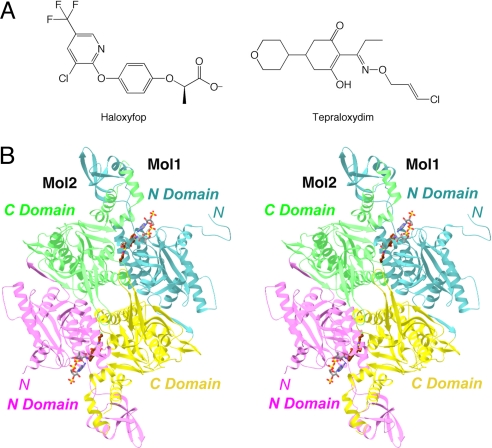
The relevance of ACCs for drug discovery is also underscored by the fact that two distinct classes of compounds, as illustrated by haloxyfop (FOPs) and tepraloxydim (DIMs) (Fig. 1A), are potent inhibitors of ACCs from sensitive plants and are in wide use as herbicides (7–12). These compounds inhibit the CT activity, as does a potent inhibitor of mammalian ACCs, CP-640186 (13), suggesting that the CT domain may be a suitable target for discovering small-molecule inhibitors against ACCs.
Fig. 1.
Crystal structure of CT domain in complex with tepraloxydim. (A) Chemical structures of the herbicides haloxyfop and tepraloxydim. (B) Schematic drawing of the structure of yeast CT domain dimer in complex with tepraloxydim. The N domains of the two monomers are colored in cyan and magenta, while the C domains are colored in yellow and green. The inhibitor is shown in stick models, in dark brown for carbon atoms. The CoA molecule is shown for reference in gray (16). The structure figures were produced with Ribbons (34), Grasp (35), and PyMOL (www.pymol.org).
We have reported the crystal structures of the CT domain of yeast ACC and its complex with CoA (14), haloxyfop (15), and CP-640186 (16). In comparison, the human CT domain has been more difficult to study, a 3.2-Å resolution structure being reported only recently (17). As the human and yeast CT domains share 50% overall amino acid sequence identity, and the sequence conservation is even higher (≈90%) in the active site region (2), the yeast CT domain remains a good surrogate for studying the molecular interactions with various inhibitors. The CT domain contains two subdomains, N and C domains (Fig. 1B), and the active site is located at the interface of a dimer of the enzyme. Haloxyfop and CP-640186 occupy different regions of the CT active site, and inhibit the enzyme by competing with the acetyl-CoA and the carboxybiotin substrate, respectively (15, 16). A large conformational change in the dimer interface is required for haloxyfop binding.
However, the molecular mechanism for the inhibitory action of the DIM herbicides is currently not known. Earlier kinetic studies showed that the FOPs and DIMs may bind at overlapping sites on ACC (12), but the significant chemical diversity between the two classes of compounds (Fig. 1A) makes it difficult to predict the binding mode of DIMs based on that of haloxyfop. We report here the crystal structure at 2.3-Å resolution of the CT domain of yeast ACC in complex with tepraloxydim. The compound has a different mechanism of inhibiting the CT domain. It probes a different region of the dimer interface as compared to haloxyfop, and requires only small, but important conformational changes in the enzyme. Comparisons to the binding mode of haloxyfop also identify two conserved interactions with the enzyme, which may be important anchoring points for the development of ACC inhibitors.
Results and Discussion
Overall Structure.
The crystal structure of the CT domain of yeast ACC in complex with tepraloxydim (Fig. 1A) has been determined at 2.3-Å resolution (Table 1, Fig. 1B). This is the highest resolution yeast CT domain structure that has been reported so far. The current model contains residues 1480–2051 and 2080–2195, 1480–2051 and 2080–2189, and 1491–2051 and 2080–2193 for the three CT domain molecules in the crystallographic asymmetric unit, three inhibitor molecules, and 1303 waters. The atomic model has excellent agreement with the crystallographic data, with an R factor of 18.7% and Rfree of 22.1%, and the expected bond lengths, bond angles, and other geometric parameters (Table 1). The majority of residues (90%) are in the most favored region of the Ramachandran plot. Residue Gln-1744 in all three CT domain molecules, with good-quality electron density, is in the disallowed region of the Ramachandran plot.
