Abstract
Mycobacterium tuberculosis (Mtb) produces a variety of methyl-branched lipids that serve important functions, including modulating the immune response during pathogenesis and contributing to a robust cell wall that is impermeable to many chemical agents. Here, we report characterization of Mtb CYP124 (Rv2266) that includes demonstration of preferential oxidation of methyl-branched lipids. Spectrophotometric titrations and analysis of reaction products indicate that CYP124 tightly binds and hydroxylates these substrates at the chemically disfavored ω-position. We also report X-ray crystal structures of the ligand-free and phytanic acid-bound protein at a resolution of 1.5 Å and 2.1 Å, respectively, which provide structural insights into a cytochrome P450 with predominant ω-hydroxylase activity. The structures of ligand-free and substrate-bound CYP124 reveal several differences induced by substrate binding, including reorganization of the I helix and closure of the active site by elements of the F, G, and D helices that bind the substrate and exclude solvent from the hydrophobic active site cavity. The observed regiospecific catalytic activity suggests roles of CYP124 in the physiological oxidation of relevant Mtb methyl-branched lipids. The enzymatic specificity and structures reported here provide a scaffold for the design and testing of specific inhibitors of CYP124.
Keywords: cytochrome P450, phytanic acid, ω-hydroxylation, X-ray structure
Mycobacterium tuberculosis (Mtb) is the causative agent of human tubercular infection that, according to the World Health Organization (1), results in more than two million deaths each year. Approximately one-third of the world's population harbors the bacterium in a latent, noninfective state, and new infections are occurring at an alarming rate. The emergence of drug-resistant and multidrug-resistant Mtb strains has made the frontline antituberculosis drugs (isoniazid, streptomycin, rifampicin, ethambutol, and pyrazinamide) less effective. Yet, no new effective antitubercular drugs have been approved since the 1960s, and there is an urgent need to identify new drug targets to help fight the spread of Mtb and quell the rising mortality rates associated with infection.
Mtb produces a rich array of lipids that, among other things, allow it to thrive in the harsh environment of the macrophage, confer resistance to a variety of chemical agents, and stimulate the host immune response during pathogenesis. A large portion of the Mtb genome encodes genes involved in lipid biosynthesis and metabolism, including 20 putative cytochrome P450 enzymes that are of interest as potential drug targets (2–5). Five of the 20 Mtb P450 enzymes (CYP51, CYP121, CYP125, CYP130, and CYP142) have been reported in purified form (2, 3, 6–14). To date, only CYP51 and CYP121 demonstrate a defined catalytic activity (14, 15). More than 10 years elapsed between the sequencing of the Mtb genome and the association of a catalytic activity with a second orphan P450 enzyme—CYP121 (15). Importantly, the recent breakthrough with CYP121 came in part from knowledge of the function of its flanking gene (15, 16). Catalytic functions are difficult to assign to the remaining Mtb P450s because they have diverged significantly from P450 enzymes of known function, and their organization within the Mtb genome provides few clues about their potential biological roles (2, 3, 17, 18).
CYP124 is found in pathogenic and nonpathogenic mycobacteria species, actinomycetes, and some proteobacteria, which suggests that it has an important catalytic activity (2). CYP124 (Rv2266) is located adjacent to a three-gene operon containing a sulfotransferase (Sft3, Rv2267c) that catalyzes the PAPS-dependent sulfation at the ω-position of menaquinone MK-9 DH-2 (19, 20). CYP128 (Rv2268c) is thought to hydroxylate the ω-position before sulfation (19). The sulfated form of the lipid, termed “S881,” is associated with the outer cell membrane of Mtb, where it acts as a negative modulator of virulence in the mouse model of infection (20). We postulated that CYP124 might have a related substrate, i.e., a lipid with repeating methyl branching due to its proximity to the Sft3 operon. We describe here the biochemical characterization of CYP124 that includes identifying a series of substrates consistent with ω-hydroxylase activity and, importantly, a marked preference for lipids containing methyl branching. We also report high-resolution structures of the ligand-free and phytanic acid-bound forms of CYP124, the first structures of a native cytochrome P450 that primarily oxidizes the chemically disfavored ω-position of a hydrocarbon chain.
Results
Spectroscopic Characterization of CYP124.
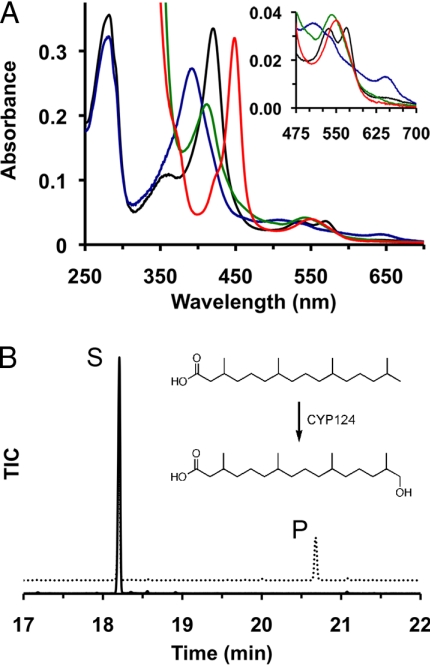
Purified CYP124 (Fig. S1 in SI Appendix) is in the ferric low-spin six-coordinated form, as judged by the UV-visible absorption spectrum that shows a large peak at 421 nm and smaller peaks at 538 and 571 nm corresponding to the β- and α-bands, respectively (Fig. 1). Ferric CYP124 underwent facile reduction to the ferrous form by treatment with sodium dithionite, which generated a spectrum with peaks at 415 and 544 nm. Ferrous CYP124 in complex with CO, however, showed the signature Soret band at 450 nm as well as a smaller peak at 555 nm. Incubating ferric CYP124 with various methyl-branched lipid substrates (see below) shifted the heme to the high-spin form, as indicated by the appearance of a dominant peak at 395 nm, small peaks at 511 and 544 nm, and a charge-transfer band at 646 nm (Fig. 1).
