Abstract
Regions of the temporal and parietal lobes are particularly damaged in Alzheimer's disease (AD), and this leads to a predictable pattern of brain atrophy. In vivo quantification of subregional atrophy, such as changes in cortical thickness or structure volume, could lead to improved diagnosis and better assessment of the neuroprotective effects of a therapy. Toward this end, we have developed a fast and robust method for accurately quantifying cerebral structural changes in several cortical and subcortical regions using serial MRI scans. In 169 healthy controls, 299 subjects with mild cognitive impairment (MCI), and 129 subjects with AD, we measured rates of subregional cerebral volume change for each cohort and performed power calculations to identify regions that would provide the most sensitive outcome measures in clinical trials of disease-modifying agents. Consistent with regional specificity of AD, temporal-lobe cortical regions showed the greatest disease-related changes and significantly outperformed any of the clinical or cognitive measures examined for both AD and MCI. Global measures of change in brain structure, including whole-brain and ventricular volumes, were also elevated in AD and MCI, but were less salient when compared to changes in normal subjects. Therefore, these biomarkers are less powerful for quantifying disease-modifying effects of compounds that target AD pathology. The findings indicate that regional temporal lobe cortical changes would have great utility as outcome measures in clinical trials and may also have utility in clinical practice for aiding early diagnosis of neurodegenerative disease.
Keywords: clinical diagnosis, clinical trials, disease-specific atrophy, image registration, MRI biomarkers
The healthy adult brain is remarkably stable structurally but undergoes gradual changes with normal aging. Structural change is accelerated in neurodegenerative disease, including Alzheimer's disease (AD). The atrophy in AD arises from neuron and synapse loss that begins in the entorhinal cortex. The pathology then spreads throughout the limbic regions of the temporal lobe, including the hippocampal formation. Subsequently, neuron loss and atrophy is observed throughout neocortical association areas in temporal, parietal, and frontal lobes (1).
The fact that atrophy associated with AD can be detected in vivo using MRI has long been known (2). Hippocampal volume loss is a consistent finding (3) and is predictive of clinical decline (4–7). However, hippocampal atrophy is not specific to AD, as it is seen in a number of psychiatric and neurodegenerative diseases (8–10). Recently, it has been shown that cortical atrophy measured on MRI parallels the spread of AD pathology (11–13). Accurate measurement of cortical thickness and subcortical volumes across multiple regions may provide a signature of the disease specific enough to be useful for early diagnosis of AD (14).
In recent studies, measures of progressive AD-related atrophy detected from serial MRI scans show promise as biomarkers in evaluating the effectiveness of disease-modifying agents. So far, these studies have focused on relatively global measures, such as whole-brain and ventricular volume change (15, 16), although some have also looked at hippocampal volume change (17, 18). In these studies, despite the known regional specificity of AD-related volumetric changes, global measures have shown greater sensitivity than local measurements, possibly because of the difficulty in obtaining accurate measurement of local brain structure change using existing methods (17). Nevertheless, these global measures of brain structure change are highly correlated with gold-standard clinical outcome measures, such as the Clinical Dementia Rating Scale Sum of Boxes and Mini Mental State Examination scores (15, 19).
The use of longitudinal anatomical quantification in multicenter clinical trials presents a number of challenges, including differences in MRI pulse sequences across scanner manufacturers, scanner-specific spatial distortions, and changes in scanner hardware and software over time that can affect image properties. In view of this, the Alzheimer's Disease Neuroimaging Initiative (ADNI) was designed to validate and compare imaging and biofluid markers of disease progression in a realistic multicenter clinical trial setting (20). The large, publicly available ADNI database thus provides a realistic setting in which to validate imaging methods aimed at assessing AD pathology. To this database, we applied a recently developed method for obtaining precise measures of interval change in cortical and subcortical regions, based on structural MRI, and determined the relative statistical power to discriminate pathology afforded by different regional measures.
Results
We examined two models of treatable effects for power calculations. The first, Model T (for “total”), assumes that the study drug modifies both disease- and aging-related changes; the second, Model D (for “disease-specific”), assumes that the study drug modifies AD- or mild cognitive impairment- (MCI) related changes but has no effect on aging-related changes. We found that multiple regional volume changes, including those of whole brain, ventricle, hippocampus, entorhinal, fusiform, inferior temporal and middle temporal cortices, provided powerful outcome measures, with several measures requiring fewer than 100 subjects per arm to detect a 25% reduction in the rate of total change in AD, with 80% power at the P < 0.05 significance level (see Methods for a description of the power calculations). Power calculations using ventricle and whole-brain volume change as outcome measures were particularly sensitive to the choice of treatable-effect model, especially in the case of MCI, where Model D required as much as six times the number of subjects per arm as Model T. When Model D was used for MCI, the best cognitive measure was as good as or outperformed these measures of more global structural change in the brain. For AD, regional cortical-volume change provided consistently superior power compared to cognitive measures regardless of choice of treatable effects model. The results indicate that volume loss in entorhinal, fusiform, inferior temporal and middle temporal cortices would serve as superior outcome measures for study drugs specifically targeting AD pathology in patients with MCI or AD.
