Abstract
Missense mutations in the cone opsins have been identified as a relatively common cause of red/green color vision defects, with the most frequent mutation being the substitution of arginine for cysteine at position 203 (C203R). When the corresponding cysteine is mutated in rhodopsin, it disrupts proper folding of the pigment, causing severe, early onset retinitis pigmentosa. While the C203R mutation has been associated with loss of cone function in color vision deficiency, it is not known what happens to cones expressing this mutant opsin. Here, we used high-resolution retinal imaging to examine the cone mosaic in two individuals with genes encoding a middle-wavelength sensitive (M) pigment with the C203R mutation. We found a significant reduction in cone density compared to normal and color-deficient controls, accompanying disruption in the cone mosaic in both individuals, and thinning of the outer nuclear layer. The C203R mosaics were different from that produced by another mutation (LIAVA) previously shown to disrupt the cone mosaic. Comparison of these mosaics provides insight into the timing and degree of cone disruption and has implications for the prospects for restoration of vision loss associated with various cone opsin mutations.
Keywords: color vision, cone mosaic, photopigment, retinal imaging, rhodopsin
Normal human color vision is trichromatic and derives from the presence of three spectrally distinct cone types: long-, middle-, and short-wavelength-sensitive (L, M, and S). Red-green color vision defects are characterized by the absence of either L or M cone function and they affect about one in 12 Caucasian males. Inherited red-green defects can be linked to disruptions at the X-chromosome opsin gene locus, where the L- and M-cone opsin genes reside in a head-to-tail array (1). Most of these disruptions involve gross gene rearrangements (2–6). However, it is becoming appreciated that missense mutations underlie a significant proportion of red-green defects (5–8). This raises the question of what impact these missense mutations have on the viability of the cones.
Some insight comes from rhodopsin. There are >130 distinct rhodopsin mutations, involving at least 89 sites within the molecule (data compiled from refs. 9–17.) With rare exception (e.g., refs. 9 and 18), each of these mutations has been associated with either retinitis pigmentosa (RP) or congenital stationary night blindness. Rhodopsin and the cone opsins have structural similarities and similar functional demands. Thus, it is reasonable to hypothesize that mutations in the cone opsins homologous to those in rhodopsin that cause retinitis pigmentosa would affect the viability of the cones.
The most common missense mutation in the cone opsins is a substitution of cytosine for thymine at nucleotide position 1101, which corresponds to a substitution of arginine for cysteine at amino acid position 203 (C203R) (Fig. S1). The corresponding mutation in rhodopsin (C187Y) disrupts proper folding of the pigment, causing severe, early onset retinitis pigmentosa (19). It has been shown that the C203R mutation also disrupts protein folding and stability (20), as the mutation of this highly conserved cysteine residue disrupts a disulfide bond that is critical for proper tertiary structure (21, 22). In rods, it has been suggested that this misfolding results in retention of the pigment in the endoplasmic reticulum (23); likewise it has been shown that C203R pigment is not successfully transported to the cone outer segment (20).
Here, we used high-resolution retinal imaging to examine the effect of the C203R mutant opsin on the structural integrity of the cone photoreceptors. We observed a significant reduction in cone density in two individuals harboring the C203R mutation, as well as a partial thinning of their outer nuclear layer (ONL). There was no difference in cone density between “typical” multigene protans lacking L genes and color-normal controls. We had previously reported a similar reduction in cone density in a dichromat whose condition was caused by the presence of an unusual combination of polymorphisms encoded by exon 3 of the M-pigment gene (“LIAVA”) (24). The same unusual sequence was associated with blue-cone monochromacy (25) and was also associated with a case of X-linked incomplete achromatopsia (26). In contrast to the C203R retina, the LIAVA retina had normal ONL thickness and reduced regularity of the remaining visible cones. Taken together, our results support a hypothesis that whether present in a rod or a cone, the C203R mutation abolishes function and results in early degeneration of the associated cell, whereas the LIAVA polymorphism leads only to a loss of function.
Results
Ophthalmic Examination and Clinical Diagnoses.
Examinations on the color deficient subjects were unremarkable, except for subtle foveal granularity in retinal pigment epithelium (RPE) pigmentation in subjects 4511 and 4515 (the individuals harboring the C203R mutation). A multifocal ERG was recorded from subject 4515 using the 103 hexagon standard protocol with the Veris Science 5.1.10X software (Electro-Diagnostic Imaging, Inc.), and all amplitudes and implicit times were normal. The two subjects 4511 (age 38 years) and 4515 (age 22 years) had uncorrected visual acuities of 20/16 and 20/12.5, respectively. The remaining seven protan subjects all were mild myopes (less than −1.75 D) and correctable to 20/25 or better (average age 22 years). Normal controls (n = 103; 47 males and 56 females) were free of any gross ocular pathology, either self-reported (n = 88) or chart review (n = 15). The normal controls ranged in age from 11 to 40 years (mean = 26 years).
Color Vision Testing.