Fig. 1.
Biochemical characterization of CYP124. (A) UV-visible absorbance spectra of CYP124 in the ferric (black), ferrous (green), ferric high-spin (blue) and ferrous-CO (red) forms. (B) Overlaid GC chromatograms showing the total ion current (TIC) versus retention time for reactions containing Fdx, Fdr, NADPH and phytanic acid incubated for 10 min in the presence (dotted line) or absence (solid line) of CYP124. The reaction scheme depicts the CYP124- and NADPH-dependent conversion of phytanic acid into ω-hydroxy-phytanic acid. Reactions were extracted, and these compounds were derivatized with BSTFA before GC analysis, yielding peaks at 18.2 min (S) and 20.7 min (P) corresponding to the trimethylsilyl-ester of phytanic acid and TMS-ω-hydroxy phytanic TMS-ester, respectively.
CYP124, like most P450 enzymes (21), binds azole drugs through coordination to the heme iron to produce characteristic Type-II spectra with a peak between 425 and 435 nm and a broad trough at 390–410 nm that reflect azole coordination to the heme iron through a nitrogen atom (22). Dissociation constants for CYP124 with azoles (clotrimazole, KD = 2.5 ± 0.1 μM; econazole, KD = 2.1 ± 0.1 μM; miconazole, KD = 1.9 ± 0.2 μM) were obtained from the concentration-dependent production of Type-II difference spectra (Fig. S2 in SI Appendix). On the other hand, cytochrome P450 enzymes typically bind to substrates by ejecting the axial heme water ligand, which gives rise to Type-I binding spectra with a peak at 385–390 nm and a trough centered at 420 nm (22). Bifonazole bound as a Type-I ligand and displayed the highest affinity toward CYP124 (KD = 494 ± 48 nM). The heme spin-state of CYP124 was not detectably perturbed when titrated with fluconazole.
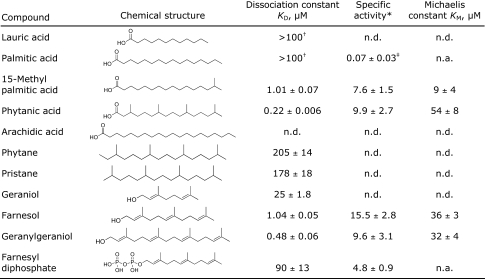
We titrated CYP124 with a variety of lipids including isoprenoids, linear and branched fatty acids, and branched alkanes (Table 1). Several of these lipids formed tight complexes with CYP124 as indicated by the resulting concentration-dependent Type-I difference spectra (Table 1 and Fig. S3 in SI Appendix). The binding affinities of CYP124 toward lauric (>100 μM), palmitic (>100 μM), 15-methyl palmitic (1.01 μM), and phytanic (0.22 μM) acid increase with methyl branching. On the other hand, the decrease in affinity toward pristane (205 μM) and phytane (178 μM) supports the argument that a polar functional group is necessary for key interactions within the active site. Moreover, the relative affinities of geraniol, farnesol, geranylgeraniol, and farnesyl diphosphate show that longer methyl-branched lipids bind tighter to the enzyme and that the type of functional group is important for binding, whereas the degree of unsaturation does not contribute significantly to binding. The proximity of the CYP124 gene to the Sft3 operon led us to also test phylloquinone and menaquinone as ligands of CYP124, but we were unable to detect binding.
Table 1.
CYP124 binds and hydroxylates methyl branchedlipids
n.d., not detected; n.a., not available, poor solubility prevented reaching saturation.
*Units of (nmol of product × min−1 × nmol of CYP124−1).
†Value indicates an estimated lower limit.
‡The reported error values are standard deviations.
CYP124 Catalyzes ω-Hydroxylation of Methyl-Branched Lipids.
Based on the Type-I spin shifts and high-affinity binding toward methyl-branched lipids, CYP124 was incubated with spinach ferredoxin, spinach ferredoxin-NADP+-reductase, various lipids, and NADPH, and the reaction products were compared by GC-MS with those obtained in control reactions in which either CYP124 or NADPH was omitted. New signals appeared in the GC chromatograms that depended on the presence of both NADPH and CYP124 in the reaction mixture (Fig. 1 and Fig. S4 in SI Appendix). In each case, the trimethylsilylated (TMS) metabolites eluted from the GC at a higher temperature than the respective substrates, which is consistent with the presence of a TMS-protected alcohol in each metabolite. The reaction products, identified by mass spectrometry using characteristic molecular ion and fragmentation patterns, confirm that CYP124 oxidation occurs primarily at the ω-position (Fig. S4 in SI Appendix). CYP124 converted phytanic (Fig. 1) and 15-methyl palmitic acid (Fig. S4 in SI Appendix) each into a single product. and their molecular ions at m/z = 472 and 430, respectively, confirm the presence of an additional TMS-protected alcohol in each. The fragment ion at m/z = 103 corresponds to the loss of -CH2OSi(CH3)3 from the ω-position of a saturated branched-lipid with a TMS-protected hydroxyl group (23), and we observed such fragments with the new phytanic and 15-methyl palmitic acid metabolites (Fig. S4 in SI Appendix).
