Abstract
Tetherin is an IFN-inducible restriction factor that inhibits HIV-1 particle release in the absence of the HIV-1 countermeasure, viral protein U (Vpu). Although ubiquitous in HIV-1 and simian immunodeficiency viruses from chimpanzees, greater spot nosed monkeys, mustached monkeys, and Mona monkeys, other primate lentiviruses do not encode a Vpu protein. Here we demonstrate that SIV from Tantalus monkeys (SIVtan) encodes an envelope glycoprotein (SIVtan Env) able to counteract tetherin from Tantalus monkeys, rhesus monkeys, sooty mangabeys, and humans, but not from pigs. We show that sensitivity to Vpu but not SIVtan Env can be transferred with the human tetherin transmembrane region. We also identify a mutation in the tetherin extracellular domain, which almost completely abolishes sensitivity of human tetherin to SIVtan Env without compromising antiviral activity or sensitivity to Vpu. SIVtan Env expression results in a reduction of surface tetherin, as well as reduction in tetherin co-localization with mature surface-associated virus. Immuno-electron microscopy reveals co-localization of SIVtan Env with tetherin in intracellular tubulo-vesicular structures, suggesting that tetherin is sequestered away from budding virions at the cell surface. Along with HIV-1 Vpu and SIV Nef, envelope glycoprotein is the third and most broadly active lentiviral-encoded tetherin countermeasure to be described. Our observations emphasize the importance of tetherin in protecting mammals against viral infection and suggest that HIV-1 Vpu inhibitors may select active envelope mutants.
Keywords: HIV, restriction, innate immunity
Mammalian cells encode restriction factors such as tetherin as a means of protecting themselves from viral infection. In turn, viruses have evolved means of overcoming restriction factors, either through variation in the target protein or through evolution of antagonistic accessory proteins such as Vpu and Nef, which act against tetherin (1–4). Tetherin is an IFN-inducible dimeric transmembrane protein with an extra-cellular coiled-coil and a predicted GPI anchor (5). It is proposed to form a protein tether linking assembled virions to infected cells, leading to their endocytosis and degradation in lysosomes (1, 6). All tetherin variants tested thus far inhibit the release of enveloped viral particles including retroviruses, filoviruses, and arena viruses (1, 2, 7–10). Many simian immunodeficiency variants do not encode a Vpu protein, suggesting the existence of an alternative means of overcoming tetherin restriction. Indeed, HIV-2 envelope protein has been reported to promote viral budding (12–15), and two recent reports demonstrate that this is the result of tetherin antagonism (3, 16). Remarkably, Ebola virus glycoprotein has also been reported to have anti-tetherin activity, suggesting that envelope glycoproteins may commonly have anti-tetherin function (17). Here we sought anti-tetherin activity for SIV from tantalus monkeys (SIVtan). We chose SIVtan as a virus without a Vpu gene and distantly related to HIV-1 and focused on its envelope protein, influenced by studies showing stimulation of HIV-2 budding by HIV-2 envelope protein. We show that SIVtan Env potently counteracts tetherin from divergent primates including humans. SIVtan Env co-localizes with tetherin in cells and results in its depletion from the cell surface and accumulation in intracellular compartments. Tetherin's sensitivity to SIVtan Env is abrogated by a point mutation at a position with evidence for positive selection.
Results
SIV Tantalus Envelope Glycoprotein Antagonizes Human, Tantalus, Rhesus, and Sooty Mangabey Monkey Tetherin Proteins.