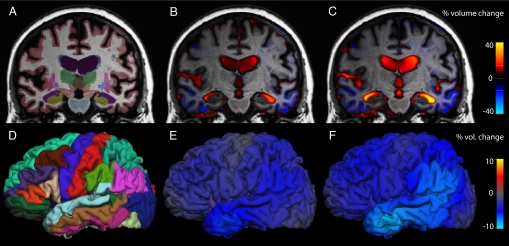
Estimated changes across the brain at 6 and 12 months, along with cortical and subcortical tissue segmentation, are shown in Fig. 1 for an individual from the MCI cohort. Fig. 2 shows the results of power calculations for imaging measures of regional change, along with the best clinical cognitive-outcome measure, based on AD subjects and healthy controls. Results for Model T are in blue, and results for Model D are in red; numerical values are in Table 1(see Fig. S1 and Table S1 for sample size estimates not incorporating random rates of change).
Fig. 1.
Tissue segmentation, with 6- and 12-month volume change fields for an MCI subject. (A) Segmentation of the baseline MRI scan, with different brain structures represented in different colors. (B) Corresponding coronal slice overlain with a heat map representation of the voxelwise estimates of volumetric change at 6 months and (C) 12 months. (D) Left hemisphere cortical parcellation of the baseline MRI scan. (E) Cortical surface overlain with a heat map representation of the estimates of cortical volumetric change at 6 months and (F) 12 months. Region-specific estimates were obtained by averaging the voxelwise change within each region of interest. In this subject, the left middle-temporal gyrus has decreased in volume by 4.7% at 6 months and by 8.2% at 12 months; the left temporal-horn lateral ventricle has increased by 17.4% at 6 months and by 35.3% at 12 months.
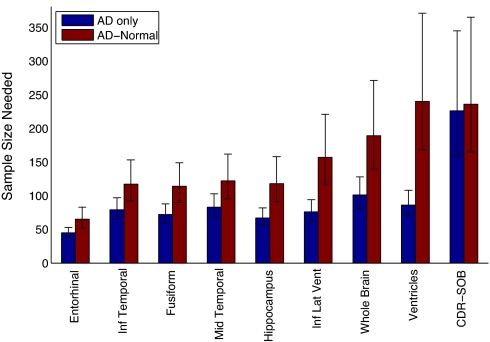
Fig. 2.
Sample size estimates for AD from a linear mixed-effects model with random slopes. The bars, with 95% confidence intervals, indicate the expected number of subjects needed per arm to detect a 25% reduction in rate of change at the P < 0.05 level with 80% power, assuming a 24-month trial with scans every 6 months. Results for Model T are in blue and results for Model D are in red; numerical values are shown in Table 1.
Table 1.
Sample size estimates (N) and annualized percent change for AD
| Measure | AD only N | AD-HC N | AD % change* | HC % change* |
|---|---|---|---|---|
| Entorhinal | 45 [39 53] | 65 [52 83] | −3.81 [−4.10 −3.52] | −0.64 [−0.85 −0.43] |
| Inf temporal | 79 [65 97] | 117 [92 153] | −3.64 [−4.00 −3.28] | −0.65 [−0.76 −0.53] |
| Fusiform | 72 [60 88] | 114 [90 149] | −2.90 [−3.17 −2.62] | −0.59 [−0.68 −0.50] |
| Mid temporal | 83 [69 103] | 122 [95 162] | −3.44 [−3.79 −3.09] | −0.60 [−0.73 −0.47] |
| Hippocampus | 67 [56 82] | 118 [91 158] | −3.28 [−3.58 −2.98] | −0.80 [−0.95 −0.65] |
| Inf lat vent | 76 [63 94] | 157 [117 221] | 15.56 [14.04 17.09] | 4.71 [3.95 5.47] |
| Whole brain | 101 [81 128] | 189 [139 271] | −1.50 [−1.67 −1.33] | −0.40 [−0.47 −0.34] |
| Ventricles | 86 [71 108] | 240 [168 371] | 11.06 [9.91 12.21] | 4.43 [3.83 5.03] |
| CDR-SOB | 226 [159 345] | 236 [165 365] | 1.76 [1.43 2.10] | 0.04 [0.00 0.07] |
| ADAS-Cog† | 324 [217 536] | 283 [192 457] | 4.84 [3.76 5.92] | −0.34 [−0.59 −0.09] |
| MMSE† | 482 [299 907] | 494 [303 948] | −2.45 [−3.11 −1.78] | −0.03 [−0.14 0.08] |
Values in brackets are 95% confidence intervals. ADAS-Cog, Alzheimer's Disease Assessment Scale—Cognitive; CDR-SOB, clinical dementia rating, sum of boxes; HC, healthy controls; MMSE, Mini-Mental State Examination.