Based on their performance on the Rayleigh match as well as pseudoisochromatic plates, Farnsworth-Munsell 100 Hue, and Medmont C100; 389, 399, and 3514 were all diagnosed as protanopes, whereas the remaining subjects were classified as protanomalous. Subject 4511's scores were 1100, 381, and 123 and subject 4515's scores were 951, 241, and 88 on the protan, deutan, and tritan axes of the Trivector test on the Cambridge Color Test (27), both consistent with a severe protan defect. Reported maximum normal values on the Trivector test are 69.3, 82.4, and 113.4 for the protan, deutan, and tritan axes, respectively (28).
Structure of the X-Chromosome Opsin Gene Array.
The results from the real-time quantitative PCR assays are expressed as the percentage of genes in the array that are downstream (ds) of the first gene and the percentage of all genes that are L. All of the protans had estimates of 0% L genes; thus, an estimate of 0% ds genes specifies an array containing a single M gene, an estimate of 50% ds genes specifies an array with two M genes, an estimate of 66% ds genes specifies an array with three M genes, and so on. Subjects 4511 and 4515 were inferred to each have three genes in their array, based on estimates of 65 and 63% ds, respectively. Subjects 0389, 0399, 3514, and 6336 had arrays with two genes (55, 45, 48, and 54% ds), while subjects 3504, 3505, and 3506 had arrays with four genes (81, 72, and 74% ds). As the number of genes in the array increases, the ability to distinguish the absolute size of the array becomes more difficult, so subject 3504 could have five genes. The inferred structure of the L/M gene array for each subject is shown in Fig. S2.
All subjects except 399, 3514, and 6336 were found to have polymorphisms at all three polymorphic sites encoded by exon 2 (65, 111, and 116) that have been predicted to introduce a small (≈1–2 nm) shift in the λmax (29), or in the optical density (30) of the associated pigment (see Fig. S1 for the position of these polymorphisms). Subjects 399 and 3514 were diagnosed as protanopes, consistent with their having only a single spectral type of M-cone pigment. Subject 389 was also diagnosed as a protanope, although he had exon 2 polymorphisms that have been associated with spectral differences sufficient to confer protanomaly. The remaining subjects with exon 2 polymorphism were diagnosed as protanomalous, although the discrimination between severe protanomaly and protanopia is often difficult. This variability is consistent with previous reports where the presence of exon 2 polymorphisms can lead to a protanomalous or protanopic phenotype (4). Sequence data on subject 6336 were not available; phenotypically, he was an extremely mild protanomalous trichromat (31). Two subjects were found to have a C203R mutation encoded by one of their M genes: 4511 (third gene in his array) and 4515 (second gene in his array).
Decreased Cone Density in Individuals Harboring C203R Mutation.
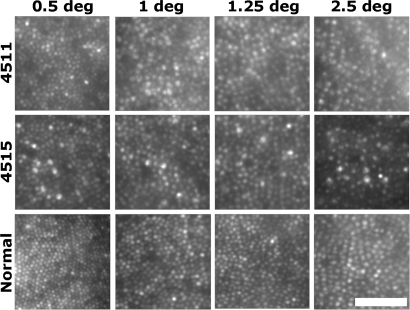
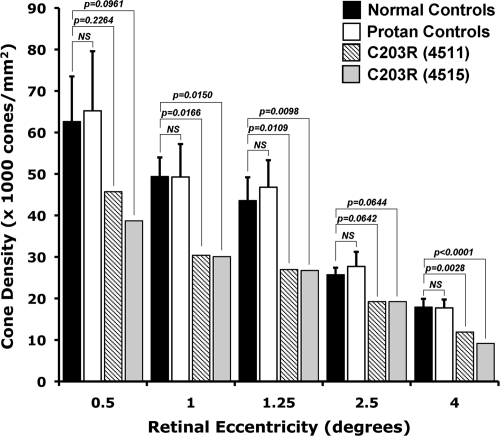
Shown in Fig. 1 are images of the cone mosaic from the two subjects with the C203R mutation. In comparison with the normal retina (Fig. 1, bottom row), these mosaics appear coarser than normal at all four locations imaged. Fig. 2 shows average density as a function of eccentricity for the normal and the C203R individuals. There is significant variability in “normal” cone density. Using a z-score analysis (number of standard deviations from the normal mean), it was found that the reduction in density was significant for both subjects at the 1, 1.25, and 4° test locations (P < 0.05). To determine whether the reduction in density was caused by the lack of an L-opsin gene, we imaged 7 other protans who had no L-opsin gene but had multiple M-opsin genes in their array (Fig. 2). There was no significant difference in cone density between the normal and protan control groups (P value at all locations was 0.28 or greater; Mann–Whitney test). Moreover, the density of the C203R retinas was significantly reduced compared to these protan controls (P < 0.005) at all but 0.5°, where P = 0.015 and P = 0.0667 for 4515 and 4511, respectively. Thus, it is the presence of the C203R mutant pigment, not the absence of an L-opsin gene, which underlies the striking disruption of the cone mosaic in subjects 4511 and 4515.
Fig. 1.
Visualizing cone mosaic disruption in the C203R retina. Shown are images from the right eyes of subject 4511, 4515, and a normal control from 0.5°, 1°, 1.25°, and 2.5° temporal to the fovea. Retinal magnification estimates are 288 μm/degree (4511), 281 μm/degree (4515), and 298 μm/degree (normal control). [Scale bar, 50 μm (≈10.5 arcmin).]