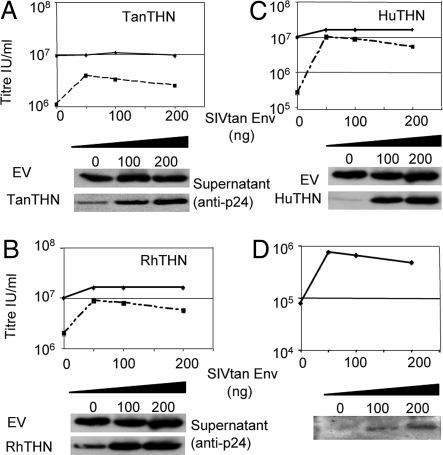
We cloned the env gene from the SIVtan full-length viral clone SIVtan1 (18). As the first third of the SIVtan nef gene overlaps the 3′ end of the env gene in an alternative reading frame, we mutated the nef ATG and tested the ability of SIVtan Env alone to antagonize tetherin from human, rhesus macaque, Tantalus monkey, and sooty mangabey. We co-transfected fixed amounts of plasmids encoding Tantalus monkey (TanTHN), rhesus monkey (RhTHN), human (HuTHN), and sooty mangabey (SmTHN) tetherins with HIV-1 vector plasmids and a titration of SIVtan envelope encoding plasmid into 293T cells. Expression of primate tetherin proteins strongly inhibited virus release as previously described (7, 8), and expression of SIVtan Env was able to partially counteract Tantalus tetherin antiviral activity (Fig. 1A). Moreover, SIVtan Env was able to potently antagonize rhesus and human tetherins (Fig. 1 B and C), as well as sooty mangabey tetherin (Fig. S1A). Importantly, expression of SIVtan Env did not significantly increase HIV-1 release in the absence of tetherin expression (Fig. 1 A–C, solid lines). As a control we measured p24 in supernatant and p55 Gag in cell lysates by Western blot using 50 ng of SIVtan Env plasmid to antagonize human and Tantalus monkey tetherins (Fig. S1B). Finally, we demonstrated that SIVtan Env was able to counteract endogenously expressed tetherin by transfecting plasmids encoding HIV vectors and SIVtan Env into HeLa cells, which constitutively express tetherin (1) (Fig. 1D). We conclude that SIVtan Env can antagonize tetherin proteins from various primates in the absence of the Nef protein.
Fig. 1.
SIV Tantalus envelope protein antagonizes human, Tantalus, and Rhesus monkey tetherin proteins. (A–C) Titers of HIV-1 released from 293T cells co-transfected with a titration of SIVtan Env plasmid lacking a Nef start codon, HIV-1 vectors and Tantalus monkey tetherin (TanTHN) (A), rhesus monkey tetherin (RhTHN) (B), or human tetherin (HuTHN) (C)-encoding plasmid (100 ng). Filled diamond and solid line denotes cells co-transfected with empty vector (EV) and ■ and broken line, cells co-transfected with tetherin expression vector. Measurement of p24 in supernatant at selected doses of SIVtan Env plasmid or empty vector (EV) were measured by Western blot. (D) HIV-1 vectors were co-transfected with a titration of SIVtan Env into HeLa cells and the titer of HIV-1 released determined. Data represent two independent experiments.
Determinants of Sensitivity to SIVtan Env Lie Outside the Transmembrane Region of Tetherin.
Tetherin's sensitivity to HIV-1 Vpu is species-specific and determined by the sequence of its transmembrane (TM) domain (7, 8). To map residues important for sensitivity to antagonism by SIVtan Env, we sought a tetherin molecule able to restrict HIV-1 but insensitive to antagonism by either Vpu or SIVtan Env. We cloned porcine tetherin as a distantly related tetherin (Fig. 2A) and found that it potently restricted HIV-1 release and was insensitive to antagonism by both Vpu and SIVtan Env (Fig. 2B and Fig. S2A). We then constructed a chimeric porcine tetherin protein with the human TM region, replacing porcine tetherin residues 20–49 with the homologous human residues 15–46. Transposition of the TM region of human tetherin was able to confer sensitivity to HIV-1 Vpu, but not SIVtan Env, on the chimera (Fig. 2C and Fig. S2B), suggesting that the determinants of tetherin sensitivity to antagonism by the envelope lie outside this region.
Fig. 2.