*Annual percent change in volume for all entries except CDR-SOB, ADAS-Cog, and MMSE.
†Not shown in Fig. 2.
Imaging measures generally outperformed the best cognitive measure, regardless of model choice. While power estimates for cognitive measures were relatively unaffected by model choice, the power estimates for the imaging measures were strongly dependent on the treatment model used. Subregional cortical measures outperformed global imaging measures and were less dependent on choice of treatment model.
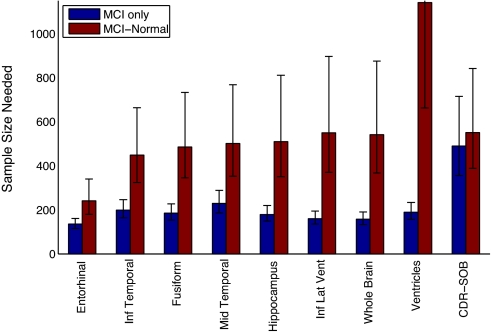
For MCI, as shown in Fig. 3 and Table 2, the dependence on model choice is even more salient than for AD. Notably, for ventricular volume, the sample size calculated using Model D is six times higher than that calculated using Model T, and exceeds that calculated for the best clinical or cognitive measure. Similar to what was found in AD, the regional temporal lobe cortical measures afforded the smallest sample sizes, regardless of model choice (see Fig. S2 and Table S2 for sample size estimates not incorporating random rates of change).
Fig. 3.
Sample size estimates for MCI (see Fig. 2 for description). Numerical values are in Table 2. Note that the Model D (red) upper bound on the 95% confidence interval for ventricles is 2,421.
Table 2.
Sample size estimates (N) and annualized percent change for MCI
| Measure | MCI only N | MCI-HC N | MCI % change* | HC† % change* |
|---|---|---|---|---|
| Entorhinal | 135 [115 161] | 241 [180 340] | −2.54 [−2.75 −2.33] | −0.64 [−0.85 −0.43] |
| Inf temporal | 199 [164 246] | 449 [324 664] | −1.93 [−2.13 −1.74] | −0.65 [−0.76 −0.53] |
| Fusiform | 185 [153 227] | 485 [345 733] | −1.54 [−1.69 −1.39] | −0.59 [−0.68 −0.50] |
| Mid temporal | 229 [186 288] | 501 [353 768] | −1.84 [−2.04 −1.64] | −0.60 [−0.73 −0.47] |
| Hippocampus | 179 [149 220] | 510 [350 811] | −1.96 [−2.15 −1.78] | −0.80 [−0.95 −0.65] |
| Inf lat vent | 160 [135 194] | 550 [371 897] | 10.23 [9.30 11.16] | 4.71 [3.95 5.47] |
| Whole brain | 158 [133 190] | 541 [367 875] | −0.88 [−0.96 −0.80] | −0.40 [−0.47 −0.34] |
| Ventricles | 189 [157 233] | 1,141 [662 2,421] | 7.47 [6.73 8.21] | 4.43 [3.83 5.03] |
| CDR-SOB | 490 [356 715] | 551 [388 842] | 0.67 [0.55 0.78] | 0.04 [0.00 0.07] |
| ADAS-Cog‡ | 1,232 [748 2,403] | 804 [500 1,502] | 1.44 [1.03 1.84] | −0.34 [−0.59 −0.09] |
| MMSE‡ | 1,214 [744 2,322] | 1,304 [751 2,800] | −0.84 [−1.08 −0.61] | −0.03 [−0.14 0.08] |
Discussion
The findings demonstrate that longitudinal volumetric change provides powerful outcome measures with which to examine putative disease-modifying medications for AD and MCI. Whole brain, ventricle, hippocampus, and cortical volumes of the entorhinal, fusiform, inferior temporal and middle temporal gyri undergo high rates of change in AD and MCI, which are quantifiable using serial MRI and the nonlinear registration procedures used here. A comparison of the current method with a standard method for quantifying global change is provided in the SI, where the analysis was restricted to a common data set of serial scans at 0, 6, and 12 months (Figs. S3 and S4 and Tables S3 and S4).
For clinical trial power calculations using longitudinal volumetric change as an outcome measure, choice of treatable-effect model influences which brain regions would be most sensitive to detect a drug effect, especially in MCI. If the drug is presumed to slow both age- and AD-related brain atrophy, then global and subregional medial temporal lobe (MTL) and cortical measures provide excellent statistical power to detect treatment effects. However, if the study drug is presumed to specifically slow AD-related brain atrophy, then subregional cortical measures provide superior power. For MCI, entorhinal cortex provided the most powerful outcome measure, which is consistent with findings suggesting that atrophy in this region is a sensitive marker of prodromal AD (11, 21).