Table 1.
Summary of crystallographic information
| Maximum resolution (Å) | 2.3 |
| Number of observations | 654,782 |
| Rmerge (%)* | 8.2 (41.4) |
| Redundancy | 3.4 (3.2) |
| I/σI | 13.8 (2.5) |
| Resolution range for refinement | 30–2.3 |
| Number of reflections | 182,528 |
| Completeness (%) | 99 (91) |
| R factor (%) | 18.7 (25.9) |
| Free R factor (%) | 22.1 (30.1) |
| rms deviation in bond lengths (Å) | 0.015 |
| rms deviation in bond angles (°) | 1.5 |
*The numbers in parentheses are for the highest resolution shell (2.36–2.3 Å).
The three CT domain monomers in the asymmetric unit have essentially the same conformation, with root-mean-square deviation (RMSD) of about 0.5 Å for their equivalent Cα atoms. The conformations of the inhibitors in the three CT domain molecules are highly similar to each other as well.
Binding Mode of Tepraloxydim.
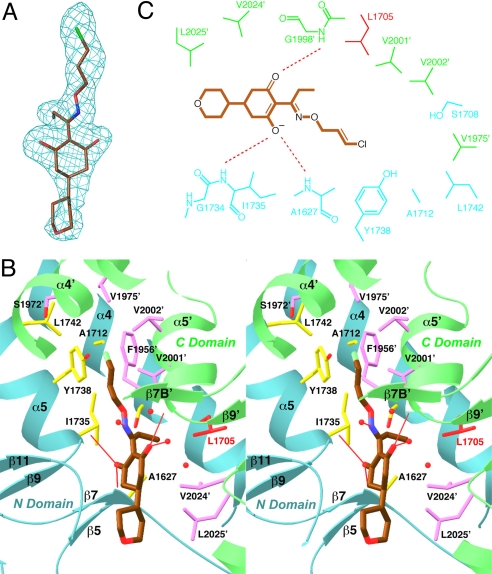
To determine the binding mode of tepraloxydim, we soaked free enzyme crystals of yeast CT domain in a solution containing 5-mM concentration of this inhibitor, at a higher pH (7.5) to enhance its solubility. The crystallographic analysis revealed well-defined electron density for the compound (Fig. 2A), in each of the three active sites in the asymmetric unit.
Fig. 2.
The binding mode of tepraloxydim. (A) Final omit Fo–Fc electron density at 2.3-Å resolution for tepraloxydim, contoured at 3σ. (B) Stereographic drawing showing the binding site for tepraloxydim. The N domain of one monomer is colored in cyan, and the C domain of the other monomer in green. The side chains of residues in the binding site are shown in yellow and magenta, respectively. The side chain of Leu-1705 is shown in red. Hydrogen bonds from the inhibitor to the protein or waters are indicated with thin red lines. Several other water molecules in the binding site are also shown as red spheres. (C) Schematic drawing of the interactions between tepraloxydim and the CT domain.
Tepraloxydim is bound in the active site region of the CT domain, at the interface between the N domain of one monomer and the C domain of the other monomer of the dimer (Fig. 1B). The central cyclohexanedione ring of the compound is situated between residues Gly-1734 and Val-2024′ (primed residue numbers indicate the C domain of the other monomer) (Fig. 2B). One of the oxygen atoms on this ring is hydrogen-bonded to the main-chain amides of Ala-1627 (with a distance of 2.6 Å) and Ile-1735 (3.1 Å) (Fig. 2B), and this oxygen is likely to be in the ionized enolate state (see below) (Fig. 2C). The other oxygen atom on this ring is hydrogen-bonded to the main-chain amide of Gly-1998′ (2.8 Å) and a water molecule (2.7 Å).