Isoprenoids (farnesol, farnesyl diphosphate, geranylgeraniol) were also efficiently oxidized by CYP124 into the respective ω-hydroxylated products (Fig. S4 in SI Appendix). These assignments were based on comparisons with the product formed in incubations of farnesol with CYP2E1, which catalyzes ω-hydroxylation of farnesol (24). Both the retention times and mass spectra of the CYP124 and CYP2E1 metabolites matched in our assays. The product from farnesyl diphosphate was treated with alkaline phosphatase before GC-MS analysis to release the farnesol structure, which then matched the mass and retention pattern seen for the metabolite of farnesol itself. The geranylgeraniol metabolites show the mass increase and fragmentation pattern characteristic of terminal oxidation adjacent to an allylic carbon (Fig. S4 in SI Appendix). Incubation of CYP124 with geranylgeraniol also produced geranylgeranic acid and ω-hydroxylated geranylgeranic acid (Fig. S4 in SI Appendix), indicating that two catalytically competent binding modes are possible with this substrate in the active site of CYP124.
Palmitic acid binds with a sharply decreased affinity and is oxidized >200-fold less effectively than analogous methyl-branched lipids (Table 1). Moreover, CYP124 oxidized the ω-1, ω-2, and ω-position of palmitic acid (Fig. S4 in SI Appendix), whereas we did not observe ω-1 metabolites with any of the methyl-branched lipids (Fig. S4 in SI Appendix), nor were any oxidized metabolites detected from incubating CYP124 with geraniol, lauric acid, phytane, pristane, phylloquinone (vitamin K1), menaquinone (vitamin K2), or bifonazole. The major reaction we observed with CYP124 is ω-oxidation of methyl-branched lipids.
Overall Structure of CYP124.
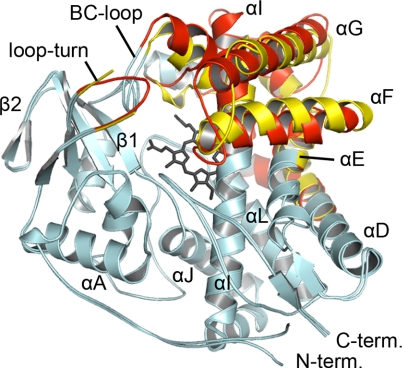
The structures of the ligand-free (1.5 Å) and substrate-bound (2.1 Å) CYP124 are similar with an RMSD value for Cα carbons of ≈0.9 Å when superimposed. The structural features commonly involved in heme binding, substrate recognition, and formation of the active site cavity are readily distinguished (Fig. 2), and are comparable with other P450 enzymes for which structures are known (7, 11, 13, 25). The “floor” of the active-site cavity is formed by heme that is positioned between the I and L helices, with the proximal Cys-379 thiolate anchoring the cofactor to the L helix. One half of the active site cavity is composed of the F, G, and C helices as well as residues from the BC and FG loop regions. The other half of the active site cavity is roughly defined by the C-terminal loop (Leu-413–His-418) that trails the L helix, as well as four antiparallel strands comprising β-sheet 1.
Fig. 2.
Superimposed structures of ligand-free and phytanic acid-bound CYP124 are shown with the common P450 secondary structure elements labeled. The protein backbone is depicted by colored ribbon and the heme by black sticks. Amino acid side chains and phytanic acid are omitted for clarity. Shown in light blue are those regions of CYP124 that do not significantly change upon substrate binding. The yellow (ligand-free) and red (phytanic acid-bound) structure elements undergo conformational change upon substrate binding.
Substrate-Induced Conformational Changes.
There are important differences between the ligand-free and phytanic acid-bound CYP124 structures. Substrate binding induces a reorganization of secondary structure elements, as shown in Fig. 2, which are largely confined to the regions typically involved in forming the active site and substrate recognition region (25). The BC-loop (Thr-100–Phe-107) moves ≈5.3 Å toward the G helix, which positions Leu-103 and Phe-107 to interact with the C-18 methyl branch of phytanic acid. The D and C helices move in concert toward the G helix, with N-terminal portions of the D helix (Lys-132–Ala 139) bending toward the active site and the proximal side of the heme. The G helix (Phe-209–Val-231) shifts 4.9 Å toward a newly ordered turn between the first and second strands of β-sheet 1, positioned across the active-site cavity that forms a lid over the active site. Importantly, Phe-209 and Phe-212 face into the active site along the hydrophobic backbone of phytanic acid. Residues of the FG loop (Gly-199–Asp-208) and F helix (Lys-184–Gly-199) are positioned toward the active site in the phytanic acid-bound structure. Substrate binding also causes the EF loop (Met-180–Lys-184) to move inward, making the structure more closed by ≈2.9–3.1 Å. Three additional regions, the GI loop, H helix, and HI loop, all relocate in unison upon substrate binding, and these movements accompany the relocation of the G helix as it closes the active site. In the ligand-free CYP124 structure, α-helical structure is missing from the middle of the I helix, whereas in the substrate-bound form, the I helix is continuous.
Structure of the CYP124 Active Site with Phytanic Acid Bound.
