Mass per-length measurements of Alzheimer's amyloid fibrils, now reported in a recent issue of PNAS (1), contradict the conviction that such fibrils should be constructed with an integral number of molecules in their ≈4.7-Å axial repeat period, which is the signature periodicity of the amyloid cross-β structure. Solid-state NMR spectra of various pathological amyloids show that their cross-β spines are built from layers of parallel in-register β-strands (2), which requires an integral number of identically bonded molecules per layer. Schmidt et al. (1) count close to 2½ Aβ molecules per layer in a single protofilament form of the 42-residue Alzheimer's Aβ peptide and in protofilaments of two distinct double-stranded forms of the more common 40-residue Aβ peptide; their image reconstructions from electron cryomicrographs suggest similar morphology for the different protofilaments. Where is the fractional molecule?
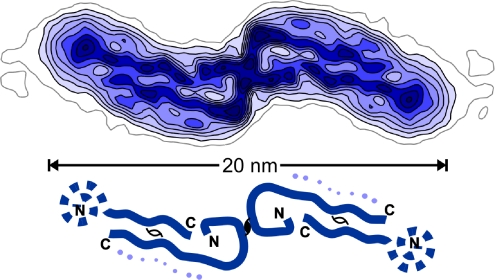
Faint density, noted last year (3) alongside the more ordered of the two β-strands per repeat identified in an ≈8-Å resolution map of the Aβ(1-40) protofilament, may mark the obscure fractional molecule. This map (Fig. 1), obtained in Grigorieff's laboratory by meticulous single-particle averaging of selected cryomicroscopy images, can be compared with the more orderly 6-Å resolution X-ray map of crystalline myoglobin, reported 50 years earlier from Kendrew's laboratory (4). Both maps revealed arrangements of secondary structural elements different from previously proposed atomic models. It took Kendrew 2 years to go from 6 Å to atomic resolution. When will atoms be seen in the orderly parts of Aβ fibrils?
Fig. 1.
Reconstructed cross-section density map at ≈8-Å resolution of the double protofilament “20-nm” Aβ(1-40) fibril (1, 3). The schematic diagram indicates the likely contour of the quasi-symmetrically paired β-strands identified in each protofilament, one traced along its 40-residue length and the other with a disordered tail. Faint density along side the long arm of the more ordered β-strand, indicated by dots in the diagram, may represent the fractional molecule per 4.7-Å repeat whose structure is very variable. N and C termini were identified in ref. 3. This figure was prepared by M. Schmidt (Brandeis University, Waltham, MA).
In this year, when Yonath, Steitz, and Ramakrishnan have been awarded the 2009 Nobel Prize for their application of protein X-ray crystallography to determine the atomic structure of the huge ribosome complex, why is the atomic structure of simple peptides in the cross-β aggregate of pathological amyloid fibrils still elusive? In 1935 Astbury and his colleagues (5) at Leeds first noted the distinctive X-ray fiber diffraction pattern later called “cross-β.” At the end of World War II, Rudall began his study of the cross-β fiber structure generated from denatured proteins, which he named in a now inaccessible report from the 1946 Leeds Symposium on Fibrous Proteins. Pauling's promulgation of his 1951 α-helix and β-sheet polypeptide models, related to Astbury's α and β X-ray fiber patterns, transformed the study of protein structures. These peptide folds were soon directly visualized in crystalline proteins but the views inside protein fibers were cruder.
Among fibrous proteins, the cross-β structures were an anomaly, mostly ill defined; however, in 1957 Parker and Rudall (6) had found a naturally occurring exemplar, the silk of the egg stalk of the green lacewing fly, that yielded a rich cross-β fiber diffraction pattern. Returning to this pattern a decade later, Rudall's student Geddes and associates (7) proposed a crystalline atomic model for the fiber core resembling Marsh, Corey, and Pauling's 1955 antiparallel β-sheet silk model (8) but with the polypeptide chain folded into eight-residue-long segments running at right angles to the fiber axis. Sutherland and her colleagues (9) have now reported the sequence of an Australian lacewing fly silk whose regularities impose the periodic antiparallel β-strand folding predicted by the crystalline fiber model.