The double bond in the oxime moiety of the inhibitor is sandwiched between the Gly-1734–Ile-1735 and Gly-1997′–Gly-1998′ amide bonds, showing π–π interactions (Fig. 2B). The nitrogen atom of the oxime moiety is hydrogen-bonded to a water molecule, while the oxygen atom does not show any hydrogen-bonding interactions. The ethyl group is placed in a hydrophobic pocket lined by the side chains of Ala-1627, Leu-1705, Ser-1708, Val-2001′, and Val-2024′. Some of the DIM herbicides have a propyl group at this position, and the structure suggests that the extra methylene group can be accommodated by the enzyme.
An unexpected discovery from the crystal structure is that the central ring of tepraloxydim and its oxime moiety are not coplanar. It has been expected that a planar arrangement of the two groups will facilitate a hydrogen-bond between the enol on the central ring and the nitrogen atom of the oxime (Fig. 1A). However, in the crystal structure, the two atoms are separated by a torsion of nearly 120° (Fig. 2B), which effectively abolishes any hydrogen-bonding interactions between the two groups.
The O-substituent on the oxime moiety, especially the chlorine atom on the ethylene group, burrows into the dimer interface, and is surrounded by the side chains of Ala-1712, Ile-1735, Tyr-1738, Leu-1742, Ser-1972′, Val-1975′, Val-2001′, and Val-2002′. The ethylene group is placed next to the side chain of Tyr-1738, having possible π–π interactions. Some of the DIM herbicides have only an ethyl group on the oxime, and such compounds are unlikely to probe as deeply into the dimer interface.
The tetrahydropyran substituent on the central cyclohexanedione ring is mostly exposed to the solvent and is partly disordered, with weaker electron density (Fig. 2A). The group has few direct interactions with the enzyme (Fig. 2C), consistent with the large variations in the chemical entities at this position among the different DIM herbicides.
The high-resolution nature of the current structure allows us to determine the location of many solvent waters. Two water molecules that mediate the interactions between the inhibitor and the protein are described above. A few other water molecules are located near the tepraloxydim binding site, held in place by interactions with the enzyme or other water molecules (Fig. 2B). On the other hand, the O-substituent on the oxime moiety is buried in the dimer interface, and there are no solvent molecules in that region of the binding site.
Small But Important Conformational Changes in the Enzyme for Tepraloxydim Binding.
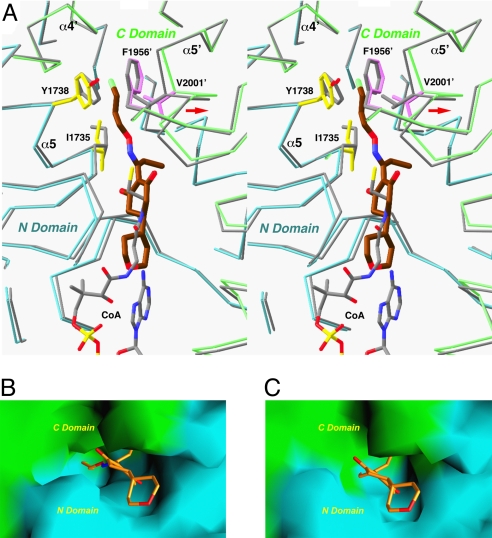
In contrast to our observations earlier with the binding of the haloxyfop herbicide (15), there are only small conformational changes in the overall structure of the CT domain upon the binding of tepraloxydim. The RMSD between equivalent Cα atoms of the entire free enzyme dimer and the tepraloxydim complex is 0.5 Å. Conformational changes in the active site region, however, are crucial for the formation of the inhibitor binding site. The largest change is seen for helix α5′, including residues 1996′–2003′, which moves by about 1 Å away from residues 1734–1738 in the other monomer (Fig. 3A). In addition, the side chains of Ile-1735 and Val-2001′ in the dimer interface assume a different rotamer in the inhibitor complex, and a small change in the main chain and side chain of Tyr-1738 is also observed (Fig. 3A). The overall effect of these changes is that they open up a small pocket in the dimer interface of the CT domain, allowing the linear substituent on the oxime moiety of the inhibitor to bind (Fig. 3B). In comparison, the pocket does not exist in the structure of the free enzyme (Fig. 3C), underscoring the functional importance of these (small) conformational changes. Without these changes, the inhibitor cannot bind to the enzyme and cannot inhibit its carboxyltransferase activity.