Fig. 2.
Quantifying cone mosaic disruption in the C203R retina. Individuals harboring a C203R mutation have lower cone densities than the normal trichromats and multigene protan controls. Density reduction was only significant at 1°, 1.25°, and 4° (using a z-score analysis). Error bars represent ± 1 SD across the respective control group at that eccentricity. Multigene protan controls did not differ from the normals (Mann–Whitney test, P > 0.28 at all eccentricities).
Retinal Thickness Topography in the C203R and LIAVA Retina.
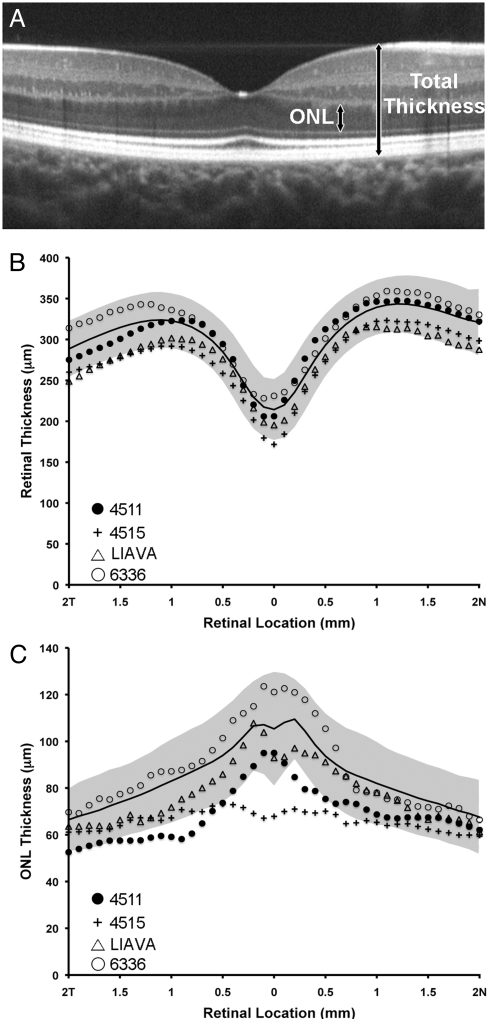
In the individuals with the C203R mutation, we were interested to know whether the reduction in cone density could be detected using optical coherence tomography (OCT). Estimates of retinal nerve fiber layer thickness around the optic disc obtained with the Stratus OCT (Carl Zeiss Meditec) “RNFL” scan protocol revealed no abnormalities in either subject (thickness values within 95th percentile). Macular thickness maps obtained with the Stratus “fast mac” protocol revealed normal retinal thickness for 4511 and slight thinning in the central and peripheral retina for 4515 (Fig. S3). However, as this scan protocol relies on substantial interpolation to derive thickness measurements in the peripheral segments of the map, we examined more closely the total retinal thickness and ONL thickness through the macula. Shown in Fig. 3 is a comparison of total retinal thickness and ONL thickness between the two C203R subjects and 93 normal controls. For the one protan control which we were able to obtain OCT images on, his total and ONL thickness was within normal limits. Averaged horizontal line scans from both C203R subjects used for analysis are shown in Fig. S4. In both subjects, the total retinal thickness was not grossly different from normal, although 4515 fell outside of 2 SD from the normal mean at a few locations along the scan. There was a slight thinning of the ONL in both subjects, falling outside 2 SD of the normal mean at the very central fovea for 4515, and just temporal to the fovea for 4511. This would be consistent with a subset of cones being missing (presumably those containing the C203R opsin). In contrast to the C203R subjects (Fig. 3), the total retinal and ONL thickness of the LIAVA patient was within normal limits at all retinal locations along the horizontal meridian (Fig. 3 B and C). This is interesting, as the C203R and LIAVA (24) retinas have approximately the same cone density, as measured in the AO images.
Fig. 3.
Assessing retinal lamination. (A) SD-OCT registered sum from the right eye of a 19-year-old female with normal color vision. Arrows illustrate the boundaries used for the ONL and total thickness measurements. Total retinal thickness (B) and ONL thickness (C) as a function of distance (n = nasal, T = temporal) from the center of the foveal pit (0 mm on abscissa) for the C203R subjects (4511 and 4515), the LIAVA subject (ref. 24), and a single protan control (6336). Average normal thickness values are plotted as black line with shaded area representing ± 2 SD.
Mosaic Geometry in the C203R and LIAVA Retina.
