Abstract
Specific sets of brain-expressed genes, such as aerobic energy metabolism genes, evolved adaptively in the ancestry of humans and may have evolved adaptively in the ancestry of other large-brained mammals. The recent addition of genomes from two afrotherians (elephant and tenrec) to the expanding set of publically available sequenced mammalian genomes provided an opportunity to test this hypothesis. Elephants resemble humans by having large brains and long life spans; tenrecs, in contrast, have small brains and short life spans. Thus, we investigated whether the phylogenomic patterns of adaptive evolution are more similar between elephant and human than between either elephant and tenrec lineages or human and mouse lineages, and whether aerobic energy metabolism genes are especially well represented in the elephant and human patterns. Our analyses encompassed ≈6,000 genes in each of these lineages with each gene yielding extensive coding sequence matches in interordinal comparisons. Each gene's nonsynonymous and synonymous nucleotide substitution rates and dN/dS ratios were determined. Then, from gene ontology information on genes with the higher dN/dS ratios, we identified the more prevalent sets of genes that belong to specific functional categories and that evolved adaptively. Elephant and human lineages showed much slower nucleotide substitution rates than tenrec and mouse lineages but more adaptively evolved genes. In correlation with absolute brain size and brain oxygen consumption being largest in elephants and next largest in humans, adaptively evolved aerobic energy metabolism genes were most evident in the elephant lineage and next most evident in the human lineage.
Keywords: Afrotheria, brain energy metabolism, Euarchontoglires, mitochondria, nucleotide substitution rates
Thousands of genes are likely to code for proteins that are important for the brain's function and structural development. A subset of these genes has coding sequences with evidence of accelerated evolution in the anthropoid primate ancestry of humans (1–10). This accelerated and likely adaptive evolution appears to correlate with the observation that both absolute and relative brain size increased much further in the anthropoid ancestry of humans than in non-anthropoid primate lineages (9). Among these genes are not only those known primarily for their importance in nervous system biology (3, 10), but also many that drive aerobic energy production (2, 6), with the latter genes playing their own crucial role in the function of neurons as attested to by the high amounts of metabolic energy that anthropoid brains consume (11). Because large brains arose convergently in different mammalian lineages (12), it is feasible to explore whether putative brain-important genes that evolved at accelerated rates during humankind's ancestry also evolved at accelerated rates in non-primate lineages where brain mass increased compared with related lineages with relatively smaller brain mass. A promising opportunity for such an exploration is provided by elephant evolution within the superordinal clade Afrotheria (13).
Among non-primate mammals now represented by genome sequences, the African savannah elephant (Loxodonta africana) and the lesser hedgehog tenrec (Echinops telfairi) differ greatly in absolute brain size but, as members of the clade Afrotheria, are phylogenetically closer to each other than to the human species. Elephants resemble humans in a number of ways, not least by having massive brains, social bonds that appear to be empathetic, long gestations, high intelligence, offspring that require an extended period of dependent care, and long life spans (Tables 1 and S1). In contrast, tenrecs have the features of insectivore-grade mammals, including small brains (≈2.6 g; see ref. 14) with a relatively small neocortex (14, 15) and short life spans.
Table 1.
Life history and phenotype variables among study taxa
| Species name | Order | Gestation, days | Age of sexual maturity, months | Life span, yr | Litter size | Brain weight, g | Body weight, g | EQ |
|---|---|---|---|---|---|---|---|---|
| Homo sapiens | Primates | 270 | 198 | 75 | 1 | 1300 | 44,000 | 8.7 |
| Mus musculus | Rodentia | 19–21 | 1.5 | 2 | 3–12 | 0.43 | 21 | 0.5 |
| Loxodonta africana | Proboscidea | 660 | 330–660 | 50–70 | 1 | 4420 | 3,505,000 | 1.6 |
| Echinops telfairi | Afrosoricida | 42–49 | 8 | 13 | 1–10 | 0.62 | 88 | 0.3 |
| Bos taurus | Cetartiodactyla | 277–290 | 18 | 20 | 1 | 473.5 | 489,900 | 0.6 |
| Canis familiaris | Carnivora | 63 | 10–24 | 12 | 3–10 | 81.14 | 12,470 | 1.3 |
Elephants have the largest absolute brain size of any land animal: 5.5 kg in Asian elephants and 6.5 kg in African savannah elephants (16–18). These values are ≈4 times the average brain mass of humans (9, 19). Concomitant with a large brain, elephants also have an extensive neocortex (20) and exhibit complex social cognitive abilities. For example, there is evidence that elephants assist injured or disabled conspecifics and recognize their own mirror reflections (18, 21). Thus, in view of the convergent similarities in brain structure, cognition, and life history between elephants and humans, we anticipated that phylogenomic analyses would reveal the patterns of adaptive evolution to be more similar between elephant and human than between elephant and tenrec lineages despite a deep phylogenetic separation between afrotherians and humans.