A point mutant in the extra-cellular domain of human tetherin renders it insensitive to SIVtan Env. (A) The amino acid sequence of pig tetherin is shown aligned to tetherin sequences from human (Hu) and Tantalus monkey (Tan). Similarity (Sim) asterisk, identical residue; colon, conserved substitution; period, semiconserved substitution; gap, no conservation. The transmembrane region is indicated with a line. (B) Pig tetherin is insensitive to SIVtan Env and HIV-1 Vpu. Fixed doses of HIV-1 Vpu or SIVtan Env were co-transfected with a titration of pig tetherin and HIV-1 vectors. Infectious titers released were plotted. (C) An identical experiment using a chimeric pig tetherin with a human transmembrane region, demonstrates that sensitivity to HIV-1 Vpu, but not SIVtan Env can be conferred by the human tetherin transmembrane region. Data represent two independently performed experiments. (D) HIV-1 vectors were co-expressed with wild-type (wt) human tetherin or mutants with empty vector (EV) (black bars) or SIVtan Env (white bars). Titers of virus released are plotted. Control (C) lane represents titer of HIV-1 released with no tetherin co-transfected. (E) Western blots indicate that released p24 levels reflect infectious titers and equal Gag expression. SIVtan Env expression was measured by Western blot detecting the C-terminal HA tag. Data represent two independently performed experiments. Error bars are standard deviations of virus titers.
A Point Mutant in the Extracellular Domain of Human Tetherin Renders It Insensitive to SIVtan Env.
We previously used positive selection analysis of tetherin to identify a key determinant of sensitivity to HIV-1 Vpu in the TM region (7). To identify determinants of sensitivity to SIVtan Env, we mutated human tetherin amino acids with evidence for positive selection to the homologous residues in the porcine protein. Having shown that the human TM region does not confer sensitivity to SIVtan Env on the porcine protein (Fig. 2C), we focused on the cytoplasmic and C-terminal extra-cellular regions of the protein. We co-expressed human tetherin mutants C9S R10P, A89T, A100D, R139K, V146A, Y153N, Q167F, and L169A with HIV-1 vectors with and without a C-terminal HA-tagged SIVtan Env. The tagged SIVtan Env protein had identical anti-tetherin activity as the untagged version (compare Fig. S1B untagged to Fig. 2D tagged). All mutants were able to effectively restrict HIV-1 release (Fig. 2D), and were fully sensitive to HIV-1 Vpu mediated rescue (Fig. S2C). Furthermore, all mutants were fully sensitive to SIVtan envelope antagonism except for human Tetherin A100D (Fig. 2D). It therefore appears that human tetherin can escape SIVtan Env and potently restrict HIV-1 if a single amino acid is changed to reflect the porcine sequence. The reciprocal mutation (D to A) in porcine tetherin did not confer sensitivity to SIVtan Env.
SIVtan Env Does Not Reduce Total Tetherin Levels but Depletes Tetherin from the Cell Surface, Probably Via Interactions Between Extracellular Domains.
Several recent studies have suggested that Vpu expression leads to tetherin degradation (6, 7, 9). We therefore examined the impact of SIVtan envelope expression on tetherin levels. We used a human tetherin construct with an HA tag in the extra cellular domain, as described (8), and co-expressed it with HIV-1 vectors in the presence and absence of HA tagged SIVtan Env (Fig. 3A). Measurement of p24 levels in the supernatant by Western blot demonstrated that tetherin inhibited p24 release as expected. SIVtan Env expression was demonstrated by Western blot detecting the C-terminal HA tag in cell lysates. Intracellular p55 Gag and tetherin were co-detected in cell extracts by Western blot, probing with both antibodies together (Fig. 3A). Importantly, SIVtan Env did not lead to tetherin depletion, despite potent antagonism of tetherin restriction (Fig. 3A). As a control we also tested the impact of SIVtan Env on the tetherin mutant A100D, which is not sensitive to SIVtan Env antagonism (Fig. 2). As expected, the A100D tetherin mutant reduced p24 in the supernatant (Fig. 3A), and SIVtan Env did not rescue suppression of virus release. This experiment also demonstrates that the A100D tetherin is expressed at a similar level to wild-type protein, both in the presence and absence of SIVtan Env.
Fig. 3.