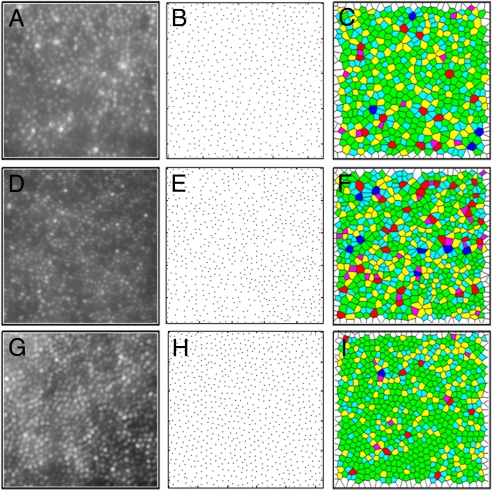
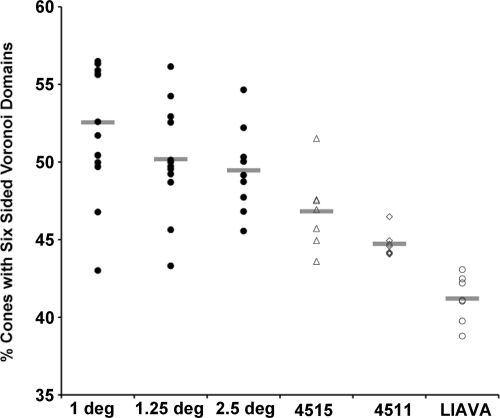
We used a Voronoi analysis to evaluate mosaic geometry. In the Voronoi analysis, each cone is identified as a point in a 2-D plane and the resultant Voronoi domain for that cone contains all points in the plane that are closer to that given cone than any other cone in the 2-D plane (Fig. 4 A–I). The Voronoi domain for each cone is a polygon, and the regularity of the cone mosaic can be assessed by examining various properties of the Voronoi domains for a given patch of retina. To compare cone packing in the LIAVA mosaic to the C203R mosaic, we looked at the percentage of cones with six-sided Voronoi domains an indicator of the overall packing geometry of the mosaic. Qualitatively, there is apparent irregularity in the C203R (Fig. 4C) and LIAVA retina (Fig. 4F) compared to normal (Fig. 4I). We compared mosaic geometry for normal controls at three different retinal locations to that for the two C203R retinas and the LIAVA retina (Fig. 5). Between 44% and 57% of the cones in the normal retina had six-sided Voronoi domains, consistent with previous estimates (32). Using a z-score analysis, the LIAVA retina had significantly fewer cones with six-sided Voronoi domains (P = 0.0155, P = 0.0117, and P = 0.0031 in comparison to 1°, 1.25°, and 2.5° normals, respectively). Conversely, both of the C203R subjects had mosaics that were not significantly reduced from normals with respect to packing geometry (assessed using a z-score analysis). There was, however, an increase in the standard deviation of the Voronoi domain areas when assessed against equivalent-density mosaics (Fig. S5), indicating that while the C203R mosaic is contiguous, there is residual noise (irregularity) in the packing.
Fig. 4.
Assessing cone mosaic packing geometry. Shown in the left column are retinal images from the right eye of subject 4515 (A), the LIAVA (24) patient (D), and a normal trichromat (G). Shown in the middle column is a plot of cone locations for the corresponding retinal image of subject 4515 (B), the LIAVA patient (E), and a normal trichromat (H). Shown in the right column are the Voronoi domains associated with each cone photoreceptor in subject 4515 (C), the LIAVA patient (F), and a normal trichromat (I). Colors in C, F, and I indicate the number of sides for each Voronoi polygon (magenta = 4, cyan = 5, green = 6, yellow = 7, red = 8, and purple = 9). Large regions of 6-sided polygons indicate a regular triangular lattice (58), whereas other colors mark points of disruptions in the hexagonal packing of the foveal mosaic. While (A) and (D) are from the same retinal eccentricity (1°), the normal mosaic was taken from 1.25° to approximately match cone density.
Fig. 5.
Evaluating regularity of the cone mosaic. Plotted is the percentage of cones with 6-sided Voronoi domains for the normal controls as well as the color defective subjects (4511, 4515, and LIAVA). Individual data points for the normals represent individual retinas (n = 15 at 1°, n = 13 at 1.25°, n = 10 at 2.5°), whereas individual data points for the color defective subjects represent measurements at seven different points within the same retina (although all were 1° from the foveal center). Horizontal bars are the mean value for each subject or group of subjects.
Discussion
Here, imaging of the living human cone mosaic has provided direct evidence that the C203R photopigment is associated with disruption of the cone mosaic. The comparison of the C203R retinas to the previously published LIAVA retina reveals a subtle but important difference in the arrangement of their remaining cones. The cones in the C203R retina are more regularly packed than those in the LIAVA retina. This coupled with a lower than normal cone density in the C203R retina, suggests that the cones expressing the C203R retina have degenerated completely whereas those in the LIAVA retina are still present (although not functioning or waveguiding). This is supported by data showing that the cone locations and cone density have not changed in the LIAVA retina over a period of 6 years (33). The partial thinning of the ONL in the C203R retina compared to a normal ONL in the LIAVA retina also supports this interpretation. We hypothesize that the C203R cones appear regularly packed because the cells expressing the C203R pigment degenerated early in foveal development, and the centripetal packing of cones that occurs during foveal development may have acted to produce contiguous and hexagonal packing of the remaining cones. The irregularity of the LIAVA mosaic is likely due to the random and partial loss of visible cones from the mosaic. The integrity of the remaining cones in color vision defects (including blue cone monochromacy and achromatopsia) takes on additional relevance given the recent results showing that trichromatic color vision can be restored in dichromatic New World primates using gene transfer techniques to deliver an additional spectrally distinct opsin gene (34).