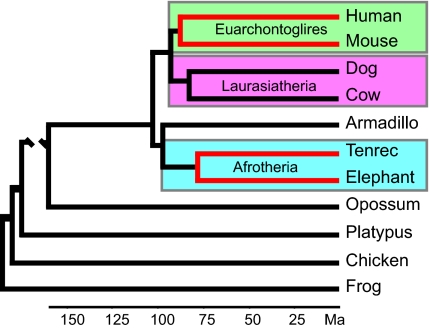
Our raw phylogenomic data consisted of the alignable coding sequences of 15 genomes from species representing all major mammalian clades (11 placental mammals, a marsupial, and a monotreme) and two other vertebrates (a bird and an amphibian). As depicted by the phylogenetic relationships of the 11 placental mammals (Fig. S1), Afrotheria (elephant, tenrec) is joined by Xenarthra (armadillo) to form Atlantogenata, and Euarchontoglires (human, chimpanzee, rhesus monkey, rabbit, rat, mouse) is joined by Laurasiatheria (dog, cow) to form Boreoeutheria (22–24). DNA evidence constrained by fossil evidence indicates that Atlantogenata and Boreoeutheria began diverging from their last common ancestor (LCA) over 100 Mega-annum (Ma; ∼million years) and that within Atlantogenata, the separation between elephant and tenrec traces back to about 78 Ma (25). By comparison, within Euarchontoglires, the human and mouse separation traces back to about 91 Ma (25). In view of the long time spans separating these lineages, many genes were poorly alignable and were excluded from our analyses, which focused on those genes showing extensive coding sequence similarity between human and afrotherian genomes (26). Also, to obtain results from the human and mouse lineages that were comparable to results from the elephant and tenrec lineages, we removed the chimpanzee, macaque, rat, and rabbit genome sequences from the multisequence alignments, leaving human and mouse as the only euarchontoglireans.
For each retained gene, we determined on the sampled branches of the 11 taxa phylogenetic tree (Fig. 1), and the original 15 taxa tree (Fig. S1), the nonsynonymous (amino acid replacing) nucleotide substitutions per nonsynonomous site, and the synonymous (amino acid unchanging) substitutions per synonymous site, dN and dS, respectively. Then, for each lineage, we indexed its genes according to their dN/dS ratios and identified those genes that belong to specific functional categories and were likely targets of positive selection.
Fig. 1.
Phylogenetic relationships of species examined based on DNA and fossil evidence (22–25). Lineages of special interest are highlighted in red. The divergence date estimated for the human/mouse LCA is 91 Ma, and the elephant/tenrec LCA is 78 Ma (25). Non-therian vertebrate branches are not drawn to scale.
We found the elephant lineage resembles the human lineage by showing slower nucleotide substitution rates than the tenrec or mouse lineage and by having more genes that exhibit nonsynonymous rates greater than synonymous. The most evident likely target of positive selection in the elephant lineage was a set of nuclear genes that code for mitochondrial functioning proteins. Previously, we have shown that these genes are highly expressed in the human brain and had evolved adaptively during an earlier period of human ancestry (27). Because mitochondria perform an essential central role in the aerobic production of energy and also because of the interdependence of the different parts of the mitochondrial molecular machinery, we call the genes whose products are found in the mitochondrion aerobic energy metabolism (AEM) genes.
Results
Lineage Similarities and Differences in Coding Sequence Evolution Rates.
Phylogenetic analyses (see Materials and Methods) provided estimates of the nonsynonymous and synonymous evolution rates of each lineage from the afrotherian LCA to present for elephant and tenrec and from the Euarchontoglires LCA to the present for human and mouse (Table S2), with each of these four lineages represented by 7,768 National Center for Biotechnology Information (NCBI) RefSeq transcripts corresponding to ≈6,000 nonredundant genes. For each RefSeq, the ones that were putative orthologues were identified in all four lineages. Per site rates of nonsynonymous (rN) or synonymous (rS) substitutions were obtained for the elephant or tenrec lineages by dividing that lineage's dN or dS by the age of the elephant/tenrec LCA and multiplying by 1,000 to yield substitutions/site/year ×10−9. Similarly, for human and mouse, the age of the human/mouse LCA was used in calculating rN or rS from dN or dS. The mean rS is 5.9–7.5 times faster than the mean rN in the elephant (rS of 1.65; rN of 0.28) and human (rS of 1.49; rN of 0.2) lineages, respectively, and approximately 8 to 10 times faster in the tenrec (rS of 3.85; rN of 0.49) and mouse (rS of 4.63; rN of 0.48) lineages, respectively. As represented by the rS values, the lineage substitution rates for selectively neutral or nearly neutral mutations are much faster in the mammals with short generation times and short life spans (fastest in the mouse lineage, next in the tenrec) than in the mammals with long generation times and long life spans (slowest in the human lineage, next slowest in the elephant). The Wilcoxon signed-rank tests (Table S3) show each lineage rS to be significantly different from each of the others (P < 2.2e−16). The mouse and tenrec lineages, i.e., the two shorter generation time lineages, also accumulated nonsynonymous substitutions at significantly faster rates than human and elephant lineages (P < 2.2e−16), which supports the view that mutation pressure, not just natural selection, shaped the course of evolution (28).
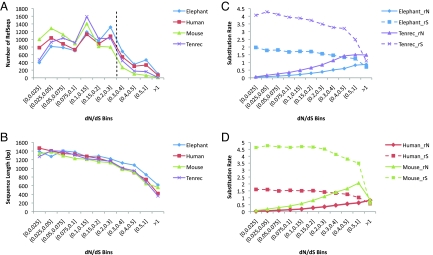
In each lineage, the vast majority of genes evolved at much faster synonymous than nonsynonymous rates (dN/dS <0.3) (Fig. 2A; Table S4). However, the human and elephant lineages have more than double the number of RefSeqs with elevated dN/dS ratios (dN/dS >0.3) than the mouse and tenrec lineages (Table S4). In each of the four lineages, the average length of the RefSeqs decreases as dN/dS increases, a trend that is pronounced for RefSeqs with dN/dS >0.3 (Fig. 2B, Table S4). We also sorted each lineage's RefSeqs according to length. On comparing the 1,000 shortest to the 1,000 longest sequences, we find that the majority of RefSeqs, both at the shortest length (150–471 bp) and longest length (2,127–7,698 bp) have low dN/dS values (i.e., dN/dS <0.3). However, the shortest-length group has 3 to 6 times more RefSeqs at high dN/dS values (i.e., dN/dS >0.3) than has the longest-length group (Table S4E). These results (Table S4E) complement those obtained from sorting the RefSeqs according to dN/dS. The data on the RefSeqs with elevated dN/dS values further indicate that increases in rN are accompanied by decreases in rS, a trend that is especially evident for RefSeqs with dN/dS >0.5 (Fig. 2 C and D, Table S4). Similar lineage trends have been observed for the coding sequences of Drosophila genomes (29).