SIVtan Env does not reduce total tetherin levels but leads to depletion of tetherin from the cell surface. (A) Empty vector (EV), wild-type human tetherin (WT), or human tetherin mutant A100D, both HA tagged in the ectodomain, were co-transfected with HIV-1 vectors and HA tagged SIVtan Env as shown. HIV-1 p24 levels released into supernatants were measured by Western blot. C terminally HA-tagged Tan Env gp160 band was detected in cell lysates by Western blot. Confocal images of immunofluorescently labeled HA ectodomain tagged human tetherin co-transfected with empty vector (B) or untagged SIVtan Env (C). Images are representative of multiple fields from two separate experiments. (D) Cell surface expression of tetherin in the presence of HIV-1 Vpu or SIVtan Env expression as shown. Black bars indicate cells negative for tetherin expression and white bars those expressing tetherin. Mean fluorescence intensities (MFI) are plotted as a percentage of empty vector control values. Error bars represent standard deviation and data are representative of two independent experiments.
As SIVtan Env does not lead to tetherin degradation, we sought an alternative mechanism of antagonism. To address this we used fluorescence microscopy to examine the distribution of tetherin in the absence (Fig. 3B) and presence (Fig. 3C) of SIVtan Env. When expressed alone in 293T cells, HA-tagged tetherin localized at the cell surface, as well as in intracellular compartments, consistent with previous observations (8, 19, 20). On co-expression of SIVtan Env the tetherin was now mostly intracellular with a perinuclear distribution. Importantly, the A100D tetherin mutant was not re-localized on co-expression of SIVtan Env (Fig. S3 A and B). To confirm and quantify the loss of wild-type tetherin at the cell surface we measured cell surface tetherin levels by flow cytometry (Fig. 3D). We transiently co-expressed the ectodomain HA-tagged human tetherin and HIV-1 Vpu or SIVtan Env and stained tetherin via the HA tag. Co-expression of either Vpu or SIVtan Env reduced cell surface tetherin levels to a similar degree to that recently demonstrated for SIV Nef (3). Using this assay cell surface levels of the A100D mutant tetherin were significantly reduced by expression of HIV-1 Vpu (Fig. S3C) but only slightly reduced by SIVtan Env expression, concordant with its insensitivity to antagonism by SIVtan Env (Fig. 3A).
To further examine the interaction between SIVtan Env and tetherin, we deleted SIVtan Env to essentially gp41 (residues 551–880) with an N-terminal HA tag. We then tested whether this mutant was able to sequester tetherin from the cell surface. HA-gp 41 was well expressed (Fig. S3D), but was not able to antagonize tetherin's restriction of viral release (Fig. S3 E–G), despite being present at the cell surface as demonstrated staining of the HA tag followed by flow cytometry (Fig. S3 H and I). Importantly, gp41 expression was unable to reduce tetherin levels at the cell surface (Fig. S3J). Together with data in Fig. 2, these data show that SIVtan Env and tetherin extracellular domains are required for relocalization of tetherin.
Co-Localization Between Tetherin and Mature HIV-1 Gag at the Cell Surface Is Abrogated by the Expression of SIVtan Env.
To test whether SIVtan Env can sequester tetherin away from budding HIV-1 virions, we expressed human tetherin with HIV-1 vector plasmids alone or with SIVtan Env. In this experiment we used an HA-tagged SIVtan Env together with an N terminally Xpress-tagged tetherin (7) which has a similar distribution to the ectodomain HA tagged protein [compare HA tetherin (Fig. 3B) to Xpress Tetherin (Fig. 4A, middle image)]. To identify mature HIV-1 virions, we used an antibody that recognizes only cleaved p17 matrix and not intact p55 Gag (21). Representative images indicate that in the absence of SIVtan Env tetherin and mature virions co-localize at the cell surface with some internal tetherin staining (Fig. 4A; further images in Fig. S4), as has been previously described (1, 9). On expression of SIVtan Env a striking re-localization of tetherin takes place and the tetherin and HIV-1 p17 signals separate (Fig. 4B; further images in Fig. S5). The SIVtan Env and tetherin proteins co-localize in intracellular compartments, suggesting that SIVtan Env antagonizes tetherin by sequestering it from the site of HIV-1 budding at the cell surface. We assume that p17 staining at the cell surface in the presence of SIVtan Env is due to restriction of HIV-1 virion release by residual tetherin. This is consistent with the observed increase in levels of cell-associated p24 in the presence of tetherin even after potent antagonism by SIVtan Env (Fig. S1B).
Fig. 4.