For understanding mechanisms that control L/M opsin gene expression, the issue of whether genes beyond the second position in the array are expressed has been of interest. Winderickx et al. (35) and Yamaguchi et al. (36) hypothesized that only the first two genes are ever expressed, as a result of increased distance from the locus control region upstream of the array. However, mRNA data from Sjoberg et al. (1998) show that a small fraction of males with normal color vision express at least three genes from their L/M array (37). It has been shown that in adults individual cones only express opsin from a single gene within the L/M array (38, 39). Thus, there are at least two possible explanations for the observed retinal phenotype of subject 4511. It could be explained by significant expression of the third gene in the array (which harbors the C203R mutation), or the disruption in cone structure was induced by one of the first two genes in the array. No missense mutations were detected in the first two genes in the array, however the same can be said for the LIAVA mutation that is now clearly associated with loss of cone function. Understanding this interesting subject awaits a more complete understanding of mechanisms responsible for gene expression and how combinations of amino acids exchanged between L and M opsins affect the function and viability of the cones.
What consequence the C203R mutation has for the health of the visual system is still unclear. It would appear that in the short term there is no measurable deficit imposed by the disruption in the cone mosaic. This reinforces the fact that many clinical tests, and indeed the visual system itself, are largely insensitive to considerable loss in cone number (40, 41). Nevertheless it is possible that there could be more long-term effects, such as an increased (or even decreased) susceptibility to age-related retinal changes (7)*. One of the advantages of in vivo imaging is that we can track retinal architecture and visual performance in the same eyes over time. Given the variability in normal L:M cone ratio in trichromats (47, 48), which is linked to variability in the relative expression of the L and M genes in the L/M array (49), we expect to see similar variability in cone loss among individuals with the C203R (and other) mutations. Psychophysical examination of individuals with these mutations will allow us to determine when and how visual function breaks down in the face of a compromised cone photoreceptor mosaic.
Materials and Methods
Subjects.
All research followed the tenets of the Declaration of Helsinki, and study protocols were approved by the institutional research boards at the Medical College of Wisconsin (retinal imaging, genetics, and color vision testing), University of Rochester (retinal imaging and clinical examination), and University of Manchester (genetics and color vision testing). Subjects provided informed consent after the nature and possible consequences of the study were explained. A complete ophthalmic examination including visual acuity measurement, slit lamp examination, and dilated funduscopic examination was performed on nine individuals with protan color vision defects. We recruited 103 individuals with normal color vision for retinal imaging with OCT (n = 86), AO (n = 10), or both AO and OCT (n = 7). We re-examined an individual (“LIAVA”) previously reported to have a patchy loss of visible cones throughout the macula (24). Axial length measurements were obtained on all subjects using an IOL Master (Carl Zeiss Meditec) for calibration of retinal images.
Adaptive Optics Retinal Imaging.
An AO ophthalmoscope was used to obtain images of the cone mosaic. Details of the imaging protocol can be found in the SI Text.
Adaptive Optics Image Processing and Analysis.
Images of the cone mosaic were registered using a MatLab-based image registration algorithm (MathWorks) and averaged for subsequent analysis. The position of cone photoreceptors were identified using a modified version of described automated Matlab software (32). Cone density was calculated using previously described methods relying on counting of the cones and spatial frequency analysis (24), and mosaic regularity and packing geometry was assessed using a previously described Voronoi analysis (50).
OCT Retinal Imaging.
High-resolution spectral domain OCT (Bioptigen, Inc.) imaging was performed in 93 normal subjects (42 male, 51 female), with a mean of 25.7 years, and one of the protan controls. Line scan sets were acquired (1,000 A-scans/B scan; 100 repeated line scans; 4-mm nominal scan length†) through the center of fixation, which was cross-referenced with a volumetric scan to ensure coincidence with the center of the foveal pit. Scan sets were directly exported and read into ImageJ for processing (52). Frames that were distorted due to large saccades or eye blinks were removed. Stratus time domain OCT (Carl Zeiss Meditec) images were acquired from the two individuals harboring the C203R mutation. Scan sets consisting of 10 6-mm horizontal line scans were acquired through the center of fixation, and custom MatLab-based software (provided by Dr. Michael Pianta, University of Melbourne) was used to align the individual A-scans (thus “flattening” the B scan), which were then read into ImageJ for processing.
Speckle noise in OCT images can be reduced by averaging multiple B-scans (53). Here, the ImageJ rigid body registration plugin “StackReg” was used to generate a stabilized frame sequence (54). Scans within each stabilized SD-OCT and Stratus scan set were averaged to produce high signal-to-noise images for subsequent segmentation. For the SD-OCT scan sets, an average of 39 scans were averaged together for any given subject, while for the two Stratus scan sets, 10 scans were average together. In both cases the resultant image quality was sufficient to resolve the requisite layers for segmentation. The internal limiting membrane (ILM), outer plexiform layer (OPL), external limiting membrane (ELM), and retinal pigment epithelium (RPE) were manually segmented. The distance between the anterior boundary of the ILM and the posterior boundary of the RPE provided total retinal thickness while the distance between the posterior boundary of the OPL and the center of the ELM provided the ONL thickness‡.