The active site of ligand-free CYP124 shows the strictly conserved Cys-379 coordinated to the heme iron (2.3 Å), water128 (2.2 Å) as the distal ligand, and the iron atom in the plane of the porphyrin ring. Many water molecules are ordered in the ligand-free active site consistent with the more open conformation favored in the absence of substrate. Cys-379 also serves as the proximal ligand (2.4 Å) with phytanic acid bound in the active site, but the iron atom moves out of the plane of the porphyrin ring toward the proximal thiolate. Importantly the terminal methyl group of phytanic acid is 3.8 Å away from the high-spin heme iron and poised for ω-oxidation (Fig. 3).
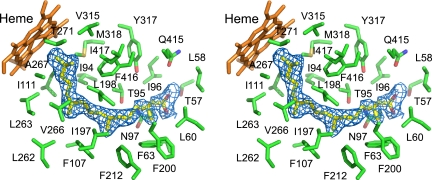
Fig. 3.
Expanded stereoscopic view of CYP124 with phytanic acid (yellow) bound in the active site. Residues within 5 Å from phytanic acid (green) are shown. The protein backbone is omitted for clarity, and the heme cofactor is shown in orange. Phytanic acid was modeled into the electron density depicted as blue wire mesh.
Residues of the active site provide hydrophobic and polar interactions to bind the methyl-branched lipid chain and carboxylic acid group, respectively. Fig. 3 shows an expanded view of phytanic acid bound in the active site of CYP124 and highlights key residues within 5 Å of the substrate. The reorganization of the BC loop and C helix provides many hydrophobic interactions to the substrate from Ile-94, Leu-103, Phe-107, and Ile-111 as well as polar interactions with the hydroxyl of Thr-95 and carboxamide of Asn-97. The newly ordered β-sheet 1 turn forms a “lid” over the active site and also binds the substrate through Ile-58, Leu-60, and Phe-63. The reorganization of the FG loop region positions several hydrophobic residues (Ile-197, Leu-198, and Phe-200) to bind the hydrophobic lipid. The movement of the G helix shifts Phe-212 to bind phytanic acid. Six other residues are in the 5- to 6-Å sphere of the substrate and stabilize substrate binding, including Val-315, Tyr-317, Met-318, Phe-416, and Ile-417. Hydrophobic residues of the I helix, including Ile-262, Leu-263, Val-266, and Ala-267, also bind phytanic acid within the active site.
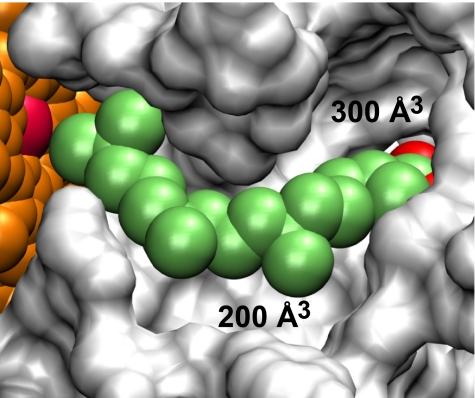
The volume of the active-site cavity with phytanic acid removed is 1,370 Å3. Two pockets of this cavity, 300 Å3 and 200 Å3, are not occupied by phytanic acid (Fig. 4), suggesting additional space available for binding the true substrate. Hydrophobic residues Leu-29, Trp-32, Ile-58, Leu-60, Leu-198, Ile-413, Ile-417, and Phe-200 and polar residues Ser-414 and Gln-415 constitute the borders of the larger 300 Å3 pocket toward the carboxylic end of the phytanic acid molecule.
Fig. 4.
Active site of CYP124 in which the surface of the cavity is shown in gray, heme is in orange with the Fe atom in magenta, and phytanic acid is highlighted in green with the carboxyl group in red. Unoccupied pockets are labeled by their volumes.
Discussion
Assigning in vivo functions or demonstrating in vitro catalytic activity with the P450 enzymes of Mtb is not trivial (2, 3, 5, 14, 15). The location of CYP124 within the genome of Mtb next to an operon coding for an important sulfotransferase (19) led us to initially explore methyl-branched lipids as substrates of CYP124 to determine whether it might have a related function. The tight binding affinity and ω-hydroxylase activity of CYP124 toward a series of lipids indicate that the enzyme preferentially metabolizes methyl-branched lipids and oxidizes the chemically disfavored ω-position. Cyp124 is among a group of differentially expressed Mtb genes during infection in the mouse lung (26), and the CYP124 gene is also conserved in many actinomycetes and proteobacteria, which suggests that the enzyme catalyzes an important reaction. Work is ongoing to more precisely address the in vivo function of CYP124 that includes using gene knockouts and lipidomics.
Regardless of whether CYP124 is involved in biosynthesis of the S881 sulfolipid, it clearly has an activity toward methyl-branched lipids, and Mtb is replete with such lipids that are involved in a variety of important and cryptic functions. The Mtb isoprenoid biosynthetic pathway is essential (27) and generates important respiratory menaquinones (28), sulfated forms of which negatively regulate the immune response in mice infected with Mtb (19, 20). In fact, CYP124 oxidizes farnesyl diphosphate (FPP), a precursor of longer-chain isoprenoids that are found in Mtb (29, 30); however, at this point, we cannot link CYP124 with in vivo activity toward FPP. Decaprenyl phosphates are essential lipid and sugar carriers in the assembly of the mycolic acid–peptidoglycan–arabinogalactan (mAGP) complex of the cell wall (31). Mtb also presents several lipids on its cell surface that have methyl branching, including sulfolipid-1 (SL-1), di- and polyacyl-treheloses (DAT and PAT), mannosyl-β-1-phosphomycoketide (MPM), phthiocerol dimycocerosate (PDIM), and mycolic acids (32–34). In addition, Mtb, like other actinomycetes, is capable of deriving energy from carbon sources with methyl branching (35, 36). In mammals, ω-oxidation of branched lipids, such as phytanic acid, shifts the register of the β-substituted carbon chain so that β-oxidation can occur (23). CYP124 could conceivably function in oxidative degradation or balancing of Mtb lipid and carbon pools.