Choice of treatment model differentially affects cognitive and MRI variables; cognitive measures often show improvements over time in healthy controls because of practice effects (22), but deterioration over time in patients. Therefore, for cognitive measures, Model T can provide more conservative power estimates and is the most commonly used model in powering clinical trials. In contrast, both normal aging and disease are associated with atrophic changes over time. Thus, Model D generally provides more conservative power estimates for imaging measures. For this reason, it is important to consider both models when comparing across cognitive and imaging measures.
One of the primary motivations for using brain volumetric changes as outcome measures in clinical trials has been the evidence for greater statistical power afforded by such measures relative to clinical and cognitive measures (23). The present results, however, demonstrate that the most commonly used global imaging measures may be less powerful than the best clinical and cognitive measures, when the more conservative, and perhaps more realistic, disease-specific model is used. These effects are magnified in the MCI cohort, which is a patient population of particular interest for drug development (24, 25). Because of the overlap in behavioral features between MCI and healthy elderly controls, MCI trials would require particularly large subject numbers when using behavioral outcome measures alone.
Another motivation for using regional volumetric changes as outcome measures in clinical trials is the desire to more directly examine the effects of therapy on the brain's AD pathology. Because AD pathology is known to be concentrated in particular cortical and subcortical gray-matter regions, it would be desirable to measure change in the specific regions where neuronal dystrophy leads to pronounced atrophy. By itself, the halting of such neuronal dystrophy would lead to a stabilization of volume loss, but other drug effects, perhaps unrelated to therapeutic effect, may also be at play. For example, a recent active immunization trial against amyloid showed greater overall brain-volume loss in subjects who generated an immune response when compared to those who did not. In this case, global volume loss was attributed to possible changes in brain hydration state related to therapy. A trial of passive immunization against amyloid showed an association between higher doses of the medication and vasogenic edema. Thus, a short-term effect of the drug might be an increase in global brain volume that could be mistaken for a neuroprotection. Further study is needed to determine whether these processes are even more salient in regions enriched for amyloid and also whether such processes eventually reach a steady state upon which a drug's neuroprotective effect may still be evaluated. Nevertheless, regional measures of volumetric change offer a finer-grained examination of these processes and the effects of a therapy on the brain, and might be proportionally less affected by global effects unrelated to regional AD pathology (17, 26).
Although not a direct measure of the molecular pathology in AD, subregional brain structural changes are a direct measure of the neurodegeneration associated with the disease, and are more directly associated with progression of clinical symptoms than are measures of amyloid (27). Imaging of amyloid protein provides a direct measure of one of the components of AD, but the sensitivity and specificity of this measure as a biomarker of AD remains an open question. There is growing evidence that amyloid protein may be elevated in some subjects who remain cognitively normal during the period of follow-up. Functional imaging measures (28–30) also show great promise as biomarkers in AD clinical trials and may be sensitive to pathology, at even earlier stages of disease.
An essential characteristic of an AD therapeutic is that it results in clinical or cognitive improvement. This improvement may be achieved through symptomatic modification (31, 32) or, preferably, through disease modification (33–36). Assessment of disease modification therefore relies on detecting a slowing of clinical decline. Clinical decline in AD occurs slowly over years, and so detecting a halt to this decline would be aided through complementary and sensitive measures. Although cognitive outcome is central to assessing therapeutic efficacy, cognitive decline is in fact a secondary effect of neuronal damage from the disease, partially reflected in regional atrophy. MRI longitudinal measures of regional volumetric change provide a valuable complement to cognitive measures in that they are not influenced by temporary symptomatic improvements, and they provide an early index of the drug's ability to reach the target organ and have an effect on AD-related atrophy.
Finally, regional volumetric measures of change show promise for eventual use in clinical practice to assist risk stratification and differential diagnosis at the earliest stages of neurodegenerative disease. These measures may be particularly powerful when combined with baseline volumetry (14) and other diagnostics, such as cerebrospinal fluid biomarkers, nuclear medicine ligands, neuropsychological tests, and genetics. The present results suggest that change in MTL cortical regions, in particular the entorhinal cortex, would provide the most sensitive and specific volumetric imaging measures early in the disease. Changes in regions such as the hippocampus, ventricles, and whole brain provide sensitive indices of disease progression but are also seen in healthy aging adults, thus reducing their specificity for the detection of AD.
Methods
ADNI.
Data used in the preparation of this article were obtained from the ADNI database (www.loni.ucla.edu/ADNI). ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies, and nonprofit organizations as a $60 million, 5-year public-private partnership. ADNI's goal is to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations. Subjects have been recruited from over 50 sites across the United States and Canada. ADNI's goal was to recruit 800 adults, ages 55 to 90, to participate in the research: ≈200 cognitively normal individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years (see www.adni-info.org). The research protocol was approved by each local institutional review board and written informed consent is obtained from each participant.
Participants.
The ADNI general eligibility criteria are described in the ADNI Protocol Summary page of the ADNI-Info Web site at adni-info.org for 2009. Briefly, subjects are not depressed, have a modified Hachinski score of 4 or less, and have a study partner able to provide an independent evaluation of functioning. Healthy control subjects have a Clinical Dementia Rating (37) of 0. Subjects with AD have a Clinical Dementia Rating of 0.5 or 1.0 and meet National Institute of Neurological Disorders and Stroke and Alzheimer's Disease and Related Disorders Association criteria for probable AD (38).