Tetherin and HIV-1 Gag co-localize in the absence of SIVtan Env and separate on its expression. (A) Double stained 293T cells co-expressing HIV-1 Gag (green), labeled with p17 matrix specific antibody, and tetherin (red), labeled via the N-terminal Xpress tag. Merged images are shown in the third row. (B) Triple stained 293T cells expressing Gag, labeled with anti-p17 matrix (green), tetherin labeled via an N-terminal Xpress tag (red), and SIVtan Env labeled via an HA tag (blue). Images are pseudocolored. Merged images are shown in the bottom panel. Images are representative of 13 fields, all of which had similar protein localizations.
Tetherin and SIVtan Env Co-Localize in Tubulo-Vesicular Structures.
To examine the distribution of tetherin and SIVtan Env at higher resolution, 293T cells were transfected with HIV vectors, N-terminally tagged human tetherin (X-THN) and HA-SIVtan Env, and were fixed and prepared for cryosectioning. Staining of semithin (0.5-μm) sections for immunofluorescence showed that X-THN and HA-SIVtan Env were both expressed, and co-localized on double-transfected cells (Fig. S6). Ultrathin (50- to 60-nm) sections were immunolabeled for electron microscopy (EM). On 293T cells expressing X-THN in the absence of SIVtan Env, tetherin was mainly seen on intracellular tubulo-vesicular membranes of 30- to 60-nm diameter that were often found in the vicinity of the Golgi apparatus (Fig. 5 A and B). Sometimes, these membranes were tightly clustered into round or oval structures of 0.3- to 1-μm diameter, which likely correspond to the brightly stained spots seen by immunofluorescence. On some of the cells, tetherin was also seen at the cell surface and on the surface associated microvilli (Fig. 5A). On cells co-expressing X-THN and the HA-SIVtan Env, tetherin was still seen in the loose or tightly clustered membranes in the Golgi region (Fig. 5 C and D), although the number of cells with surface tetherin staining was significantly reduced, consistent with Fig. 3. The HA-SIVtan Env protein was likewise localized on tightly clustered or more loosely arranged tubulo-vesicular membranes (Fig. 5 E–G). These membranes were again often observed in the vicinity of the Golgi apparatus, and some gold particles could be seen over the Golgi stacks (Fig. 5F). Double staining revealed that in cells expressing both tetherin and HA-SIVtan Env, the two proteins co-localized in the clusters of tubulo-vesicular membranes (Fig. 5 H–J), some of which were prominently expanded. Gold particles, identifying tetherin or the HA-SIVtan Env protein, could sometimes be observed in the same small vesicles (Fig. 5 I and J). The proximity to the Golgi apparatus suggests that this tubulo-vesicular compartment may be related to the trans-Golgi network (TGN), although recycling endosomes, which are also located in this region, cannot be excluded.
Fig. 5.
Tetherin and SIVtan Env co-localize specifically in tubulo-vesicular structures. (A and B) 293T cells expressing HIV-1 Gag and X-THN were stained with anti-tetherin antibodies and 10 nm PAG. Tetherin is seen at the cell surface and on intracellular membranes which can be arranged in tight clusters of >0.5-μm diameter. (C and D) In 293T cells co-expressing HIV-1 Gag, X-THN, and HA-SIVtan Env, tetherin is still found on membranes in the Golgi area and on tight clusters of tubulo-vesicular membranes. (E–G) Staining with mouse anti-HA and 10 nm PAG shows that the HA-SIVtan Env protein is also on Golgi stacks, membranes in the Golgi area, and on tight clusters of tubulo-vesicular membranes, which can be expanded into large clusters (E). (H–J) Double labeling shows co-localization of HA-SIVtan Env and tetherin in tubulo-vesicular membranes. (I) Shows an enlargement of the area marked in H. Sometimes tetherin (PAG 10 nm, e.g., at the black arrows) can be seen in vesicles containing the HA-SIVtan Env (PAG 5 nm). (J) Tetherin (PAG 5 nm, e.g., at the black arrows) co-stains with HA-SIVtan Env (PAG 10 nm). No gold labeling was seen on untransfected cells. G, Golgi apparatus; m, mitochondrion; PM, plasma membrane. (Scale bars, 200 nm.) All images are representative of at least eight cells.