Fig. 3.
Conformational changes in the CT domain upon inhibitor binding. (A) Structural overlay of the CT domain free enzyme (in gray) and the tepraloxydim complex (in cyan and green for the N and C domains) near the inhibitor binding site. Side chains in the binding site with large conformational changes are also shown. Phe-1956′ is shown for reference. The position of CoA is also shown (in gray). The shift in the position of the α5′ helix is indicated with the red arrow. (B) Molecular surface of the binding site in the tepraloxydim complex, colored in cyan for the N domain and green for the C domain. (C) Molecular surface of the active site of the free enzyme. The model of tepraloxydim is included for reference, and the O-substituent of the oxime moiety is in steric clash with the enzyme. For panels B and C, residues 1759–1772 and 2026′–2098′ have been removed to give a better view of the binding site.
A Different Mechanism of Inhibition by Tepraloxydim.
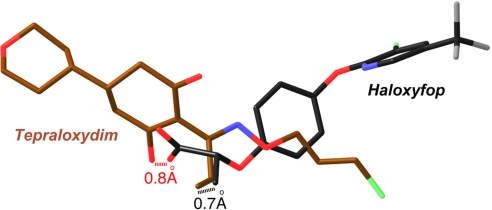
Tepraloxydim inhibits CT activity by blocking the acetyl-CoA substrate from binding to the enzyme. Both the central cyclohexanedione ring and the tetrahydropyran group have severe steric clash with the linear portion of the CoA molecule in the active site (Fig. 3A). The crystal structure reveals that tepraloxydim has a different mechanism of inhibiting the CT domain as compared to that of haloxyfop (15) and CP-640186 (16), even though haloxyfop also inhibits CT by blocking acetyl-CoA from binding to the active site. Tepraloxydim is chemically very distinct from haloxyfop (Fig. 1A), and the compounds have only limited overlap in their bound positions in the active site of the CT domain (Fig. 4). The aryl group at the end of haloxyfop has no equivalent in tepraloxydim (Fig. 4), and the two compounds probe different regions of the CT dimer interface. In fact, this aryl group is located near Phe-1956′ in the haloxyfop complex, causing a large conformational change in this region of the CT domain (15). In comparison, binding of tepraloxydim does not produce any significant disturbance to the conformation of Phe-1956′ (Fig. 3A).
Fig. 4.
Tepraloxydim has a different mechanism of inhibiting the CT domain. Overlay of the binding modes of tepraloxydim (in dark brown) and haloxyfop (in black). The distances between the two oxygen atoms and two methyl groups in the inhibitors are indicated.
Similarly, most of the central cyclohexanedione ring and the tetrahydropyran group of tepraloxydim have no counterpart in haloxyfop (Fig. 4). The structural comparison suggests that the cyclohexanedione ring is probably as important for generating an enolate oxyanion as for establishing strong interactions with the enzyme. One of the carboxylate oxygen atoms of the propionate group in haloxyfop is located within 0.8 Å of an oxygen atom on the cyclohexanedione ring of tepraloxydim (Fig. 4), and both are involved in hydrogen-bonding interactions with the main-chain amides of Ala-1627 and Ile-1735. This suggests that tepraloxydim is likely bound to CT in the enolate form (Fig. 2C). The main-chain amides of Ala-1627 and Ile-1735 may also have an important role in substrate recognition and catalysis by CT.