In this study, we used baseline and follow-up data collected before August 27, 2009 from the ADNI database. Group clinical and demographic baseline data for the 169 healthy control, 299 MCI subjects, and 129 AD subjects in this study are presented in Table 3.
Table 3.
Group demographics at baseline
| Group | HC subjects (n = 169) | MCI subjects (n = 299) | AD subjects (n = 129) |
|---|---|---|---|
| Age* (years) | 76.2 ± 5.2 | 74.6 ± 7.4 | 74.6 ± 7.8 |
| Female† | 83 (49.1%) | 111 (37.1%) | 63 (48.8%) |
| Years of Education | 16.0 ± 2.8 | 15.8 ± 3.0 | 15.0 ± 3.0 |
| CDR-SOB | 0.03 ± 0.12 | 1.56 ± 0.88 | 4.23 ± 1.54 |
| ADAS-Cog | 6.0 ± 2.8 | 11.6 ± 4.3 | 18.5 ± 6.2 |
| MMSE | 29.1 ± 1.1 | 27.0 ± 1.8 | 23.4 ± 2.0 |
| APOE high risk | 47 (27.8%) | 170 (56.8%) | 90 (69.8%) |
APOE is apolipoprotein E gene; APOE risk was defined as the presence of the APOE e4 allele.
*Data are mean ± standard deviation
†Data are numbers of subjects, and numbers in parentheses are percentages.
Data Acquisition and Preparation.
Raw Digital Imaging and Communications in Medicine MRI scans, including two three-dimensional T1-weighted volumes per subject per visit, were downloaded from the public ADNI site (www.loni.ucla.edu/ADNI/Data/index.shtml). These data were collected across a variety of scanners with protocols individualized for each scanner, as defined at www.loni.ucla.edu/ADNI/Research/Cores/index.shtml. In our laboratory, MRI data were reviewed for quality and automatically corrected for spatial distortion caused by gradient nonlinearity (39). For each subject at each visit, the two three-dimensional T1-weighted images were rigid-body aligned to each other, averaged to improve signal-to-noise ratio, and resampled to isotropic 1-mm voxels. Baseline volumetric segmentation (40, 41) and cortical surface reconstruction (42–45) and pacrellation (46, 47) were performed using a data analysis pipeline based on the FreeSurfer software package and customized Matlab code, optimized for use on large multisite data sets. The automated whole-brain segmentation procedure uses a probabilistic atlas and applies a Bayesian classification rule to assign a neuroanatomic label to each voxel. The atlas consists of a manually derived training set created by the Center for Morphometric Analysis (Massachusetts General Hospital, Harvard Medical School) from 40 non-ADNI subjects across the adult age range, including individuals with AD. Automated volumetric segmentation required only qualitative review to ensure that there was no technical failure of the application.
The cortical surface was reconstructed to measure thickness at each surface location, or vertex, to allow visualization of group differences at each vertex. The surface was parceled into distinct regions of interest (ROIs). The cortical-surface model was manually reviewed and edited for accuracy. Minimal editing was performed according to standard, objective rules, including correction of errors in removal of nonbrain areas and inclusion of white-matter areas of hypointensity adjacent to the cortical ribbon. Qualitative review and editing were performed, with blinding to the diagnostic status, by one of three technicians trained and supervised by an expert neuroanatomist with more than 10 years of experience (C.F.-N.). The technicians had a minimum of 4 months of experience reviewing brain MR images before their involvement in this project.
Qualitative review and editing required ≈45 min per subject. Baseline image construction was carried out on a Linux cluster composed of dual quad-core 2.5 GHz CPUs (Xeon E5420; Intel) with 16 GB RAM; each image reconstruction was run as an independent process and took ≈24 h of computational time.
Estimation of ROI Volumentric Interval Change.
For each subject, follow-up images were fully affine-registered to the baseline image, and their intensities brought into local agreement (i.e., corrected for relative B1-induced intensity distortion). Nonlinear registration of the images was then performed, where voxel centers are moved about until a good match between the images is made. Several methods exist for causing this to happen, including fluid deformation (48–50) and tensor-based morphometry (51). For the results presented here, however, we developed and applied a method (52) based on linear elasticity and closer in spirit to tensor-based morphometry. This method proceeds essentially as follows. The images are heavily blurred (smoothed), making them almost identical, and a merit or potential function calculated. This merit function expresses the intensity difference between the images at each voxel, and depends on the displacement field for the voxel centers of the image being transformed; it is also regularized to keep the displacement field spatially smooth. The merit function by design will have a minimum when the displacement field induces a good match between the images. The displacement field in general will turn cubic voxels into displaced, irregular hexahedra whose volumes (53) give the volume-change field. The merit function is minimized efficiently using standard numerical methods. Having found a displacement field for the heavily blurred pair of images, the blurring is reduced and the procedure repeated, thus iteratively building up a better displacement field. Two important additions to this are: (i) applying the final displacement field to the image being transformed, then nonlinearly registering the resultant image to the same target, and finally tracing back through the displacement fields, thus calculated to find the net displacement field; and (ii) restricting to ROIs and zooming when tissue structures are separated by only a voxel or two. These additional features enable very precise registration involving large or subtle deformations, even at small spatial scales with low boundary contrast.