Fig. 2.
Numbers, lengths, and rates of the RefSeqs distributed according to increase in their dN/dS ratios. (A) Number of RefSeqs with given dN/dS ratios for the elephant, human, tenrec, and mouse lineages. Elephant and human have more RefSeqs with elevated dN/dS than tenrec and mouse when dN/dS >0.3 (black dashed line). (B) Length (base pairs) of RefSeqs with given dN/dS ratios for the elephant, human, tenrec, and mouse lineages. Average length of RefSeq decreases as dN/dS increases. (C) Nonsynonymous and synonymous evolution rates (rN and rS, respectively) comparison of elephant and tenrec. Tenrec rN and rS are significantly higher than elephant rN (WST, P < 2.2 × 10−16) and rS (WST, P < 2.2 × 10−16). (D) Nonsynonymous and synonymous evolution rates comparison (rN and rS, respectively) of human and mouse. Mouse rN and rS are significantly higher than human rN (WST, P < 2.2 × 10−16) and rS (WST, P < 2.2 × 10−16).
Gene ontology analysis (30) of the 7768 RefSeqs in each of the four mammalian lineages revealed that the products of 501 RefSeqs localized to the mitochondrial cellular component (GO:0005739). Sorting these AEM genes according to dN/dS revealed (Table S5) that bins at dN/dS >0.2 for elephant and human lineages have more than double the number of RefSeqs than tenrec and mouse lineages. Indeed, in the dN/dS >1 bin, there are 15 elephant and 4 human RefSeqs but 0 tenrec and 0 mouse sequences (Table S5). The mean dN/dS ratios of the AEM genes of the large-brained mammals, but not the small-brained mammals, were larger than the mean dN/dS ratios of the total lineage genes, significantly so for the elephant lineage [Wilcoxon signed-rank test (WST) P = 5.257 × 10−12] and for the human lineage (WST P = 1.205 × 10−5) (Table S5).
The More Prevalent Distinct Sets of Genes.
Having subdivided each of the four lineage gene lists into bins based on dN/dS intervals, we tested the gene content of each bin for statistically significant enrichment of specific gene ontology functional categories (30). Each enriched functional category consisted of genes with gene ontology annotations that, when compared with the annotations of the bin's other genes, had higher frequencies in the bin than in the human genome. The overrepresented gene sets from low dN/dS bins could be considered the main targets of sustained purifying selection, whereas the overrepresented gene sets from high dN/dS bins could represent functional gene categories targeted by positive selection during their evolutionary histories.
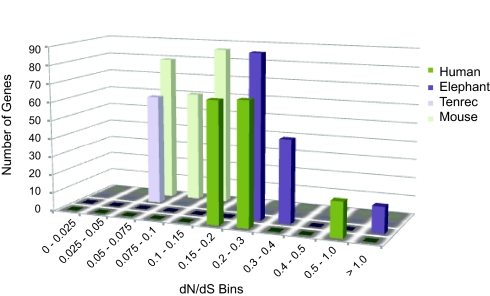
AEM genes were overrepresented at the higher dN/dS bins for elephant and human but at the lower bins for tenrec and mouse (Table S6). These genes were overrepresented in bins with dN/dS >0.15 in elephant and human and overrepresented in bins with dN/dS <0.15 in tenrec and mouse (Fig. 3). These results suggest that a subset of AEM genes has indeed undergone accelerated coding sequence evolution in the larger-brained species but remained under strong purifying selection in the smaller-brained species.
Fig. 3.
Number of clustered overrepresented mitochondrion-related genes (GO:0005739; at enrichment scores >2) per nonoverlapping dN/dS bin. Lower dN/dS values indicate evidence suggestive of purifying selection, whereas higher dN/dS values indicate evidence suggestive of positive selection. The clustered mitochondrion-related genes are present in higher dN/dS bins in the human and elephant lineages and in lower bins in mouse and tenrec lineages.
In contrast to AEM genes, immunity-related genes (including GO:0045087 innate immune response, GO:0006959 humoral immune response, GO:0002682 regulation of immune system process, GO:0002253 activation of immune response, GO:0006952 defense response) were generally found in bins with dN/dS >0.2 in all four lineages. This finding would be expected from pathogen-driven pressures resulting in positive selection on immunity genes in multiple evolutionary lineages (31, 32).
Genes involved in molecular transduction and sensory perception (particularly olfactory) were also enriched in all four lineages. Lineage-specific positive selection and pseudogenization have shaped these genes in recent evolution (33, 34). Olfactory receptor and functionally related genes (including GO:0004984 olfactory receptor activity, GO:0007608 sensory perception of smell, GO:0007606 sensory perception of chemical stimulus, GO:0007600 sensory perception, GO:0060089 molecular transducer activity) had higher dN/dS ratios in human than in mouse and higher dN/dS ratios in elephant than in tenrec. In human, these genes are located in bins with dN/dS 0.15–0.4, whereas in mouse, they are located in bins with dN/dS 0.075–0.2. In elephant, these genes are located in bins with dN/dS 0.2–1.0, whereas in tenrec, they are located in bins with dN/dS 0.075–0.3. Thus, these gene groups may also have undergone greater adaptive evolution in the more encephalized lineages. All functional annotation clusters prevalent in specific dN/dS bins have been made available (Dryad Accession Nos. 926 and 929).