P450 oxidation of hydrocarbons at the ω-position is more difficult than at the internal positions. Experimental and theoretical studies of the P450 reaction cycle show that H-atom abstraction is the highest energy barrier in the pathway, and differences in chemical reactivity between positions are generally a function of the relative strengths of the C–H bonds that undergo homolytic cleavage, and thus of the relative stabilities of the radical intermediates resulting from homolysis (37–40). The catalytic core of P450 enzymes is highly conserved (41), and the oxidizing species (Compound I, [FeIV = O]+·) should also be equivalent for P450 enzymes with different regiospecificities. The parameters that govern the hydroxylation of thermodynamically disfavored sites have been explored (42–54). The model resulting from these studies suggests that P450 ω-hydroxylases restrict substrate conformation within the active site to position the terminal methyl group near the heme while preventing more reactive positions on the substrate from encountering the heme.
The structures presented here are in accord with this model, and provide important insight into how CYP124 catalyzes ω-hydroxylation. The terminal methyl group of phytanic acid is positioned 3.8 Å away from the heme, consistent with the ω-regiospecificity we observed. The active site with phytanic acid bound in it is both hydrophobic and conformationally restrictive near the heme, consistent with a single phytanic acid metabolite. Other P450 ω-hydroxylases typically also have a patch of polar amino acid side-chain residues that bind the substrate “tail” (55). CYP124 substrates also require anchoring in the active site by a polar functional group (hydroxy or carboxylic acid) and either removing it or replacing it by a large diphosphate moiety results in a large decrease in binding affinity. The CYP124 structure reveals that the carboxylate group of phytanic acid is bound in the active site near a patch of amino acid side chains with polar functional groups; however, the spacing suggests that the native substrate has a slightly longer lipid chain. This polar region, although important for initial binding, does not define the regiospecificity of CYP124. Isoprenoids (farnesol, farnesyl diphosphate, and geranylgeraniol) differing in chain length, binding affinity, and specific activity were all converted into ω-hydroxylated products. Geranylgeraniol was oxidized at the ω-position and further oxidized to ω-hydroxy geranylgeranic acid as well as directly to geranylgeranic acid (Fig. S4 in SI Appendix).
The CYP124 crystal structure with phytanic acid bound suggests that regiospecific hydroxylation is the result of many weak interactions that tightly bind and position the substrate within the geometry of the active site. Indeed, the side chains of Val-315 and Ile-417 are more than 4 Å away from the terminal branch, creating enough space for ambiguous positioning of the terminal carbon of the nonbranched lipids. Lack of methyl branching in palmitic acid lends flexibility such that it can sample conformations amenable for ω-1 and ω-2 oxidation. The chemically more preferable positions become oxidized, but ω-oxidation is still observed. Introducing a single methyl branch, as in 15-methyl palmitic acid, results in ≈100-fold higher specific activity and at least the same increase in binding affinity. Most importantly, ω-1 hydroxylation is now excluded in favor of the ω-hydroxylation. Binding of one of the two methyls of the branched hydrocarbon terminus in a specific active-site cavity apparently locks the other methyl in the position required for ω-hydroxylation. It is likely that in unbranched hydrocarbons, the terminal methyl is bound in the same cavity, making substrate oxidation inefficient and, when it occurs, nonregiospecific.
The results presented here are of a cytochrome P450 of the CYP124 family, and, importantly, this ortholog is one of 20 P450 enzymes that are potential drug targets in Mtb. Our demonstration that CYP124 has ω-hydroxylase activity toward methyl-branched lipids provides important clues about its in vivo function and suggests physiologically relevant substrates for the enzyme. The substrate specificity defined here, together with the high-resolution structures, provides a scaffold for design and testing of specific CYP124 inhibitors.
Materials and Methods
CYP124 Cloning, Overexpression, and Purification.
CYP124 (Rv2266) was amplified from Mtb H37Rv genomic DNA, as described in ref. 7, by using a sense-oligo (TTT TTT CAT ATG CAT CAC CAT CAC CAT CAC GGC CTC AAC ACG GCG ATC) to introduce a unique NdeI restriction site and a six-histidine affinity tag. The antisense DNA oligo (AAA AAA AAG CTT ATT ATT AGG ACC ACG TAA CTG GCA GCG TC) added a unique HindIII restriction site. The CYP124 gene was verified and then ligated into the pCW expression vector (6, 7), and the enzyme was overexpressed in Escherichia coli DH5α as described in ref. 7. CYP124 was purified by using Ni-NTA2+ followed by ion-exchange chromatography steps as described in ref. 7.
Spectrophotometry.
UV-visible spectra were recorded on a Cary dual beam spectrophotometer using 1-cm path length quartz cuvettes. The CYP124 holoenzyme was quantified by using the Soret band by subtracting the signal of the ferrous-deoxy form of CYP124 from the ferrous CO-bound enzyme generated with 1 mM sodium dithionite. An extinction coefficient (450–490 nm) of 91 mM−1cm−1 was used (56). CYP124 ligands were assessed by using a dual-beam spectrophotometer, and difference spectra were recorded at 25 °C between 350 and 500 nm. Compounds that induced typical Type-I or Type-II difference spectra were titrated to establish their affinities toward the enzyme (7).