All available healthy controls, MCI subjects, and subjects with AD who passed the qualitative baseline review described above were thus registered. From the deformation field, a volume-change field was calculated; an example is shown in Fig. 1. For each subject, the volume-change field was averaged over each ROI, including those of the cortical surface (change in cortical volume to first-order results from change in thickness), to give the percentage change from baseline. Further visual quality control, blind to diagnosis, was carried out by a technician on the volume-change field to exclude cases where there was significant degradation in meaningful registration for at least one ROI because of artifacts or major changes in scanner hardware between visits (e.g., change of scanner model or type of RF coil). The most common form of artifact, affecting approximately half of the rejected scans, was caused by within-scan subject motion. In future clinical trials, the loss of scans caused by motion artifacts may be greatly reduced by using real-time motion-correction procedures (54, 55). Artifacts resulting from change in scanner models between visits typically include differential contrast or spatial blurring, mostly affecting the fine-scale estimates of change (e.g., within the cortical ROIs). Artifacts resulting from change in RF coil, specifically from a traditional quadrature head coil to a phased-array coil, primarily resulted in dramatic changes in blood inflow effects, which in turn predominantly affected MTL measures. The combination of artifacts affecting the volume change field reduced the number of healthy control follow-up scans by 14.2%, the number of MCI follow-up scans by 14.5%, and the number of AD follow-up scans by 15.8%.
For a subject to be included in our statistical analyses, several criteria needed to be satisfied: the baseline cortical parcellation and subcortical segmentation had to pass review, as described above; for a tight comparison between cognitive and volumetric measures, a subject's follow-up was eliminated unless both volumetric and cognitive data, including a clinical diagnosis, existed for that follow-up; there was at least one good follow-up, along with the good baseline; a healthy control needed to remain such at all follow-ups; and finally, the volume-change field had to pass review. Quality control on the volume-change field reduced the number of healthy controls by 8.6% to 169, the number of MCI subjects by 8.5% to 299, and the number of AD subjects by 12.2% to 129.
Power Calculations.
We examined two models of treatable effects for power calculations: Model T assumes that the study drug modifies both disease- and aging-related changes; Model D assumes that the study drug modifies only AD- or MCI-related changes.
Power calculations were performed using a mixed-effects regression model for the outcome variable (absolute cognitive measure or subregional percent-volume change) as a linear function of time, with random (individual-specific) slope or trend term and, for the cognitive measures, random intercept (baseline value). Sample sizes per arm were estimated based on a z-test (56) for absolute mean slopes for AD and MCI subjects (Model T), and the difference in mean slopes for AD and MCI subjects from healthy controls (Model D). The sample size required to detect 25% slowing in mean rate of decline for a hypothetical disease-modifying treatment versus placebo was estimated for a 24-month, two-arm, equal-allocation trial, with a 6-month assessment interval. Power calculations were performed with the requirement that the trial have 80% power to detect the treatment effect using a two-sided significance level of 5%. The sample size per arm scales with the variance of the within-group rate of change (slope), which has both between-subject and within-subject (residual error variance of the mixed-effects model) components. Thus, for Model T, the treatment-effect size of interest was 25% of the rate of change in the patient population (MCI or AD), and for Model D it was 25% of the difference between the rates of change in the patient and normal populations. Confidence intervals of 95% for sample sizes were based on 95% confidence intervals for the treatment-effect size of interest. Power calculations were implemented in Matlab version 2008b, using the nlme function in the Statistics Toolbox. Sample size estimates based on a linear random-effects model ignoring between-subject variance in the rate of change (i.e., taking the group-specific rate of change as a fixed effect) are provided in Figs. S1 and S2, and Tables S1 and S2.
Supplementary Material
Acknowledgments.
Thanks to Alan Koyama, Robin Jennings, and Chris Pung for downloading and processing the Alzheimer's Disease Neuroimaging Initiative MRI data. This research was supported by National Institute of Health Grants R01AG031224, R01AG22381, U54NS056883, P50NS22343, and P50MH081755 (to A.M.D.). Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institute of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck and Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles.
Footnotes
Conflict of interest statement: A.M.D. is a founder and holds equity in CorTechs Labs, Inc, and also serves on the Scientific Advisory Board. The terms of this arrangement have been reviewed and approved by the University of California at San Diego in accordance with its conflict of interest policies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906053106/DCSupplemental.