Color Vision Testing.
Color vision in the protan subjects was assessed using a variety of tests, including the Rayleigh match, pseudoisochromatic plates (AO-HRR, Dvorine, Ishihara), C100 Color Vision tester, the Farnsworth-Munsell 100-Hue Test, and the Neitz Test of Color Vision (57). The C203R subjects were also tested using the Cambridge Color Test (27). Normal controls had normal color vision as assessed with the Neitz Test of Color Vision and/or the AO-HRR.
Molecular Genetics.
Details of the genetic analysis performed on the L/M gene array can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank M. Chung, D. Conklyn, A. Dubra, R. F. Itzhaki, M. Pianta, J. Porter, P. Summerfelt, and M. A. Wozniak for their assistance. This work was supported by The National Eye Institute (J.C. J.N., M.N., and D.R.W.), Research to Prevent Blindness (J.C., J.N., and M.N.), The Wellcome Trust (D.H.F. and R.C.B.), The Research Council of Norway (R.C.B.), The Harry J. Heeb Foundation (J.N. and M.N.), a Kirchgessner Foundation Vision Research Grant (J.C.), The E. Matilda Ziegler Foundation for the Blind (J.C.), The R.D. and Linda Peters Foundation (J.C.), Hope for Vision (J.C.), and a Fight-for-Sight grant-in-aid (to J.C.). J.C. is the recipient of a Career Development Award from Research to Prevent Blindness. This work has also been supported in part by the National Science Foundation Science and Technology Center for Adaptive Optics, managed by the University of California at Santa Cruz under Cooperative Agreement No. AST-9876783.
Footnotes
Conflict of interest statement: D.R.W. declares financial support from Bausch and Lomb. D.R.W. has licensed a patent relating to adpative optics imaging to Optos.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910128106/DCSupplemental.
There have been a few reports suggesting cone degeneration associated with a color vision defect (42–46), though in most of these cases the color vision defect was not proposed to be causative, nor was the genotype thoroughly described.
The scan length of each OCT scan sets was corrected for inter-individual differences in axial length (51). Axial length in our normal OCT subjects (n = 93) ranged from 21.49 to 26.62 mm, as such the nominal 4-mm horizontal SD-OCT scans had actual scan lengths ranging from 3.60 to 4.46 mm. The two C203R subjects had axial lengths of 23.75 (4511) and 23.19 (4515), resulting in adjusted Stratus OCT scan lengths of 5.83 and 5.69 mm, respectively.
References
- 1.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: The genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 2.Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 3.Deeb SS, et al. Genotype-phenotype relationships in human red/green color-vision defects: Molecular and psychophysical studies. Am J Hum Genet. 1992;51:687–700. [PMC free article] [PubMed] [Google Scholar]
- 4.Jagla WM, Jägle H, Hayashi T, Sharpe LT, Deeb SS. The molecular basis of dichromatic color vision in males with multiple red and green visual pigment genes. Hum Mol Genet. 2002;11:23–32. doi: 10.1093/hmg/11.1.23. [DOI] [PubMed] [Google Scholar]
- 5.Ueyama H, et al. An A-71C substitution in a green gene at the second position in the red/green visual-pigment gene array is associated with deutan color-vision deficiency. Proc Natl Acad Sci USA. 2003;100:3357–3362. doi: 10.1073/pnas.0637437100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neitz M, et al. Variety of genotypes in males diagnosed as dichromatic on a conventional clinical anomaloscope. Vis Neurosci. 2004;21:205–216. doi: 10.1017/s0952523804213293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winderickx J, et al. Defective colour vision associated with a missense mutation in the human green visual pigment gene. Nat Genet. 1992;1:251–256. doi: 10.1038/ng0792-251. [DOI] [PubMed] [Google Scholar]
- 8.Ueyama H, et al. Analysis of L-cone/M-cone visual pigment gene arrays in Japanese males with protan color-vision deficiency. Vision Res. 2004;44:2241–2252. doi: 10.1016/j.visres.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Dryja TP, McEvoy JA, McGee TL, Berson EL. Novel rhodopsin mutations Gly114Val and Gln184Pro in dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:3124–3127. [PubMed] [Google Scholar]
- 10.Neidhardt J, Barthelmes D, Farahmand F, Fleischhauer JC, Berger W. Different amino acid substitutions at the same position in rhodopsin lead to distinct phenotypes. Invest Ophthalmol Vis Sci. 2006;47:1630–1635. doi: 10.1167/iovs.05-1317. [DOI] [PubMed] [Google Scholar]
- 11.Preising M, Lorenz B, editors. Mutations of the Rhodopsin Gene. Retinal International's Scientific Newsletter. n.d. Available at http://www.retina-international.org/sci-news/rhomut.htm.