The methyl group of the propionate in haloxyfop is located within 0.7 Å of that in the ethyl group of tepraloxydim (Fig. 4). Therefore, these two methyl groups share the same binding site, which includes residue Leu-1705 (Fig. 2C), equivalent to one of the resistance mutations in ACC (see below). In addition, the oxime moiety of tepraloxydim partly overlays with the central phenyl ring of haloxyfop.
Overall, tepraloxydim has a different mechanism of inhibiting the CT domain, probing a different region of the dimer interface. At the same time, the compound shares two binding interactions with haloxyfop, despite their chemical diversity. These two binding sites may be important anchoring points for developing CT inhibitors. In addition, hybrid compounds that contain features of both the FOPs and DIMs are more potent inhibitors of ACC (18).
Molecular Basis for Distinct Sensitivity to Resistance Mutations.
Both the FOP and the DIM compounds are potent inhibitors of ACCs from sensitive plants. Over the past years, resistance mutations in these enzymes have been characterized (9, 10, 19–22). Interestingly, the data show that the FOP and DIM compounds can have distinct sensitivity to these mutations. Most of the herbicide sensitive ACCs contain an Ile residue at a position equivalent to Leu-1705 of yeast ACC. A mutation of Ile to Leu at this position confers resistance to both FOP and DIM herbicides. This is consistent with the structural observations that this residue is located in the binding site of both compounds (Fig. 2C). Although the CT domain of yeast ACC, with a Leu residue at this position, is much less sensitive to the herbicides (14), binding of the compounds was observed because of the high concentration (5-mM) used in the soaking experiments.
In comparison, mutation at another site, equivalent to residue 1967 of yeast ACC, strongly affects the potency of FOP inhibitors while having only small effects on the potency of DIM inhibitors (10). Our structures provide an explanation for the molecular basis of this observation. The FOP herbicides cause a major conformational change for Phe-1956′ and other residues in the dimer interface, and the 1967′ residue is located in the binding site for the aryl group at the end of the compounds (15). In contrast, the DIM herbicides probe a different region of the dimer interface and do not perturb the conformation of Phe-1956′. Residue 1967′ is located far from the tepraloxydim binding site, and the compound is not as sensitive to mutations at this position.
The multidomain eukaryotic ACCs have highly conserved sequences, and most of the residues in the herbicide binding site are conserved among all eukaryotic ACCs (2). Nonetheless, the FOP and DIM herbicides exhibit remarkable selectivity, and are poorly active toward fungal and animal ACCs. The molecular mechanism for this selectivity is not fully understood. Amino acid variations in the herbicide binding pocket itself, as illustrated by the 1705 and 1967 residues, are only partly responsible for the differences in herbicide sensitivity (15). Amino acid substitutions in other parts of the protein (long-range effects), as well as conformational flexibility in the binding site region (as conformational changes are required for herbicide binding), may be additional factors that contribute to the selectivity. Finally, the current structural information is based on the complex with the CT domain of yeast ACC, which is mostly insensitive to the herbicides (Ki of 250 μM for haloxyfop) (14). There could also be differences in the binding modes of the herbicides to ACCs from sensitive plants, although such differences will probably be small due to the sequence conservation.
Crystal structures of the CT domain in complex with three different classes of inhibitors (haloxyfop, CP-640186, and tepraloxydim) have now been reported (15, 16). The binding modes of these compounds are highly different from each other, suggesting that there are many different molecular mechanisms for inhibiting this catalytic activity. A large number of potent and chemically diverse inhibitors of mammalian ACCs have been reported recently (23–29). It will be very interesting to characterize whether any of these compounds also function at the CT active site.
Materials and Methods
Protein Production and Crystallization.
The expression, purification, and crystallization of the CT domain (residues 1476–2233) of yeast ACC followed protocols described earlier (15). Crystals of the free enzyme were produced at 4 °C using the hanging-drop vapor diffusion method. The reservoir solution contains 0.1 M sodium citrate (pH 5.5), 8% (wt/vol) PEG8000, and 10% (vol/vol) glycerol. The protein is at 10 mg/mL concentration.