Adaptively Evolved Gene Sets.
Often after natural selection increases the frequency of beneficial alleles in an evolving species, it then maintains the high frequency of these alleles by rejecting harmful or less beneficial alleles. This transformation of positive selection into purifying selection (35) is more likely to occur at nonsynonymous sites than at synonymous sites, where theory predicts synonymous substitutions steadily accumulate over time. Thus, considering the many millions of years of time involved in the descent of the elephant or tenrec lineage (about 78 Ma) and the human or mouse lineage (about 91 Ma), it is not surprising that <1.4% of the thousands of genes examined have dN/dS >1 (Table S4) and that the percentage of such likely positively selected genes is smallest for mouse (0.12%). To increase our chances of detecting gene sets that were likely targets of positive selection, we examined by the functional annotation clustering tool (30) the 5% of RefSeqs (389) with the more elevated dN/dS ratios, these RefSeqs having dN/dS >0.61, >0.38, >0.51, and >0.32 in elephant, tenrec, human, and mouse lineages, respectively. The functional annotation clusters with the enrichment scores (ES) >2 for these genes (Table S7) verify the gene-set results for mitochondrial-related gene ontology categories based on nonoverlapping dN/dS bins. AEM genes are in the enriched clusters of the two large-brained lineages (elephant, human), but not in the small-brained lineages (tenrec, mouse). On extending our analyses to include the paired lineages to cow and dog, we obtained results resembling those for tenrec and mouse. In both cow and dog lineages, immunity genes have high enrichment scores but energy metabolism genes have quite low scores (Table S7). It may be noted that cow and dog have much smaller brain weights than elephant and human, although the cow's body weight is an order of magnitude larger than human's (Table 1 and Table S1). Thus, the adaptive evolution of AEM genes appears to be associated with enlarged brains but not enlarged body size.
In the elephant lineage, the specific gene category with the cellular component annotation mitochondrion (GO:0005739) comprises the cluster with the highest enrichment score in the dN/dS >1 bin (Table S6; 14 genes with this annotation) and in the dN/dS >0.61 bin (Table S7; 37 genes with this annotation). Among human lineage genes with significantly elevated dN/dS ratios (>0.51), one of the most enriched gene sets has the annotation mitochondrion (Table S7; 21 genes). It is noteworthy that 6 human mitochondrion genes, which appear to have evolved adaptively, have elephant orthologues that also appear to have evolved adaptively (Table S8). These particular AEM genes code for an ATP synthase (mitochondrial FO complex) subunit, the cytochrome oxidase 8A subunit, a mitochondrial ribosomal protein, a chaperone protein, a CoA epimerase, and an aminotransferase.
We assumed that not only genes with dN/dS >1 had evolved adaptively, but also that a majority of the 21 human lineage mitochondrial functioning genes at dN/dS >0.51 had so evolved. We verified this assumption by testing it empirically using codeml results from multisequence alignments that encompassed all alignable, putative orthologues from 15 vertebrate taxa and thus partitioned the descent to the human species into three time periods labeled primate stem (91–25 Ma) (25), ape stem (25–6 Ma) (36), and human terminal (6–0 Ma) (36). In the ape stem, the most enriched cluster for genes at dN/dS >1 consisted of 52 genes with the annotation mitochondrion (ES = 5.16); in the human terminal, there was also an annotation cluster of 48 genes localized to the mitochondrion but at an enrichment score of 1.64; and in the primate stem, at a quite low enrichment score (ES = 0.85), the mitochondrion cluster consisted of 10 genes. Of the 21 genes localized to the mitochondrion at dN/dS >0.51 in the unpartitioned lineage from the rodent-primate LCA to the human species, most (17 genes) were found among the mitochondrial functioning genes showing evidence of positive selection (dN/dS >1) during one or another of the successive evolutionary periods in descent to the human species; 5 of the 17 showed this evidence in the primate stem, 13 in the ape stem, and 7 in the human terminal, with 8 of the 17 genes showing dN/dS >1 in more than one of these three successive evolutionary periods (Table S8). Most noteworthy, 4 of the 6 genes adaptively evolved in both human and elephant lineages show dN/dS >1 in the human terminal, the period of human ancestry when brain size increased dramatically.
Discussion
Our phylogenomic study has yielded two key results. The first result reveals that the rate of DNA coding sequence evolution has slowed in elephant ancestry as in human ancestry (Table S2). A slower pace of nucleotide substitutions would be expected to ensue from the lengthening of generation times and life spans (Table 1 and Table S1). Convergent evolution in elephant and human ancestries most likely produced reductions in the rate of occurrence of replication-dependent mutations (37) and possibly, to hypothesize, improvements in the mechanisms for preventing or repairing injuries to DNA (38). Such mechanisms help preserve the integrity of the high-energy-consuming neurons of enlarged brains. The second key result is the evidence of convergent patterns of adaptive evolution in nuclear genes that code for mitochondrial functioning proteins. These AEM genes are critical for the increased energy production needed to support a large brain, a trait convergently shared by both elephants and humans. To preface our case for adaptive evolution, we start by considering the role of neutral evolution.
The Case for Adaptive Evolution.