- 12.Stone EM. Finding and interpreting genetic variations that are important to ophthalmologists. Trans Am Ophthalmol Soc. 2003;101:437–484. [PMC free article] [PubMed] [Google Scholar]
- 13.Horn F, et al. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster A, et al. Novel rhodopsin mutations and genotype-phenotype correlation in patients with autosomal dominant retinitis pigmentosa. Br J Ophthalmol. 2005;89:1258–1264. doi: 10.1136/bjo.2004.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sohocki MM, et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001;17:42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan LS, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: A screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–3064. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Zhang Q, Shen H, Li S, Xiao X. Analysis of rhodopsin and peripherin/RDS genes in Chinese patients with retinitis pigmentosa. Yan Ke Xue Bao. 1998;14:210–214. in Chinese. [PubMed] [Google Scholar]
- 18.Macke JP, et al. Identification of novel rhodopsin mutations responsible for retinitis pigmentosa: Implications for the structure and function of rhodopsin. Am J Hum Genet. 1993;53:80–89. [PMC free article] [PubMed] [Google Scholar]
- 19.Richards JE, Scott KM, Sieving PA. Disruption of conserved rhodopsin disulfide bond by Cys187Tyr mutation causes early and severe autosomal dominant retinitis pigmentosa. Ophthalmology. 1995;102:669–677. doi: 10.1016/s0161-6420(95)30972-4. [DOI] [PubMed] [Google Scholar]
- 20.Kazmi MA, Sakmar TP, Ostrer H. Mutation of a conserved cysteine in the X-linked cone opsins causes color vision deficiencies by disrupting protein folding and stablilty. Invest Ophthalmol Vis Sci. 1997;38:1074–1081. [PubMed] [Google Scholar]
- 21.Karnik SS, Sakmar TP, Chen H-B, Khorana HG. Cysteine residues 110 and 187 are required for the formation of correct structure in bovine rhodopsin. Proc Natl Acad Sci USA. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwa J, Reeves PJ, Klein-Seetharaman J, Davidson F, Khorana HG. Structure and function in rhodopsin: Further elucidation of the role of the intradiscal cysteines, Cys-110, -185, and -187, in rhodopsin folding and function. Proc Natl Acad Sci USA. 1999;96:1932–1935. doi: 10.1073/pnas.96.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: Implications for therapy. Trends in Mol Med. 2005;11:177–185. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Carroll J, Neitz M, Hofer H, Neitz J, Williams DR. Functional photoreceptor loss revealed with adaptive optics: An alternate cause for color blindness. Proc Natl Acad Sci USA. 2004;101:8461–8466. doi: 10.1073/pnas.0401440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathans J, et al. Molecular genetics of human blue cone monochromacy. Science. 1989;245:831–838. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- 26.Crognale MA, et al. Characterization of a novel form of X-linked incomplete achromatopsia. Vis Neurosci. 2004;21:197–203. doi: 10.1017/s0952523804213384. [DOI] [PubMed] [Google Scholar]
- 27.Regan BC, Reffin JP, Mollon JD. Luminance noise and the rapid determination of discrimination ellipses in color deficiency. Vision Res. 1994;34:1279–1299. doi: 10.1016/0042-6989(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 28.Ventura DF, et al. In: Preliminary norms for the Cambridge Color Test. Normal & Defective Color Vision. Mollon JD, Pokorny J, Knoblauch K, editors. Oxford: Oxford Univ Press; 2003. [Google Scholar]
- 29.Sharpe LT, et al. Red, green, and red-green hybrid pigments in the human retina: Correlations between deduced protein sequences and psychophysically measured spectral sensitivities. J Neurosci. 1998;18:10053–10069. doi: 10.1523/JNEUROSCI.18-23-10053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neitz J, Neitz M, He JC, Shevell SK. Trichromatic color vision with only two spectrally distinct photopigments. Nat Neurosci. 1999;2:884–888. doi: 10.1038/13185. [DOI] [PubMed] [Google Scholar]
- 31.Neitz J, Neitz M, Kainz PM. Visual pigment gene structure and the severity of color vision defects. Science. 1996;274:801–804. doi: 10.1126/science.274.5288.801. [DOI] [PubMed] [Google Scholar]
- 32.Li KY, Roorda A. Automated identification of cone photoreceptors in adaptive optics retinal images. J Opt Soc Am A Opt Image Sci Vis. 2007;24:1358–1363. doi: 10.1364/josaa.24.001358. [DOI] [PubMed] [Google Scholar]
- 33.Rha J, et al. Spectral domain optical coherence tomography and adaptive optics: Imaging photoreceptor layer morphology to interpret preclinical phenotypes. In: Anderson RE, Hollyfield JG, LaVail MM, editors. Retinal Degenerative Diseases: Laboratory and Therapeutic Investigations, Advances in Experimental Medicine and Biology. New York: Springer; In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mancuso K, et al. Gene therapy for red-green colour blindness in adult primates. Nature. 2009;461:784–787. doi: 10.1038/nature08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winderickx J, Battisti L, Motulsky AG, Deeb SS. Selective expression of human X chromosome-linked green opsin genes. Proc Natl Acad Sci USA. 1992;89:9710–9714. doi: 10.1073/pnas.89.20.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi T, Motulsky AG, Deeb SS. Visual pigment gene structure and expression in the human retinae. Hum Mol Genet. 1997;6:981–990. doi: 10.1093/hmg/6.7.981. [DOI] [PubMed] [Google Scholar]