The same year (1968) the cross-β silk model was published, Eanes and Glenner (10) reported that pathological amyloid specimens gave the characteristic cross-β X-ray diffraction pattern; and the following year Bonner, Cohen, and Skinner (11) showed that it was the ultra thin fibrils, which they imaged by electron microscopy from such specimens, that produced the cross-β pattern. Since then, fibrils that stain like amyloid and generate a cross-β diffraction pattern have come under increasingly intense scrutiny, now all called amyloid, whether or not they are associated with a disease.
How do proteins, which have better things to do, aggregate in aggravating amyloid fibrils? Waugh (12), in 1957, kinetically characterized the self-nucleated assembly process of fibrous insulin, now evident as a prototype of how the varieties of pathological amyloid fibrils are formed. Under conditions that did not allow spontaneous nucleation, he could seed growth with fragments of preformed insulin fibrils; furthermore, by changing the solvent, he could transform one self-propagating fibril strain into another. If these events had occurred in a living organism rather than in test tubes, fibrous insulin would now be called a prion. Ironically, Waugh did not accept X-ray diffraction evidence indicating a conformational switch to a cross-β structure in his assembled fibrils (13); because he could recover native insulin crystals from fibrils dissolved in alkali, he thought the critical nucleating switch involved no significant conformational change.
Self-controlled conformational switching appears to be fundamental in biological self-assembly (14). Asakura (15) showed, in a 1968 experiment similar to Waugh's, that fragments of bacterial flagella filaments in his test tubes acted like an enzyme to convert the unsociable flagellin monomer conformation into the firmly associated α-helical subunit structure of the polymer. Unlike the erratic accretion of pathological amyloid fibrils, the conformational switching in functional filament formation is governed by often-elaborate biological machinery to ensure that growth starts and stops at the right place and at the right time for purposeful actions, as exemplified by the way flagellar filament self-assembly is regulated in living bacteria.
A pathological amyloid can be defined as an adventitious cross-β fibril structure of no use to the organism producing it. Nature appears to have selected few cross-β fibril designs for purposeful use. Lacewing fly silk (6, 7, 9) and the fungal HET-s prion fibril (16) have happily functional, well-ordered cross-β architecture. Some other functional cross-β fibril structures are now under study. In contrast to the [HET-s] prion (which functions in self/nonself recognition when different fungal nuclei connect) the yeast prions [URE3] and [PSI+] are classed as diseases (17) and are interesting as amyloid models for mammalian prion diseases.
Polymorphism is a hallmark of pathological amyloid structures. Yeast prion strains are identified as self-propagating amyloid polymorphs, with, as yet, ill-defined structural differences. At the atomic level, variations in the side-chain packing of the parallel in-register β-strands of pathological amyloids (2) appear large compared with the orderly packing in segments of the purposeful HET-s β solenoid structure (16), as judged by differences in the sharpness and detail of their solid-state NMR spectra. At the level of fibril morphology, great variety has been observed in the electron microscope. All of this variability contributes unwelcome entropic stability to pathological amyloid fibrils.
Parallel in-register hydrogen bonding can faithfully propagate a particular pattern of β-strand segments arranged in a layer of a nucleating aggregate, even if there are large lateral fluctuations. Such fibrous aggregates should have an integral number of molecules per layer if all of the molecules are part of the cross-β spine. Rounding off mass per-length measurements to the nearest integer indicated that the number of units per repeat should be one for yeast prion fibrils and two or three for different Alzheimer's Aβ protofilaments.
Different Aβ protofilaments have similar structures with ≈2.5 molecules per repeat.
Carefully calibrated mass per-length measurements have shown that fibrils propagating three different strains of the yeast prion [PSI+] do not all obey the one molecule per repeat rule (18); in fact, fibrils with ≈1.0 and 1.2 prion molecules per repeat coexist in the most infectious amyloid strain. Unlike this errant polymorphism, the measurements by Schmidt et al. (1) indicate that different Aβ protofilaments have similar structures with ≈2.5 molecules per repeat. How can these contradictions with the NMR evidence for an integral number be resolved? The 8-Å resolution map of the “20-nm” Aβ fibril (Fig. 1) suggests an answer: the faint density attributed to the fractional molecule per repeat indicates large structural variations (3). This density might have escaped notice were it not for the mass measurements, and the disordered molecules signaled by this density have not yet been noticed in NMR spectra. This answer is unlikely to satisfy anyone who believes that the polymorphism of pathological amyloids must be restricted to structures in which all of the molecules are aligned in the cross-β spine.