Discussion
We have shown that SIVtan Env is able to antagonize tetherin from Tantalus monkey, rhesus monkey, sooty mangabey and humans (Fig. 1 and Fig. S1). Mammalian tetherin sequences are divergent and this variability leads to species-specific sensitivity to tetherin antagonists (3, 4, 7–9) (Figs. 1 and 2). Indeed, SIVtan Env is unable to antagonize tetherin from pigs allowing us to use a chimeric porcine tetherin with a human transmembrane region to show that sensitivity to HIV-1 Vpu but not SIVtan envelope can be transferred with the human transmembrane region (Fig. 2). Since beginning this work SIV and HIV-2 Nef proteins have also emerged as species-specific tetherin antagonists (3, 4). The broad anti-tetherin activity of SIVtan Env is therefore in contrast to the more restricted activities of HIV-1 Vpu or SIV Nef (3, 4, 7, 8).
Tetherin has been under significant selective pressure throughout mammalian evolution, presumably from viral antagonists such as those encoded by primate lentiviruses (Vpu, Nef, Env), Ebola virus (Env), or herpes viruses (K5) (1–4, 7–9, 17, 22). Supporting the notion that Env proteins might have provided selective pressure we found that changing a single human tetherin amino acid, which is under adaptive selection, almost completely abolished its sensitivity to antagonism by SIVtan Env without compromising its antiviral activity. Notably, this change did not impact on sensitivity to the alternative antagonist HIV-1 Vpu (7), suggesting that different sites on tetherin interact with SIVtan Env and HIV-1 Vpu. In the experiments presented we have exogenously expressed both tetherin and SIVtan Env, and it is not certain that the protein levels expressed represent those found naturally during infection in vivo. However, the decrease in HIV-1 viral release in the absence of Vpu is similar to that observed in IFN treated Jurkat T cells and human primary cells derived from peripheral blood (1, 23). This implies similar effective endogenous protein levels in relevant cells. Moreover, only a small amount of transfected SIVtan Env plasmid was required to overcome transfected tetherin both in HeLa and 293T cells, and thus we believe that our conclusions are reasonable.
It is remarkable that primate lentiviruses, which only encode nine or so genes, have evolved the ability to use three proteins to antagonize tetherin. It is also striking that all of these proteins have been described as down regulating CD4, albeit through diverse mechanisms (24, 25). The mechanism of tetherin inhibition remains incompletely characterized but again, there are clearly significant mechanistic differences between the various viral antagonists. HIV-1 Vpu appears to lead to a reduction of steady-state tetherin levels (6, 7, 9) and exclusion from sites of particle assembly at the plasma membrane (2, 26). Recent data suggest that Nef may remove tetherin from the cell surface (3) as it does other cell surface molecules (27). Fluorescence microscopy and flow cytometry herein show that SIVtan Env also reduces steady state cell surface expression of tetherin (Figs. 3 and 4) and electron microscopy suggests that this is the result of sequestration of tetherin in peri-nuclear tubulo-vesicular structures, likely the TGN and/or recycling endosomes (Fig. 5). It is currently unclear whether tetherin is directly sequestered from the cell surface, or whether it is prevented from reaching the plasma membrane by SIVtan Env.
It seems paradoxical that SIVtan Env antagonizes human tetherin more effectively than it antagonizes tetherin from its native species, Tantalus monkey. This may be due to anti-tetherin activity of the SIVtan Nef protein (4), although it may also be because the virus was isolated by growth in the human T-cell line MOLT4 (18). Such an isolation protocol may select for mutations that improve tropism for human cells. Point mutations in a virus can influence sensitivity to restriction factors including TRIM5 (28) and APOBEC3G (29), and therefore species-specific tropism. It is clear that SIVtan Env can antagonize tetherin and it is unlikely that this property developed entirely during in vitro isolation. It is noteworthy that SIVmac grows well in human T cells isolated from peripheral blood (30) although it does not appear to be able to antagonize human tetherin (3, 4), and it may be that these culture systems do not express high levels of tetherin unless induced by IFN.