References
- 1.Braak H, Braak E. Evolution of the neuropathology of Alzheimer's disease. Acta Neurol Scand Suppl. 1996;165:3–12. doi: 10.1111/j.1600-0404.1996.tb05866.x. [DOI] [PubMed] [Google Scholar]
- 2.Jack CR, Jr, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atiya M, Hyman BT, Albert MS, Killiany R. Structural magnetic resonance imaging in established and prodromal Alzheimer disease: A review. Alzheimer Dis Assoc Disord. 2003;17:177–195. doi: 10.1097/00002093-200307000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Jack CR, Jr, et al. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Leon MJ, et al. Imaging and CSF studies in the preclinical diagnosis of Alzheimer's disease. Ann N Y Acad Sci. 2007;1097:114–145. doi: 10.1196/annals.1379.012. [DOI] [PubMed] [Google Scholar]
- 6.Devanand DP, et al. Hippocampal and entorhinal atrophy in mild cognitive impairment: Prediction of Alzheimer disease. Neurology. 2007;68:828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- 7.Hampel H, et al. Core candidate neurochemical and imaging biomarkers of Alzheimer's disease. Alzheimers Dement. 2008;4:38–48. doi: 10.1016/j.jalz.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Frisoni GB, et al. Hippocampal and entorhinal cortex atrophy in frontotemporal dementia and Alzheimer's disease. Neurology. 1999;52:91–100. doi: 10.1212/wnl.52.1.91. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto M, et al. Medial temporal and whole-brain atrophy in dementia with Lewy bodies: A volumetric MRI study. Neurology. 1998;51:357–362. doi: 10.1212/wnl.51.2.357. [DOI] [PubMed] [Google Scholar]
- 10.Boccardi M, et al. The MRI pattern of frontal and temporal brain atrophy in fronto-temporal dementia. Neurobiol Aging. 2003;24:95–103. doi: 10.1016/s0197-4580(02)00045-3. [DOI] [PubMed] [Google Scholar]
- 11.Killiany RJ, et al. Use of structural magnetic resonance imaging to predict who will get Alzheimer's disease. Ann Neurol. 2000;47:430–439. [PubMed] [Google Scholar]
- 12.Dickerson BC, et al. MRI-derived entorhinal and hippocampal atrophy in incipient and very mild Alzheimer's disease. Neurobiol Aging. 2001;22:747–754. doi: 10.1016/s0197-4580(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 13.McDonald CR, et al. Regional rates of neocortical atrophy from normal aging to early Alzheimer's disease. Neurology. 2009;73:457–465. doi: 10.1212/WNL.0b013e3181b16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEvoy LK, et al. Alzheimer disease: Quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251:195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: Power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000;57:339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- 16.Schott JM, et al. Measuring atrophy in Alzheimer disease: A serial MRI study over 6 and 12 months. Neurology. 2005;65:119–124. doi: 10.1212/01.wnl.0000167542.89697.0f. [DOI] [PubMed] [Google Scholar]
- 17.Fox NC, et al. Effects of Abeta immunization (AN1792) on MRI measures of cerebral volume in Alzheimer disease. Neurology. 2005;64:1563–1572. doi: 10.1212/01.WNL.0000159743.08996.99. [DOI] [PubMed] [Google Scholar]
- 18.Ridha BH, et al. Volumetric MRI and cognitive measures in Alzheimer disease: Comparison of markers of progression. J Neurol. 2008;255:567–574. doi: 10.1007/s00415-008-0750-9. [DOI] [PubMed] [Google Scholar]
- 19.Jack CR, Jr, et al. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer's disease: Implications for sequence of pathological events in Alzheimer's disease. Brain. 2009;132:1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jack CR, Jr, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killiany RJ, et al. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology. 2002;58:1188–1196. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- 22.Duff K, et al. Short-term practice effects in amnestic mild cognitive impairment: Implications for diagnosis and treatment. Int Psychogeriatr. 2008;20:986–999. doi: 10.1017/S1041610208007254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciumas C, Montavont A, Ryvlin P. Magnetic resonance imaging in clinical trials. Curr Opin Neurol. 2008;21:431–436. doi: 10.1097/WCO.0b013e3283056a3c. [DOI] [PubMed] [Google Scholar]
- 24.Grundman M, et al. Brain MRI hippocampal volume and prediction of clinical status in a mild cognitive impairment trial. J Mol Neurosci. 2002;19:23–27. doi: 10.1007/s12031-002-0006-6. [DOI] [PubMed] [Google Scholar]
- 25.Thal LJ. Therapeutics and mild cognitive impairment: Current status and future directions. Alzheimer Dis Assoc Disord. 2003;17(Suppl 2):S69–S71. doi: 10.1097/00002093-200304002-00012. [DOI] [PubMed] [Google Scholar]
- 26.Weller RO, Boche D, Nicoll JA. Microvasculature changes and cerebral amyloid angiopathy in Alzheimer's disease and their potential impact on therapy. Acta Neuropathol. 2009;188:87–102. doi: 10.1007/s00401-009-0498-z. [DOI] [PubMed] [Google Scholar]