CYP124 Catalytic Assays.
Catalytic assays were carried out at 25 °C in glass tubes in a volume of 0.5 mL. CYP124 (120 pmol) was preincubated with substrates in 50 mM potassium phosphate (pH 7.4) containing spinach ferredoxin and spinach ferredoxin-NADP+ reductase. All reactions contained 10 μg/mL catalase and an NADPH-regenerating system consisting of glucose 6-phosphate dehydrogenase (2.4 units) and 1 mM glucose-6-phosphate. Reactions were started by adding 0.5 mM NADP+, control reactions contained all of the components except CYP124 or NAPDH, and reactions were quenched by adding 1 mL of 1 M HCl, 5 ml methyl tert-butyl ether (MTBE), mixed, and centrifuged (4 °C at 1,500 × g for 15 min). The organic phases were evaporated and converted to the respective trimethylsilyl (TMS) derivatives by resuspension in 50 μL of BSTFA (Pierce) for 2 h at 25 °C. Reaction mixtures were analyzed by using an HP5790 gas chromatography system fitted with a DB5-MS column (30 m × 0.25 mm × 0.25 μm) as described in ref. 57. Separation was achieved by using a column temperature of 70 °C for 1 min, increased by 10 °C/min up to 300 °C, and finally held at 300 °C for 1 min. Specific activities were determined at saturating concentrations of substrate, typically 5 × Km.
Crystallization, Data Collection, and Crystal Structure Determination.
The initial screening of crystallization conditions was performed by using commercial high-throughput screening kits (Hampton Research), a nanoliter drop-setting Mosquito robot (TTP; Labtech) operating with 96-well plates, and a hanging-drop crystallization protocol. Optimization of conditions was carried out manually in 24-well plates. The protein was from 0.5 mM frozen stock in 50 mM potassium phosphate (pH 7.5), and before crystallization, the protein was diluted to 0.1–0.2 mM by mixing with 10 mM Tris·HCl (pH 7.5) alone or supplemented with 2 mM phytanic acid. Crystals of the ligand-free CYP124 grew from 1% PEG MME 2000, 0.1 M Na cacodylate (pH 6.8), and 0.9 M succinic acid. Crystals of the CYP124-phytanic acid complex grew from 21% PEG 4000, 0.1 M Bis-Tris (pH 5.5) and 0.2 M Ca acetate. Before data collection, the crystals were cryoprotected by plunging them into a drop of reservoir solution supplemented with 20% glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected at 100–110 K at beamline 8.3.1, Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, CA. Data indexing, integration, and scaling were conducted by using MOSFLM (58), HKL2000 (59), and ELVES (60) software suites. Crystal structures were determined by molecular replacement using the atomic coordinates of Mtb CYP125 as a search model (Table 1).
Supplementary Material
Acknowledgments.
We thank Andrew Munro and David Leys for providing the CYP125 coordinates before publication, Sylvie Kandel for CYP2E1, Hugues Ouellet for helpful advice, Chiung-Kuang Chen and the staff members of beamline 8.3.1, James Holton, George Meigs, and Jane Tanamachi (the Advanced Light Source at Lawrence Berkeley National Laboratory) for assistance with data collection. This work was supported by National Institutes of Health Grants grants RO1AI07824 (to P.R.O.d.M.) and RO1GM078553 (to L.M.P.). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2WM4 and 2WM5).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907398106/DCSupplemental.
References
- 1.World Health Organization. World Health Organization Facts Sheets on Tuberculosis. 2009 [Google Scholar]
- 2.Ouellet H, Johnston JB, Ortiz de Montellano PR. The Mycobacterium tuberculosis cytochrome P450 system. Arch Biochem Biophys. doi: 10.1016/j.abb.2009.07.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLean KJ, Munro AW. Structural biology and biochemistry of cytochrome P450 systems in Mycobacterium tuberculosis. Drug Metab Dispos. 2008;40:427–446. doi: 10.1080/03602530802186389. [DOI] [PubMed] [Google Scholar]
- 4.Aguero F, et al. Genomic-scale prioritization of drug targets: The TDR Targets database. Nat Rev Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLean KJ, Dunford AJ, Neeli R, Driscoll MD, Munro AW. Structure, function and drug targeting in Mycobacterium tuberculosis cytochrome P450 systems. Arch Biochem Biophys. 2007;464:228–240. doi: 10.1016/j.abb.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Ouellet H, Lang J, Couture M, Ortiz de Montellano PR. Reaction of Mycobacterium tuberculosis cytochrome P450 enzymes with nitric oxide. Biochemistry. 2009;48:863–872. doi: 10.1021/bi801595t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouellet H, Podust LM, Ortiz de Montellano PR. Mycobacterium tuberculosis CYP130. Crystal structure, biophysical characterization, and interactions with antifungal azole drugs. J Biol Chem. 2008;283:5069–5080. doi: 10.1074/jbc.M708734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLean KJ, et al. Characterization of active site structure in CYP121. A cytochrome P450 essential for viability of Mycobacterium tuberculosis H37Rv. J Biol Chem. 2008;283:33406–33416. doi: 10.1074/jbc.M802115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seward HE, Roujeinikova A, McLean KJ, Munro AW, Leys D. Crystal structure of the Mycobacterium tuberculosis P450 CYP121-fluconazole complex reveals new azole drug-P450 binding mode. J Biol Chem. 2006;281:39437–39443. doi: 10.1074/jbc.M607665200. [DOI] [PubMed] [Google Scholar]