Certain aspects of the results on coding sequence evolution indicate that many nonsynonymous substitutions originated from selectively neutral or nearly neutral (39) mutations. Not only were the rates of synonymous substitutions, i.e., likely selectively neutral substitutions, faster in the lineages with shorter generation times but the rates of nonsynonymous substitutions were also faster. In each lineage, on ordering the genes according to increasing dN/dS ratios, genes with higher ratios have decreased synonymous rates (Fig. 2 C and D) and shorter coding sequences (Fig. 2B), suggesting that the pool of nucleotide sites where neutral or nearly neutral mutations occurred included nonsynonymous sites as well as synonymous and that asymmetrical distributions of mutations, overly concentrated at these nonsynonymous sites, occurred more often in the shorter coding sequences.
Nevertheless, without disputing the inference that many nonsynonymous substitutions were initially selectively neutral or nearly neutral, we can still make a case from the results of the gene ontology analyses that certain clusters of genes at the higher dN/dS ratios have evolved adaptively. The initially neutral mutations that genetic drift had spread could later in a descendant population have acquired beneficial synergistic effects, perhaps due to change in the ecological environment or to some positively selected regulatory changes. If so, these genes would then be preserved by purifying selection. However, the descendant population with these genes would out-compete other populations with less synergistic genes. From this perspective of competing groups, such synergistic genes could be considered positively selected.
The case for adaptive evolution is supported by the finding that in all mammalian lineages examined in our study (elephant, tenrec, human, mouse, dog, and cow), immune system genes were overrepresented in the higher dN/dS bins. As noted, it is well established that pathogen-driven pressures have made immune system genes the targets of positive selection in multiple mammalian lineages (31, 32). The overrepresentation of AEM genes that occurred in the higher dN/dS bins for the larger-brained and long-lived mammalian lineages further supports the case for adaptive evolution (Fig. 3, Tables S6 and S7). Indeed, the elephant results are most striking. In the elephant lineage at the higher dN/dS bins, the top annotation clusters consisted of AEM genes, and the number of genes in these mitochondrial clusters was larger than for any other lineage with a long unpartitioned evolutionary history. This finding correlates closely with the elephant's massive brain and microstructural evidence of increased energetic requirements for neocortical neurons as well as other life history variables that are similar in humans and elephants (Table 1 and Table S1).
The Elephant's Adaptively Evolved Brain and Chemosensory Behavior.
With its extremely large mass, the elephant brain consumes 200 ml of O2/min (40). This value exceeds the adult human brain's rate of oxygen consumption (49 ml of O2/min) (40). Furthermore, several lines of indirect evidence suggest that the elephant neocortex is likely to require a great deal of energy on a neuron-specific basis. The elephant neocortex has a relatively low density of very large-sized neurons (41, 42) with extensive dendritic arborization (16), suggesting that each receives a relatively large number of synapses from interconnecting axons as compared with a large-brained primate (43). Accordingly, the interneuronal space in the elephant neocortex is occupied by a remarkably high density of glial cells (42). Astrocytes, one of the major glial cell types, are thought to play a critical role in transporting energy substrates to neurons in response to synaptic activity (44). From our previous observations (ref. 45 and Butti et al., unpublished), the ratio of glia to neurons in the anterior cingulate cortex of African savannah elephants is 4.7. Because the number of glia relative to neurons in the neocortex tends to increase with larger brain size (46–49), the glia-neuron ratio in elephants is comparable with the values reported for other very large-brained mammals such as 1.6 for humans (46), 3.1 for the bottlenose dolphin (50), 4.5 for the fin whale (48), and 7.7 for the minke whale (51). It has been suggested that interspecific variation in the neocortical glia-neuron ratio correlates with the size of the dendritic tree and the length of axons, because the increased scale of these structures would require a larger number of astrocytes to regulate glucose uptake and more oligodendrocytes to actively synthesize the myelin for long-range projecting axon fibers (46, 52, 53). Indeed, the volume of white matter underlying the neocortex gray matter has been demonstrated to increase disproportionately across species with larger brain sizes because axons must become either thicker or more heavily myelinated to sustain a high velocity of axon potentials over long distances, consequently leading to a relatively larger energetic investment in axonal maintenance (43). Elephants follow this trend by having one of the most extreme ratios of white matter to neocortical gray matter among mammals (20, 54). These neurobiological observations strengthen the case that many of the elephant's aerobic energy metabolism genes had evolved adaptively, as indicated by the data in Tables S6 and S7. Presumably, such evolution helped perfect the mitochondrial machinery that gives the elephant's brain the ability to efficiently use huge amounts of metabolic energy.
The evidence of adaptive evolution of olfactory and chemosensory genes in the elephant lineage is congruent with a range of experimental and field studies indicating the importance of chemosensory perception in the social signaling of elephants (55, 56). For example, musth bulls communicate their reproductive status to other elephants via temporal gland secretions and continuous dribbling of strong-smelling urine (55, 56). Furthermore, elephants' exquisite capacity for olfactory discrimination is expressed in their chemosensory memory. Individual Asian elephants separated from their mothers for decades, for instance, showed selectively higher flehmen responses to their mother's urine than to urine from unrelated, but familiar, females (57).
Toward Further Dissecting the Convergence Between Elephant and Human Ancestries.
In agreement with this biological and behavioral data on elephants, our genomic data indicate that AEM genes and chemosensory genes have been preferred targets of positive selection in large-brained mammals but not in small-brained mammals. Indeed, the most striking finding is that higher dN/dS bins were found to be enriched for genes that code for mitochondrial proteins in both elephant and human lineages: A pattern not observed in their more closely related sister taxa (tenrec and mouse respectively) and the cow and dog lineages (Tables S6 and S7). These results correlate with absolute brain size and brain oxygen consumption, which are also convergent traits shared by elephants and humans. It would be beneficial to a large-brained, long-lived mammal if the positively selected amino acid replacements had so improved the machinery for aerobic energy production that, with minimum production of reactive oxygen species (ROS), this machinery could then completely satisfy the large brain's huge energy needs. Minimizing ROS production would help protect the integrity of long-lived neurons by reducing DNA damage. Biochemical experiments could test if these amino acid replacements affect the efficiency of aerobic energy production. Such study could examine the functional consequences of specific replacements that occurred in the mitochondrial functioning proteins of large-brained mammals, especially any convergent replacements between the elephant and human proteins.