To prepare the tepraloxydim complex, the free enzyme crystals were soaked overnight in a solution containing 0.1 M sodium citrate (pH 7.5), 8% (wt/vol) PEG8000, 10% (vol/vol) glycerol, 5 mM tepraloxydim, and 5% (vol/vol) dimethyl sulfoxide. A higher pH of the solution was used as the inhibitor has very low solubility at acidic pH. The crystals were cryoprotected by the introduction of 25% (vol/vol) glycerol and flash-frozen in liquid nitrogen for data collection at 100K.
Data Collection and Structure Determination.
X-ray diffraction data were collected at the X25 beamline of the National Synchrotron Light Source (NSLS). The diffraction images were processed with the HKL package (30). The crystal belongs to space group C2, with unit cell parameters of a = 247.3 Å, b = 124.8 Å, c = 145.4 Å, and β = 94.3°. There are three CT molecules in the asymmetric unit, forming a noncrystallographic dimer and a crystallographic dimer. The structure refinement was carried out with the programs CNS (31) and Refmac (32). Clear electron density for the herbicide was observed from the crystallographic analysis (Fig. 2A). The atomic model was built with the program O (33). The crystallographic information is summarized in Table 1.
Acknowledgments.
We thank Neil Whalen for setting up the X25 beamline and Linda Yu for help with data collection at the synchrotron source. This research is supported in part by the National Institutes of Health Grant DK67238 (to L.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in Protein Data Bank, www.pdb.org (PDB ID code 3K8X).
References
- 1.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 2.Tong L. Acetyl-coenzyme A carboxylase: Crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: Target for metabolic syndrome. J Lipid Res. 2009;50:S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu-Elheiga L, Matzuk MM, Abo-Hashema KAH, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- 5.Harwood HJ., Jr Treating the metabolic syndrome: Acetyl-CoA carboxylase inhibition. Expert Opin Ther Targets. 2005;9:267–281. doi: 10.1517/14728222.9.2.267. [DOI] [PubMed] [Google Scholar]
- 6.Tong L, Harwood HJ., Jr Acetyl-coenzyme A carboxylases: Versatile targets for drug discovery. J Cell Biochem. 2006;99:1476–1488. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gronwald JW. Lipid biosynthesis inhibitors. Weed Sci. 1991;39:435–449. [Google Scholar]
- 8.Devine MD, Shukla A. Altered target sites as a mechanism of herbicide resistance. Crop Protection. 2000;19:881–889. [Google Scholar]
- 9.Zagnitko O, Jelenska J, Tevzadze G, Haselkorn R, Gornicki P. An isoleucine/leucine residue in the carboxyltransferase domain of acetyl-CoA carboxylase is critical for interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors. Proc Natl Acad Sci USA. 2001;98:6617–6622. doi: 10.1073/pnas.121172798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delye C, Zhang X.-Q., Chalopin C, Michel S, Powles SB. An isoleucine residue within the carboxyl-transferase domain of multidomain acetyl-coenzyme A carboxylase is a major determinant of sensitivity to aryloxyphenoxypropionate but not to cyclohexanedione inhibitors. Plant Physiol. 2003;132:1716–1723. doi: 10.1104/pp.103.021139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi A, Yamada S, Tanaka K. Mechanism of herbicidal activity of a new cyclohexane-1,3-dione, tepraloxydim to Poa annua L. Weed Biol Management. 2002;2:84–91. [Google Scholar]
- 12.Rendina AR, Craig-Kennard AC, Beaudoin JD, Breen MK. Inhibition of acetyl-coenzyme A carboxylase by two classes of grass-selective herbicides. J Agric Food Chem. 1990;38:1282–1287. [Google Scholar]