- 10.McLean KJ, et al. Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin, and their interactions. Biochemistry. 2006;45:8427–8443. doi: 10.1021/bi0601609. [DOI] [PubMed] [Google Scholar]
- 11.Leys D, et al. Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450. J Biol Chem. 2003;278:5141–5147. doi: 10.1074/jbc.M209928200. [DOI] [PubMed] [Google Scholar]
- 12.McLean KJ, et al. Expression, purification and spectroscopic characterization of the cytochrome P450 CYP121 from Mycobacterium tuberculosis. J Inorg Biochem. 2002;91:527–541. doi: 10.1016/s0162-0134(02)00479-8. [DOI] [PubMed] [Google Scholar]
- 13.Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14alpha-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc Natl Acad Sci USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellamine A, Mangla AT, Nes WD, Waterman MR. Characterization and catalytic properties of the sterol 14-alpha-demethylase from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1999;96:8937–8942. doi: 10.1073/pnas.96.16.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belin P, et al. Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2009;106:7426–7431. doi: 10.1073/pnas.0812191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 17.McLean KJ, et al. The preponderance of P450s in the Mycobacterium tuberculosis genome. Trends Microbiol. 2006;14:220–228. doi: 10.1016/j.tim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Kelly SL, Kelly DE, Jackson CJ, Warrilow AGS, Lamb DC. The diversity and importance of microbial cytochromes P450. In: Ortiz de Montellano PR, editor. Cytochrome P450. Structure, Mechanism, and Biochemistry. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 585–617. [Google Scholar]
- 19.Holsclaw CM, et al. Structural characterization of a novel sulfated menaquinone produced by Sft3 from Mycobacterium tuberculosis. ACS Chem Biol. 2008;3:619–624. doi: 10.1021/cb800145r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mougous JD, et al. A sulfated metabolite produced by sft3 negatively regulates the virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:4258–4263. doi: 10.1073/pnas.0510861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balding PR, et al. How do azoles inhibit cytochrome P450 enzymes? A density functional study. J Phys Chem. 2008;112:12911–12918. doi: 10.1021/jp802087w. [DOI] [PubMed] [Google Scholar]
- 22.Jefcoate CR. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 1978;52:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- 23.Xu F, Ng VY, Kroetz DL, Ortiz de Montellano PR. CYP4 isoform specificity in the omega-hydroxylation of phytanic acid, a potential route to elimination of the causative agent of Refsum's disease. J Pharm Exp Ther. 2006;318:835–839. doi: 10.1124/jpet.106.104976. [DOI] [PubMed] [Google Scholar]
- 24.DeBarber AE, Bleyle LA, Roullet J-BO, Koop DR. Omega-hydroxylation of farnesol by mammalian cytochromes P450. Biochim Biophys Acta. 2004;1682:18–27. doi: 10.1016/j.bbalip.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Poulos TL, Johnson EF. Structures of cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450. Structure, Mechanism, and Biochemistry. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 87–114. [Google Scholar]
- 26.Dubnau E, Chan J, Mohan VP, Smith I. Response of Mycobacterium tuberculosis to growth in the mouse lung. Infect Immun. 2005;73:3754–3757. doi: 10.1128/IAI.73.6.3754-3757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohdich F, Bacher A, Eisenreich W. Isoprenoid biosynthetic pathways as anti-infective drug targets. Biochem Soc Trans. 2005;33:785–791. doi: 10.1042/BST0330785. [DOI] [PubMed] [Google Scholar]
- 28.Dhiman RK, et al. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol Microbiol. 2009;72:85–97. doi: 10.1111/j.1365-2958.2009.06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulbach MC, et al. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J Biol Chem. 2001;276:11624–11630. doi: 10.1074/jbc.M007168200. [DOI] [PubMed] [Google Scholar]
- 30.Crick DC, et al. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol. 2000;182:5771–5778. doi: 10.1128/jb.182.20.5771-5778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis. 2003;83:91–97. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 32.Gokhale RS, Saxena P, Chopra T, Mohanty D. Versatile polyketide machinery for the biosynthesis of complex mycobacterial lipids. Nat Prod Rep. 2007;24:267–277. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- 33.Minnikin DE, Kremer L, Dover LG, Besra GS. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem Biol. 2002;9:545–553. doi: 10.1016/s1074-5521(02)00142-4. [DOI] [PubMed] [Google Scholar]
- 34.Kumar P, et al. PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis virulence factor Sulfolipid-1. Proc Natl Acad Sci USA. 2007;104:11221–11226. doi: 10.1073/pnas.0611649104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Geize R, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Visser SP, Kumar D, Cohen S, Shacham R, Shaik S. A predictive pattern of computed barriers for C-H hydroxylation by Compound I of cytochrome P450. J Am Chem Soc. 2004;126:8362–8363. doi: 10.1021/ja048528h. [DOI] [PubMed] [Google Scholar]