The translational implications of our findings for HIV therapeutics are two-fold. Firstly, pharmacological blockade of HIV-1 Vpu anti-tetherin function may lead to viral escape through acquisition of anti-tetherin activity by HIV-1 Env or Nef. Secondly, the discovery of antagonist resistant tetherin mutants, for example tetherin A100D, suggests tetherin binding drugs might be found that protect it from viral encoded countermeasures by mimicking this and similar mutations (7, 8).
Methods
Cell Lines, Plasmids, and Viral Infection Assays.
SIVtan envelope was PCR cloned from a full-length SIVtan clone (18), obtained via the NIH AIDS reagents program, into the pCAGGS expression vector. An N-terminal HA-tagged SIVtan Env deletion mutant consisting of amino acids 551–880, essentially gp41, was also constructed. Human and Tantalus monkey tetherins have been described (7). Rhesus tetherin was cloned from rhesus macaque kidney LLC-MK2 cells as described (7). Identical porcine tetherin cDNAs were cloned from ST IOWA and MPK porcine cell lines. Sooty mangabey tetherin was a gift from Welkin Johnson. Wild-type human tetherin was N-terminal Xpress epitope tagged by cloning into pCDNA4 (Invitrogen), a gift from Gordon Perkins. Ectodomain HA-tagged human tetherin was a gift from Stuart Neil (8). Preparation of VSV-G pseudotyped, YFP encoding HIV-1 has been described (7). One hundred nanograms tetherin constructs were co-transfected along with 0–200 ng HIV-1 Vpu or SIVtan Env constructs or empty vector (pCDNA3.1, Invitrogen).
Western Blots, Immunofluorescence, and Flow Cytometry.
Western blots were performed as described (7, 31). For immuno-fluorescence, cells seeded on coverslips were transfected as above. After 48 h cells were fixed, quenched, blocked as described (32), and permeabilized with 0.1% TX-100. Primary antibodies used were: rat FITC anti-HA antibody (Roche), mouse anti-Xpress IgG1 (Invitrogen), and mouse anti-p17 antibody 4C9 ARP342 (Centre for AIDS Reagents, NIBSC, Potters Bar, UK). Secondary antibodies were: goat anti-mouse Alexa Fluor 488 or goat anti-mouse-Alexa Fluor 594 (Invitrogen). Confocal microscopy was performed using a Leica TCS SPE, DM2500 Microscope (Leica Microsystems). For cell surface staining, cells were transfected with ectodomain HA tagged tetherin (8) and either empty vector, HIV-1 Vpu or SIVtan Env. After 48 h, cells were trypsinized and incubated with primary anti-HA antibody (Covance) followed by a secondary anti-mouse FITC conjugated antibody (Dako). Tetherin staining was also performed using anti-tetherin primary antibody (MaxPab, Abnova) with the same secondary (Fig. S3J).
Immuno-Electron Microscopy.
Transfected 293T cells were fixed, embedded, and frozen for cryosectioning as described (32). Ultrathin (50- to 60-nm) cryosections were stained with anti-tetherin (MaxPab, Abnova) or mouse anti-HA (Covance), a rabbit anti-mouse bridging antibody (DakoCytomation), and 10 nm protein A-gold (PAG, the EM Lab, Utrecht University, The Netherlands). For double staining, sections were first labeled with rat FITC anti-HA antibody (Roche), rabbit anti-rat bridging antibodies (Dako), and 5 or 10 nm PAG. Sections were fixed, quenched, and incubated with anti-tetherin, rabbit anti-mouse antibodies, and a second PAG probe of different size (10 or 5 nm, respectively). Sections were embedded in uranyl acetate in methylcellulose and examined with a Technai G2 Spirit transmission electron microscope (FEI).
Supplementary Material
Acknowledgments.