Another avenue of study should be directed at discovering positively selected changes in the regulatory circuits that control the expression of AEM and other brain-important genes. We found that the coding sequences of many brain-important genes have been under pervasive, strong, purifying selection in multiple mammalian lineages (Dryad Accession Nos. 926 and 929). Thus, if these genes played a role in the adaptive evolution that produced greatly enlarged brains, that role may have been mediated by positively selected regulatory mutations in promoters or other noncoding DNA sequences with regulatory functions. After genomes from the diverse clades within Afrotheria and within Euarchontoglires are completely sequenced, it will be possible to carry out a much more thorough phylogenomic search for converging patterns of adaptive evolution in elephant and human evolutionary histories.
Materials and Methods
We used OCPAT (26) to align putative gene orthologues among the following 15 vertebrate draft sequenced genomes: Homo sapiens (NCBI Build 36.1), Pan troglodytes (v2.1), Macaca mulatta (v1), Mus musculus (NCBI m36), Rattus norvegicus (v3.4), Oryctolagus cuniculus (v1, http://www.broadinstitute.org/science/projects/mammals-models/rabbit/rabbit-genome-sequencing-project), Canis familiaris (v2), Bos taurus (v3.1, http://www.hgsc.bcm.tmc.edu/projects/bovine/), Dasypus novemcinctus (v1, Broad Institute Mammalian Genome Project), Loxodonta africana (v2, http://www.broadinstitute.org/mammals/elephant), Echinops telfairi (v2, Broad Institute Mammalian Genome Project), Monodelphis domestica (v4, http://www.broadinstitute.org/mammals/opossum), Ornithorhynchus anatinus (v5, http://genome.wustl.edu/genomes/view/ornithorhynchus_anatinus/), Gallus gallus (v2.1), and Xenopus tropicalis (v4.1). If no homologous gene sequence was identified for a given species, this species was not included in the gene alignment. Alignments with <50 codons were discarded. Evolutionary information about the retained RefSeqs was then obtained using the Fig. S1 phylogenetic tree and codeml (58) under the free-ratio model. Because the time traversed by the elephant or tenrec lineage in descent from the afrotherian LCA was longer by an order of magnitude than that of the human lineage in descent from the chimpanzee/human LCA, we removed the chimpanzee, macaque, rabbit, and rat sequences from the alignments and, with an 11 taxa phylogenetic tree (Fig. 1), again obtained evolutionary information using codeml under the free-ratio model. The resulting codeml data (such as N, S, dN, dS, dN/dS values) for the genes of each lineage under study were curated to reduce errors. Genes were discarded if they had any one of the following values: dS >1, N greater than the RefSeq length, N + S > RefSeq length by 50 or more bp, and (just for codeml result from the 11 taxa phylogenetic tree) N*dN or S*dS <1. For each RefSeq discarded in any one of the four lineages under intensive study (human, mouse, elephant, tenrec), its putative orthologues in the other three lineages were also discarded. Each of these four lineages was then represented by 7,768 RefSeqs from ≈6,000 genes. All codeml data have been deposited at http://www.datadryad.org/repo/ (Dryad Accession No. 921).
The retained genes of each lineage under study were sorted into nonoverlapping bins according to their dN/dS values. Adaptive evolution was inferred not only for genes having dN/dS >1, but also for genes having dN/dS >0.61, >0.38, >0.51, >0.32, >0.41, and >0.38 for elephant, tenrec, human, mouse, dog, and cow genes, respectively, in each case the top 5% of a distribution according to increasing dN/dS values. Computation of dN/dS values for human and mouse genes were based on divergence from the human/mouse LCA, for elephant and tenrec genes on divergence from the elephant/tenrec LCA, and for dog and cow on divergence from the dog/cow LCA. Human NCBI RefSeq (NM no. only) GenBank accession numbers were used in all gene ontology analyses. Non-human orthologues of the human genes were retrieved from Ensembl Biomart. For uniformity of functional annotation enrichment results, we used human NM no. RefSeq accession numbers to refer both to human genes and to their putative non-human orthologues.
Wilcoxon signed-ranked tests were used to test whether dN/dS, rN, and rS differ significantly by lineage, and whether the mean dN/dS for the mitochondria-related RefSeqs (501 RefSeqs) differ significantly from the mean dN/dS for nonmitochondrial-related RefSeqs (7,267 alignments). All statistical analyses were done using R.
Gene Symbol Retrieval.
We retrieved gene symbols from Ensembl Biomart (http://www.ensembl.org/biomart/indexhtml). We used the Ensembl 52 database and the Homo sapiens genes (NCBI36) database. We used the “ID list limit” filter. Under “Attributes,” we chose “Associated Gene Name.” We eliminated multiple redundant accession numbers if they corresponded to the same gene symbol.
Functional Annotation Clustering.