- 38.Korzekwa KR, Jones JP, Gillette JR. Theoretical studies on cytochrome P-450 mediated hydroxylation: A predictive model for hydrogen atom abstractions. J Am Chem Soc. 1990;112:7042–7046. [Google Scholar]
- 39.Olsen L, Rydberg P, Rod TH, Ryde U. Prediction of activation energies for hydrogen abstraction by cytochrome P450. J Med Chem. 2006;49:6489–6499. doi: 10.1021/jm060551l. [DOI] [PubMed] [Google Scholar]
- 40.Feenstra KA, Starikov EB, Urlacher VB, Commandeur JNM, Vermeulen NPE. Combining substrate dynamics, binding statistics, and energy barriers to rationalize regioselective hydroxylation of octane and lauric acid by CYP102A1 and mutants. Prot Sci. 2007;16:420–431. doi: 10.1110/ps.062224407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortiz de Montellano PR, editor. Cytochrome P450. Structure, Mechanism, and Biochemistry. 3rd Ed. New York: Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- 42.CaJacob CA, Chan WK, Shephard E, Ortiz de Montellano PR. The catalytic site of rat hepatic lauric acid omega-hydroxylase. Protein versus prosthetic heme alkylation in the omega-hydroxylation of acetylenic fatty acids. J Biol Chem. 1988;263:18640–18649. [PubMed] [Google Scholar]
- 43.Alterman MA, Hanzlik RP. Hydroxylation of fatty acids by microsomal and reconstituted cytochrome P450 2B1. FEBS Lett. 2002;512:319–322. doi: 10.1016/s0014-5793(02)02260-3. [DOI] [PubMed] [Google Scholar]
- 44.He X, Cryle MJ, De Voss JJ, Ortiz de Montellano PR. Calibration of the channel that determines the omega-hydroxylation regiospecificity of cytochrome P4504A1. J Biol Chem. 2005;280:22697–22706. doi: 10.1074/jbc.M502632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bambal RB, Hanzlik RP. Active site structure and substrate specificity of cytochrome P450 4A1: Steric control of ligand approach perpendicular to heme plane. Biochem Biophys Res Commun. 1996;219:445–449. doi: 10.1006/bbrc.1996.0253. [DOI] [PubMed] [Google Scholar]
- 46.Fisher MB, Zheng Y-M, Rettie AE. Positional specificity of rabbit CYP4B1 for omega-hydroxylation of short-medium chain fatty acids and hydrocarbons. Biochem Biophys Res Commun. 1998;248:352–355. doi: 10.1006/bbrc.1998.8842. [DOI] [PubMed] [Google Scholar]
- 47.Cook BR, Reinert TJ, Suslick KS. Shape selective alkane hydroxylation by metalloporphyrin. J Am Chem Soc. 1986;108:7281–7286. [Google Scholar]
- 48.White RE, McCarthy M-B, Egeberg KD, Sligar SG. Regioselectivity in the cytochromes P-450: Control by protein constraints and by chemical reactivities. Arch Biochem Biophys. 1984;228:493–502. doi: 10.1016/0003-9861(84)90015-8. [DOI] [PubMed] [Google Scholar]
- 49.Bambal RB, Hanzlik RP. Effects of steric bulk and conformational rigidity on fatty acid omega hydroxylation by a cytochrome P450 4A1 fusion protein. Arch Biochem Biophys. 1996;334:59–66. doi: 10.1006/abbi.1996.0429. [DOI] [PubMed] [Google Scholar]
- 50.Oliver CF, et al. A single mutation in cytochrome P450 BM3 changes substrate orientation in a catalytic intermediate and the regiospecificity of hydroxylation. Biochemistry. 1997;36:1567–1572. doi: 10.1021/bi962826c. [DOI] [PubMed] [Google Scholar]
- 51.Adas F, et al. Requirement for omega and (omega-1)-hydroxylations of fatty acids by human cytochromes P450 2E1 and 4A11. J Lipid Res. 1999;40:1990–1997. [PubMed] [Google Scholar]
- 52.Hoch U, Falck JR, Ortiz de Montellano PR. Molecular basis for the omega-regiospecificity of the CYP4A2 and CYP4A3 fatty acid hydroxylases. J Biol Chem. 2000;275:26952–26958. doi: 10.1074/jbc.M004841200. [DOI] [PubMed] [Google Scholar]
- 53.Rock DA, Perkins BNS, Wahlstrom J, Jones JP. A method for determining two substrates binding in the same active site of cytochrome P450BM3: An explanation of high energy product formation. Arch Biochem Biophys. 2003;416:9–16. doi: 10.1016/s0003-9861(03)00228-5. [DOI] [PubMed] [Google Scholar]
- 54.Hoch U, Zhang Z, Kroetz DL, Ortiz de Montellano PR. Structural determination of the substrate specificities and regioselectivities of the rat and human fatty acid omega-hydroxylases. Arch Biochem Biophys. 2000;373(1):63–71. doi: 10.1006/abbi.1999.1504. [DOI] [PubMed] [Google Scholar]
- 55.Coon MJ. Omega oxygenases: nonheme-iron enzymes and P450 cytochromes. Biochem Biophys Res Commun. 2005;338:378–385. doi: 10.1016/j.bbrc.2005.08.169. [DOI] [PubMed] [Google Scholar]
- 56.Omura T, Sata R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1965;239:2370–2378. [PubMed] [Google Scholar]
- 57.Jiang Y, Ortiz de Montellano PR. Cooperative effects on radical recombination in CYP3A4-catalyzed oxidation of the radical clock beta-thujone. ChemBioChem. 2009;10:650–653. doi: 10.1002/cbic.200800772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EAMCB Newslett. Protein Crystallogr. 1992:26. [Google Scholar]
- 59.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 60.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci USA. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.