We thank Paul Bieniasz (Aaron Diamond AIDS Research Center and The Rockefeller University, New York), Stuart Neil (Department of Infectious Disease, King's College London, Guy's Hospital, London), Gordon Perkins (Department of Immunology, University College London), Frank Kirchhoff (Institute of Molecular Virology, Universitätsklinikum, Ulm, Germany), Welkin Johnson (Department of Microbiology and Molecular Genetics, Harvard Medical School, New England Primate Research Center, Southborough, MA), and Stephane Hue (Medical Research Council Centre for Medical Molecular Virology, University College London) for reagents, and Bruce Cheesebro, Marcelo Soares, and Beatrice Hahn for reagents via the National Institutes of Health AIDS Research and Reference Reagent Program. This work was supported by Wellcome Trust fellowships to R.K.G. (WT081772MA) and G.J.T. (WT076608), European Commission Sixth Framework Programme Project Grant LSHB-CT-2006-037377 (to G.M. and Y.T.), the Medical Research Council, and HIV Anti-Capsid Assembly and Envelope Incorporation research network Grant HEALTH-F3–2008-201095 (to P.M., A.P.-M., and M.M.), and NIHR UCLH/UCL Comprehensive Biomedical Research Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. FJ527910 (pig tetherin), FJ345303 (tantalus tetherin), and GQ304749 (rhesus tetherin)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0907075106/DCSupplemental.
References
- 1.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 2.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia B, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kupzig S, et al. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell RS, et al. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RK, et al. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 2009;5:e1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNatt MW, et al. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffinet C, et al. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. Inhibition of Lassa and Marburg virus production by tetherin. J Virol. 2008;83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchhoff F. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat Rev Microbiol. 2009;7:467–476. doi: 10.1038/nrmicro2111. [DOI] [PubMed] [Google Scholar]
- 12.Bour S, Strebel K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J Virol. 1996;70:8285–8300. doi: 10.1128/jvi.70.12.8285-8300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bour S, Schubert U, Peden K, Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: A Vpu-like factor? J Virol. 1996;70:820–829. doi: 10.1128/jvi.70.2.820-829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abada P, Noble B, Cannon PM. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J Virol. 2005;79:3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noble B, Abada P, Nunez-Iglesias J, Cannon PM. Recruitment of the adaptor protein 2 complex by the human immunodeficiency virus type 2 envelope protein is necessary for high levels of virus release. J Virol. 2006;80:2924–2932. doi: 10.1128/JVI.80.6.2924-2932.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Tortorec A, Neil SJ. Antagonism and intracellular sequestration of human tetherin by the HIV-2 envelope glycoprotein. J Virol. 2009 doi: 10.1128/JVI.01515-09. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soares MA, et al. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology. 1997;228:394–399. doi: 10.1006/viro.1996.8387. [DOI] [PubMed] [Google Scholar]
- 19.Dube M, et al. Suppression of tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J Virol. 2009;83:4574–4590. doi: 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollason R, Korolchuk V, Hamilton C, Jepson M, Banting G. A CD317/tetherin-RICH2 complex plays a critical role in the organization of the subapical actin cytoskeleton in polarized epithelial cells. J Cell Biol. 2009;184:721–736. doi: 10.1083/jcb.200804154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferns RB, Tedder RS, Weiss RA. Characterization of monoclonal antibodies against the human immunodeficiency virus (HIV) gag products and their use in monitoring HIV isolate variation. J Gen Virol. 1987;68(Pt 6):1543–1551. doi: 10.1099/0022-1317-68-6-1543. [DOI] [PubMed] [Google Scholar]
- 22.Bartee E, McCormack A, Fruh K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006;2:e107. doi: 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levesque K, Finzi A, Binette J, Cohen EA. Role of CD4 receptor down-regulation during HIV-1 infection. Curr HIV Res. 2004;2:51–59. doi: 10.2174/1570162043485086. [DOI] [PubMed] [Google Scholar]
- 25.Lindwasser OW, Chaudhuri R, Bonifacino JS. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr Mol Med. 2007;7:171–184. doi: 10.2174/156652407780059177. [DOI] [PubMed] [Google Scholar]
- 26.Jouvenet N, et al. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindler M, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 28.Ylinen L, Keckesova Z, Wilson SJ, Ranasinghe S, Towers GJ. Differential restriction of HIV-2 and SIVmac by TRIM5alpha alleles. J Virol. 2005;79:11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci USA. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniel MD, et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 31.Bouamr F, et al. The C-terminal portion of the Hrs protein interacts with Tsg101 and interferes with human immunodeficiency virus type 1 Gag particle production. J Virol. 2007;81:2909–2922. doi: 10.1128/JVI.01413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol. 2007;177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.