- 27.Engler H, et al. Two-year follow-up of amyloid deposition in patients with Alzheimer's disease. Brain. 2006;129:2856–2866. doi: 10.1093/brain/awl178. [DOI] [PubMed] [Google Scholar]
- 28.Sperling R. Functional MRI studies of associative encoding in normal aging, mild cognitive impairment, and Alzheimer's disease. Ann N Y Acad Sci. 2007;1097:146–155. doi: 10.1196/annals.1379.009. [DOI] [PubMed] [Google Scholar]
- 29.Wu W, Small SA. Imaging the earliest stages of Alzheimer's disease. Curr Alzheimer Res. 2006;3:529–539. doi: 10.2174/156720506779025161. [DOI] [PubMed] [Google Scholar]
- 30.Petrella JR, Prince SE, Wang L, Hellegers C, Doraiswamy PM. Prognostic value of posteromedial cortex deactivation in mild cognitive impairment. PLoS ONE. 2007;2:e1104. doi: 10.1371/journal.pone.0001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodda J, Morgan S, Walker Z. Are cholinesterase inhibitors effective in the management of the behavioral and psychological symptoms of dementia in Alzheimer's disease? A systematic review of randomized, placebo-controlled trials of donepezil, rivastigmine and galantamine. Int Psychogeriatr. 2009;21:813–824. doi: 10.1017/S1041610209990354. [DOI] [PubMed] [Google Scholar]
- 32.Persson CM, Wallin AK, Levander S, Minthon L. Changes in cognitive domains during three years in patients with Alzheimer's disease treated with donepezil. BMC Neurol. 2009;9:7–19. doi: 10.1186/1471-2377-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuno N. Donepezil in the treatment of patients with Alzheimer's disease. Expert Rev Neurother. 2009;9:591–598. doi: 10.1586/ern.09.23. [DOI] [PubMed] [Google Scholar]
- 34.Pepeu G, Giovannini MG. Cholinesterase inhibitors and beyond. Curr Alzheimer Res. 2009;6:86–96. doi: 10.2174/156720509787602861. [DOI] [PubMed] [Google Scholar]
- 35.Assal F, van der Meulen M. Pharmacological interventions in primary care: Hopes and illusions. Front Neurol Neurosci. 2009;24:54–65. doi: 10.1159/000197884. [DOI] [PubMed] [Google Scholar]
- 36.Franke AG, Lieb K, Fellgiebel A. [From symptomatic to disease modifying therapy? Recent developments in the pharmacotherapy of Alzheimer's disease] Fortschr Neurol Psychiatr. 2009;77:326–333. doi: 10.1055/s-0028-1109378. (Translated from German) (in German). [DOI] [PubMed] [Google Scholar]
- 37.Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- 38.McKhann G, et al. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 39.Jovicich J, et al. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 40.Fischl B, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 41.Fischl B, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23(Suppl 1):S69–84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Dale AM, Sereno MI. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: A linear approach. J Cognitive Neurosci. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- 43.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 44.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 45.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischl B, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 47.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 48.Christensen GE, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Trans Image Process. 1996;5:1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- 49.Miller MI, Christensen GE, Amit Y, Grenander U. Mathematical textbook of deformable neuroanatomies. Proc Natl Acad Sci USA. 1993;90:11944–11948. doi: 10.1073/pnas.90.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freeborough PA, Fox NC. Modeling brain deformations in Alzheimer disease by fluid registration of serial 3D MR images. J Comput Assist Tomogr. 1998;22:838–843. doi: 10.1097/00004728-199809000-00031. [DOI] [PubMed] [Google Scholar]
- 51.Ashburner J, Andersson JL, Friston KJ. High-dimensional image registration using symmetric priors. Neuroimage. 1999;9:619–628. doi: 10.1006/nimg.1999.0437. [DOI] [PubMed] [Google Scholar]
- 52.Holland D, Hagler DJ, Fennema-Notestine C, Dale AM. Longitudinal nonlinear registration and quantitative analysis of change in whole brain and regions of interest. Alzheimers & Dementia: The Journal of the Alzheimer's Association. 2008;4:T288. [Google Scholar]
- 53.Grandy J. Efficient Computation of Volume of Hexahedral Cells. Livermore, CA: Lawrence Livermore National Laboratory; 1997. [Google Scholar]
- 54.van der Kouwe AJ, Benner T, Dale AM. Real-time rigid body motion correction and shimming using cloverleaf navigators. Magn Reson Med. 2006;56:1019–1032. doi: 10.1002/mrm.21038. [DOI] [PubMed] [Google Scholar]
- 55.White NS, et al. PROMO–Real-time prospective motion correction in MRI using image-based tracking. Magn Reson Med. 2009 doi: 10.1002/mrm.22176. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. Hoboken, NJ: Wiley-Interscience; 2004. p. 406. Eqn. (15.2) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.