We used a functional annotation clustering tool (30) for each “species and dN/dS bin” combination to group genes with shared annotations. The algorithm assigned a significance P value, corrected for multiple testing, to each subgroup representing a gene ontology annotation within the cluster and an enrichment score to the whole cluster. The clusters with the higher enrichment scores consisted of subgroups with the higher significance values, and thus these clusters provided an integrated view of the more significantly enriched or overrepresented gene functional categories within each dN/dS bin. All functional annotation clustering data have been deposited at http://www.datadryad.org/repo/ (Dryad Accession Nos. 926, 929, and 930).
The enrichment score for each annotation cluster was based on the geometric mean of p- values of the cluster's assorted annotations. The k− similarity threshold was set to 0.7 for the genes with these assorted annotations, and other options were set to their default values. Our criterion for inclusion of clustered results for Fig. 3 was a cluster enrichment score >2.
Supplementary Material
Acknowledgments.
We thank Camilla Butti for help in determining the elephant glia-neuron ratio, Chris Kuzawa for insightful discussion, and Richelo Soliven and M. Gopi Chand for providing research support. This research was supported by the National Science Foundation (Grants BCS0550209, BCS0827546, BCS0515484, BCS0549117, BCS0827531, DGE0801634) and the James S. McDonnell Foundation (Grant 22002078).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Dryad Global Wood Density Database, http://hdl.handle.net/10255/dryad.908.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911239106/DCSupplemental.
References
- 1.Kouprina N, et al. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:E126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doan JW, et al. Coadaptive evolution in cytochrome c oxidase: 9 of 13 subunits show accelerated rates of nonsynonymous substitution in anthropoid primates. Mol Phylogenet Evol. 2004;33:944–950. doi: 10.1016/j.ympev.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Dorus S, et al. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 4.Evans PD, et al. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum Mol Genet. 2004;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- 5.Wang YQ, Su B. Molecular evolution of microcephalin, a gene determining human brain size. Hum Mol Genet. 2004;13:1131–1137. doi: 10.1093/hmg/ddh127. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt TR, et al. Rapid electrostatic evolution at the binding site for cytochrome c on cytochrome c oxidase in anthropoid primates. Proc Natl Acad Sci USA. 2005;102:6379–6384. doi: 10.1073/pnas.0409714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossman LI, Wildman DE, Schmidt TR, Goodman M. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 2004;20:578–585. doi: 10.1016/j.tig.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Goodman M, Grossman LI, Wildman DE. Moving primate genomics beyond the chimpanzee genome. Trends Genet. 2005;21:511–517. doi: 10.1016/j.tig.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Sherwood CC, Subiaul F, Zawidzki TW. A natural history of the human mind: Tracing evolutionary changes in brain and cognition. J Anat. 2008;212:426–454. doi: 10.1111/j.1469-7580.2008.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert SL, Dobyns WB, Lahn BT. Genetic links between brain development and brain evolution. Nat Rev Genet. 2005;6:581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]
- 11.Sokoloff L. In: Handbook of Physiology, Section I, Neurophysiology. Field J, Magoun HW, Hall VE, editors. Washington, DC: American Physiological Society; 1960. pp. 1843–1864. [Google Scholar]
- 12.Finarelli JA, Flynn JJ. Brain-size evolution and sociality in Carnivora. Proc Natl Acad Sci USA. 2009;106:9345–9349. doi: 10.1073/pnas.0901780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roca AL, O'Brien SJ. Genomic inferences from Afrotheria and the evolution of elephants. Curr Opin Genet Dev. 2005;15:652–659. doi: 10.1016/j.gde.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Stephan H, Baron G, Frahm HD. Comparative Brain Res in Mammals, Vol. 1: Insectivora: With a Stereotaxic Atlas of the Hedgehog Brain. New York: Springer Verlag; 1991. [Google Scholar]
- 15.Krubitzer L, Kunzle H, Kaas J. Organization of sensory cortex in a Madagascan insectivore, the tenrec (Echinops telfairi) J Comp Neurol. 1997;379:399–414. [PubMed] [Google Scholar]
- 16.Cozzi B, Spagnoli S, Bruno L. An overview of the central nervous system of the elephant through a critical appraisal of the literature published in the XIX and XX centuries. Brain Res Bull. 2001;54:219–227. doi: 10.1016/s0361-9230(00)00456-1. [DOI] [PubMed] [Google Scholar]
- 17.Shoshani J, Kupsky WJ, Marchant GH. Elephant brain Part 1: Gross morphology, functions, comparative anatomy, and evolution. Brain Res Bull. 2006;70:124–157. doi: 10.1016/j.brainresbull.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Hart BL, Hart LA, Pinter-Wollman N. Large brains and cognition: Where do elephants fit in? Neurosci Biobehav Rev. 2008;32:86–98. doi: 10.1016/j.neubiorev.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Hofman MA. Size and shape of the cerebral cortex in mammals. I. The cortical surface. Brain Behav Evol. 1985;27:28–40. doi: 10.1159/000118718. [DOI] [PubMed] [Google Scholar]
- 20.Hakeem AY, et al. Brain of the African elephant (Loxodonta africana): Neuroanatomy from magnetic resonance images. Anat Rec. 2005;287:1117–1127. doi: 10.1002/ar.a.20255. [DOI] [PubMed] [Google Scholar]
- 21.Plotnik JM, de Waal FBM, Reiss D. Self-recognition in an Asian elephant. Proc Natl Acad Sci USA. 2006;103:17053–17057. doi: 10.1073/pnas.0608062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wildman DE, et al. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci USA. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallstrom BM, Kullberg M, Nilsson MA, Janke A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol. 2007;24:2059–2068. doi: 10.1093/molbev/msm136. [DOI] [PubMed] [Google Scholar]