- 37.Sjoberg SA, Neitz M, Balding SD, Neitz J. L-cone pigment genes expressed in normal color vision. Vision Res. 1998;38:3213–3219. doi: 10.1016/s0042-6989(97)00367-2. [DOI] [PubMed] [Google Scholar]
- 38.Hagstrom SA, Neitz J, Neitz M. Variations in cone populations for red-green color vision examined by analysis of mRNA. NeuroReport. 1998;9:1963–1967. doi: 10.1097/00001756-199806220-00009. [DOI] [PubMed] [Google Scholar]
- 39.Hagstrom SA, Neitz M, Neitz J. Cone pigment gene expression in individual photoreceptors and the chromatic topography of the retina. J Opt Soc Am A Opt Image Sci Vis. 2000;17:527–537. doi: 10.1364/josaa.17.000527. [DOI] [PubMed] [Google Scholar]
- 40.Geller AM, Sieving PA. Assessment of foveal cone photoreceptors in Stargardt's macular dystrophy using a small dot detection task. Vision Res. 1993;33:1509–1524. doi: 10.1016/0042-6989(93)90144-l. [DOI] [PubMed] [Google Scholar]
- 41.Makous W, et al. Retinal microscotomas revealed with adaptive-optics microflashes. Invest Ophthalmol Vis Sci. 2006;47:4160–4167. doi: 10.1167/iovs.05-1195. [DOI] [PubMed] [Google Scholar]
- 42.Kellner U, Sadowski B, Zrenner E, Foerster MH. Selective cone dystrophy with protan genotype. Invest Ophthalmol Vis Sci. 1995;36:2381–2387. [PubMed] [Google Scholar]
- 43.Reichel E, Bruce AM, Sandberg MA, Berson EL. An electroretinographic and molecular genetics study of X-linked cone degeneration. Am J Ophthalmol. 1989;108:540–547. doi: 10.1016/0002-9394(89)90431-5. [DOI] [PubMed] [Google Scholar]
- 44.Scholl HPN, Kremers J, Besch D, Zrenner E, Jägle H. Progressive cone dystrophy with deutan genotype and phenotype. Graefes Arch Clin Exp Ophthalmol. 2006;244:183–191. doi: 10.1007/s00417-005-0022-7. [DOI] [PubMed] [Google Scholar]
- 45.Scholl HPN, Kremers J, Wissinger B. Macular dystrophy with protan genotype and phenotype studied with cone type specific ERG's. Curr Eye Res. 2001;22:221–228. doi: 10.1076/ceyr.22.3.221.5514. [DOI] [PubMed] [Google Scholar]
- 46.Michaelides M, et al. X-linked cone dysfunction syndrome with myopia and protanopia. Ophthalmology. 2005;112:1448–1454. doi: 10.1016/j.ophtha.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 47.Carroll J, Neitz M, Neitz J. Estimates of L:M cone ratio from ERG flicker photometry and genetics. J Vis. 2002;2:531–542. doi: 10.1167/2.8.1. [DOI] [PubMed] [Google Scholar]
- 48.Hofer H, Carroll J, Neitz J, Neitz M, Williams DR. Organization of the human trichromatic cone mosaic. J Neurosci. 2005;25:9669–9679. doi: 10.1523/JNEUROSCI.2414-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunther KL, Neitz J, Neitz M. Nucleotide polymorphisms upstream of the X-chromosome opsin gene array tune L:M cone ratio. Vis Neurosci. 2008;25:265–271. doi: 10.1017/S0952523808080280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baraas RC, et al. Adaptive optics retinal imaging reveals S-cone dystrophy in tritan color-vision deficiency. J Opt Soc Am A Opt Image Sci Vis. 2007;24:1438–1446. doi: 10.1364/josaa.24.001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett AG, Rudnicka AR, Edgar DF. Improvements on Littmann's method of determining the size of retinal features by fundus photography. Graefes Arch Clin Exp Ophthalmol. 1994;232:361–367. doi: 10.1007/BF00175988. [DOI] [PubMed] [Google Scholar]
- 52.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 53.Jørgensen TM, Thomadsen J, Christensen U, Soliman W, Sander B. Enhancing the signal-to-noise ratio in ophthalmic optical coherence tomography by image registration-method and clinical examples. J Biomed Opt. 2007;12 doi: 10.1117/1.2772879. 041208. [DOI] [PubMed] [Google Scholar]
- 54.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 55.Jacobson SG, et al. Human cone photoreceptor dependence on RPE65 isomerase. Proc Natl Acad Sci USA. 2007;104:15123–15128. doi: 10.1073/pnas.0706367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacobson SG, et al. Identifying photoreceptors in blind eyes caused by RPE65 mutations: Prerequisite for human gene therapy success. Proc Natl Acad Sci USA. 2005;102:6177–6182. doi: 10.1073/pnas.0500646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neitz M, Neitz J. A new mass screening test for color-vision deficiencies in children. Color Res Appl. 2001;26:S239–S249. [Google Scholar]
- 58.Pum D, Ahnelt PK, Grasl M. Iso-orientation areas in the foveal cone mosaic. Vis Neurosci. 1990;5:511–523. doi: 10.1017/s0952523800000687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.