- 13.Harwood HJ, Jr, et al. Isozyme-nonselective N-substituted bipiperidylcarboxamide acetyl-CoA carboxylase inhibitors reduce tissue malonyl-CoA concentrations, inhibit fatty acid synthesis, and increase fatty acid oxidation in cultured cells and in experimental animals. J Biol Chem. 2003;278:37099–37111. doi: 10.1074/jbc.M304481200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Yang Z, Shen Y, Tong L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science. 2003;299:2064–2067. doi: 10.1126/science.1081366. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Tweel B, Tong L. Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by haloxyfop and diclofop. Proc Natl Acad Sci USA. 2004;101:5910–5915. doi: 10.1073/pnas.0400891101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Tweel B, Li J, Tong L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase in complex with CP-640186. Structure. 2004;12:1683–1691. doi: 10.1016/j.str.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Madauss KP, et al. The human ACC2 CT-domain C-terminus is required for full functionality and has a novel twist. Acta Crystallogr. 2009;D65:449–461. doi: 10.1107/S0907444909008014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rendina AR, et al. Overlap between herbicidal inhibitors of acetyl-coenzyme A carboxylase: Enhanced binding of cyclic triketones, a novel class of graminicide. Pestic Sci. 1995;43:368–371. [Google Scholar]
- 19.Delye C, Zhang X-Q, Michel S, Matejicek A, Powles SB. Molecular bases for sensitivity to acetyl-coenzyme A carboxylase inhibitors in black-grass. Plant Physiol. 2005;137:794–806. doi: 10.1104/pp.104.046144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XQ, Powles SB. Six amino acid substitutions in the carboxyl-transferase domain of the plastidic acetyl-CoA carboxylase gene are linked with resistance to herbicides in a Lolium rigidum population. New Phytol. 2006;172:636–645. doi: 10.1111/j.1469-8137.2006.01879.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, et al. Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Natl Acad Sci USA. 2007;104:3627–3632. doi: 10.1073/pnas.0611572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alarcon-Reverte R, Moss SR. Resistance to ACCase-inhibiting herbicides in the weed Lolium multiflorum. Commun Agric Appl Biol Sci. 2008;73:899–902. [PubMed] [Google Scholar]
- 23.Cheng D, Chu, et al. Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes. Prot Expr Purif. 2007;51:11–21. doi: 10.1016/j.pep.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Kim KW, Yamane H, Zondlo J, Busby J, Wang M. Expression, purification, and characterization of human acetyl-CoA carboxylase 2. Prot Expr Purif. 2007;53:16–23. doi: 10.1016/j.pep.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Clark RF, et al. Structure-activity relationships for a novel series of thiazolyl phenyl ether derivatives exhibiting potent and selective acetyl-CoA carboxylase 2 inhibitory activity. Bioorg Med Chem Lett. 2006;16:6078–6081. doi: 10.1016/j.bmcl.2006.08.100. [DOI] [PubMed] [Google Scholar]
- 26.Clark RF, et al. Phenoxy thiazole derivatives as potent and selective acetyl-CoA carboxylase 2 inhibitors: Modulation of isozyme selectivity by incorporation of phenyl ring substituents. Bioorg Med Chem Lett. 2007;17:1961–1965. doi: 10.1016/j.bmcl.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Corbett JW, Harwood HJ., Jr Inhibitors of mammalian acetyl-CoA carboxylase. Recent Pat Cardiovasc Drug Discov. 2007;2:162–180. doi: 10.2174/157489007782418928. [DOI] [PubMed] [Google Scholar]
- 28.Shinde P, et al. Synthesis of spiro[chroman-2,4′-piperidin]-4-one derivatives as acetyl-CoA carboxylase inhibitors. Bioorg Med Chem Lett. 2009;19:949–953. doi: 10.1016/j.bmcl.2008.11.099. [DOI] [PubMed] [Google Scholar]
- 29.Corbett JW. Review of recent acetyl-CoA carboxylase inhibitor patents: Mid-2007–2008. Expert Opin Ther Pat. 2009;19:943–956. doi: 10.1517/13543770902862180. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Brunger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Carson M. Ribbon models of macromolecules. J Mol Graphics. 1987;5:103–106. [Google Scholar]
- 35.Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]