- 24.Prasad AB, et al. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammalian phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, et al. OCPAT: An online codon-preserved alignment tool for evolutionary genomic analysis of protein coding sequences. Source Code for Biology and Medicine. 2007;2:5. doi: 10.1186/1751-0473-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uddin M, et al. Distinct genomic signatures of adaptation in pre- and postnatal environments during human evolution. Proc Natl Acad Sci USA. 2008;105:3215–3220. doi: 10.1073/pnas.0712400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci USA. 2007;104:12235. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larracuente AM, et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Sherman BT, et al. DAVID Knowledgebase: A gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BioMed Central Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellegren H. Comparative genomics and the study of evolution by natural selection. Mol Ecol. 2008;17:4586–4596. doi: 10.1111/j.1365-294X.2008.03954.x. [DOI] [PubMed] [Google Scholar]
- 32.Marques JT, Carthew RW. A call to arms: Coevolution of animal viruses and host innate immune responses. Trends Genet. 2007;23:356–364. doi: 10.1016/j.tig.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Mundy NI. Genetic basis of olfactory communication in primates. Am J Primatol. 2006;68:559–567. doi: 10.1002/ajp.20252. [DOI] [PubMed] [Google Scholar]
- 34.Gilad Y, Man O, Glusman G. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 2005;15:224–230. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodman M. Positive selection causes purifying selection. Nature. 1982;295:630. doi: 10.1038/295630a0. [DOI] [PubMed] [Google Scholar]
- 36.Goodman M, et al. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 37.Kim S-H, Elango N, Warden C, Vigoda E, Yi SV. Heterogeneous genomic molecular clocks in primates. PLOS Genet. 2006;2:e163. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britten RJ. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986;231:1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- 39.Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 40.Grande F. In: Assessment of Energy Metabolism in Health and Disease. Calvert S, editor. Columbus, OH: Ross Laboratories; 1980. pp. 88–92. [Google Scholar]
- 41.Tower DB. Structural and functional organization of mammalian cerebral cortex: The correlation of neurone density with brain size. J Comp Neurol. 1954;101:19–52. doi: 10.1002/cne.901010103. [DOI] [PubMed] [Google Scholar]
- 42.Haug R. Brain sizes, surfaces, and neuronal sizes of the cortex cerebri: A stereological investigation of man and his variability and a comparison with some mammals (primates, whales, marsupials, insectivores, and one elephant) Am J Anat. 1987;180:126–142. doi: 10.1002/aja.1001800203. [DOI] [PubMed] [Google Scholar]
- 43.Wang SS, et al. Functional trade-offs in white matter axonal scaling. J Neurosci. 2008;28:4047–4056. doi: 10.1523/JNEUROSCI.5559-05.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsacopoulos M, Magistretti PJ. Metabolic coupling between glia and neurons. J Neurosci. 1996;16:877–885. doi: 10.1523/JNEUROSCI.16-03-00877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakeem AY, et al. Von Economo neurons in the elephant brain. Anat Rec. 2009;292:242–248. doi: 10.1002/ar.20829. [DOI] [PubMed] [Google Scholar]
- 46.Sherwood CC, et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci USA. 2006;103:13606–13611. doi: 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friede RL. Quantitative share of the glia in development of the cortex. Acta Anat. 1954;20:290–296. [PubMed] [Google Scholar]
- 48.Hawkins A, Olszewski J. Glia/nerve cell index for cortex of the whale. Science. 1957;126:76–77. doi: 10.1126/science.126.3263.76. [DOI] [PubMed] [Google Scholar]
- 49.Tower DB, Young OM. Interspecies correlations of cerebral cortical oxygen consumption, acetylcholinesterase activity and chloride content: Studies on the brains of the fin whale (Balaenoptera physalus) and the sperm whale (Physeter catodon) J Neurochem. 1973;20:253–267. doi: 10.1111/j.1471-4159.1973.tb12125.x. [DOI] [PubMed] [Google Scholar]
- 50.Garey LJ, Leuba G. A quantitative study of neuronal and glial numerical density in the visual cortex of the bottlenose dolphin: Evidence for a specialized subarea and changes with age. J Comp Neurol. 1986;247:491–496. doi: 10.1002/cne.902470408. and erratum (1986) 250–263. [DOI] [PubMed] [Google Scholar]
- 51.Eriksen N, Pakkenberg B. Total neocortical cell number in the mysticete brain. Anat Rec. 2007;290:83–95. doi: 10.1002/ar.20404. [DOI] [PubMed] [Google Scholar]
- 52.Friede RL. The relationship of body size, nerve cell size, axon length, and glial density in the cerebellum. Proc Natl Acad Sci USA. 1963;49:187–193. doi: 10.1073/pnas.49.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friede RL, Van Houten WH. Neuronal extension and glial supply: Functional significance of glia. Proc Natl Acad Sci USA. 1962;48:817–821. doi: 10.1073/pnas.48.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K, Sejnowski TJ. A universal scaling law between gray matter and white matter of cerebral cortex. Proc Natl Acad Sci USA. 2000;97:5621–5626. doi: 10.1073/pnas.090504197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hollister-Smith JA, Alberts SC, Rasmussen LEL. Do male African elephants, Loxodonta africana, signal musth via urine dribbling? Anim Behav. 2008;76:1829–1841. [Google Scholar]
- 56.Rasmussen LEL, Riddle HS, Krishnamurthy V. Mellifluous matures to malodorous in musth. Nature. 2002;415:975–976. doi: 10.1038/415975a. [DOI] [PubMed] [Google Scholar]
- 57.Rasmussen LEL, Krishnamurthy V. How chemical signals integrate Asian elephant society: The known and the unknown. Zoo Biol. 2000;19:405–423. [Google Scholar]
- 58.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.