Naturally occurring pathological amyloid fibrils have not been restrained by natural selection to obey orderly rules of behavior. Their stable, self-propagating one dimensionally periodic cross-β spine characterizes these structures. Within the spine significant lateral fluctuations can occur through transient hydration (19) and variation in side-chain interactions (2). Now it appears that the same molecules that build the spine can also stick on the sides of the regimented β-strands in disorderly conformations (1, 3, 18). Clever experimental manipulation may restrain pathological amyloid molecules to behave more like lacewing fly silk or HET-s prion fibrils, but would these still be pathological amyloids?
Much attention has been focused on the structure of Alzheimer's Aβ fibrils because they are associated with the devastating disease. However, amyloid fibrils do not appear to directly cause the cognitive problems initiating Alzheimer's dementia. Selkoe and his colleagues (20) have shown that detergent stable Aβ dimers isolated from Alzheimer's brains specifically impair synaptic plasticity and memory. A synthetic covalently linked dimer has similar toxicity and stable synaptotoxic dimers can also be isolated from solubilized amyloid plaques. Are the toxic dimers lurking in amyloid fibrils to be found in the orderly cross-β spine or among the disorderly hangers-on identified by Grigorieff and his colleagues (1, 3)?
Footnotes
The author declares no conflict of interest.
See companion article on page 19813 in issue 47 of volume 106.
References
- 1.Schmidt M, et al. Comparison of Alzheimer's Aβ(1-40) and Aβ(1-42) amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci USA. 2009;106:19813–19818. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tycko R. Molecular structure of amyloid fibrils: Insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 3.Sachse C, Fandrich M, Grigorieff N. Paired β-sheet structure of an Aβ(1-40) amyloid fibril revealed by electron microscopy. Proc Natl Acad Sci USA. 2008;105:7462–7466. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kendrew JC, et al. A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 5.Astbury WT, Dickinson S, Bailey K. The X-ray interpretation of denaturation of the seed globulins. Biochem J. 1935;29:2351–2360. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker KD, Rudall KM. The silk of the egg stalk of the green lace-wing fly: Structure of the silk of Chrysopa egg stalks. Nature. 1957;179:905–906. [Google Scholar]
- 7.Geddes AJ, Parker KD, Atkins EDT, Beighton E. “Cross-β” conformation in proteins. J Mol Biol. 1968;32:343–344. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 8.Marsh RE, Corey RB, Pauling L. The structure of Tussah silk fibroin. Acta Cystallogr. 1955;8:710–715. doi: 10.1016/0006-3002(55)90178-5. [DOI] [PubMed] [Google Scholar]
- 9.Weisman S, et al. Fifty years later: The sequence, structure, and function of lacewing cross-beta silk. J Struct Biol. 2009;168:467–475. doi: 10.1016/j.jsb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Eanes E, Glenner G. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 11.Bonnar I, Cohen AS, Skinner M. Characterization of the amyloid fibril as a cross-β protein. Proc Soc Exp Biol Med. 1969;131:1373–1375. doi: 10.3181/00379727-131-34110. [DOI] [PubMed] [Google Scholar]
- 12.Waugh DF. A mechanism for the formation of fibrils from protein molecules. J Cell Comp Physiol. 1957;49:145–164. doi: 10.1002/jcp.1030490415. [DOI] [PubMed] [Google Scholar]
- 13.Koltun WL, Waugh DF, Bear RS. An X-ray diffraction investigation of selected types of insulin fibrils. J Am Chem Soc. 1954;76:413–417. [Google Scholar]
- 14.Caspar DLD. Self-control of self-assembly. Curr Biol. 1991;1:30–32. doi: 10.1016/0960-9822(91)90119-h. [DOI] [PubMed] [Google Scholar]
- 15.Asakura S. A kinetic study of in vitro polymerization of flagellin. J Mol Biol. 1968;35:237–239. doi: 10.1016/s0022-2836(68)80051-8. [DOI] [PubMed] [Google Scholar]
- 16.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 17.Nakayashiki N, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz-Avalos R, King CY, Wall J, Simon M, Caspar DLD. Strain-specific morphologies of yeast prion amyloid fibrils. Proc Natl Acad Sci USA. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR provides evidence for mobile water molecules in β-amyloid fibrils. Proc Natl Acad Sci USA. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankar GM, et al. Amyloid-β protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]