Abstract
Growing evidence shows that microRNAs (miRNAs) regulate various developmental and homeostatic events in vertebrates and invertebrates. Osteoblast differentiation is a key step in proper skeletal development and acquisition of bone mass; however, the physiological role of non-coding small RNAs, especially miRNAs, in osteoblast differentiation remains elusive. Here, through comprehensive analysis of miRNAs expression during osteoblast differentiation, we show that miR-206, previously viewed as a muscle-specific miRNA, is a key regulator of this process. miR-206 was expressed in osteoblasts, and its expression decreased over the course of osteoblast differentiation. Overexpression of miR-206 in osteoblasts inhibited their differentiation, and conversely, knockdown of miR-206 expression promoted osteoblast differentiation. In silico analysis and molecular experiments revealed connexin 43 (Cx43), a major gap junction protein in osteoblasts, as a target of miR-206, and restoration of Cx43 expression in miR-206-expressing osteoblasts rescued them from the inhibitory effect of miR-206 on osteoblast differentiation. Finally, transgenic mice expressing miR-206 in osteoblasts developed a low bone mass phenotype due to impaired osteoblast differentiation. Our data show that miRNA is a regulator of osteoblast differentiation.
Keywords: Connexin43, miR-206
The osteoblast, a cell type of a mesenchymal origin, plays a major role in skeletal development and bone formation (1, 2). Understanding the regulatory mechanism of osteoblast differentiation is a prerequisite for developing strategies to treat bone loss diseases such as osteoporosis (3–5). In the last two decades, progress in molecular and genetic research has uncovered various regulatory processes of osteoblast differentiation (1, 2, 4). Central to this regulation are transcription factors; Runx2, Osterix, and β-catenin are, to date, the transcription factors known to be essential for osteoblast differentiation (2). In addition, while some transcription factors, including C/EBPβ, Smad1, and Smad5, bind to Runx2 and enhance its transcriptional activity, others, such as Twist, inhibit Runx2 transcriptional activity (6). However, given the fact that the number of coding genes in vertebrates and invertebrates (which lack a skeleton) is comparable (7), there must be additional mechanisms for controlling skeletal development other than transcriptional regulation of gene expression.
Recently, miRNAs have emerged as important regulators in various developmental, physiological, and pathological conditions such as tumorigenesis, viral infection, and cell differentiation and function (8–10). miRNAs are single-stranded small RNA molecules that are approximately 21 or 22 nucleotides long (9). They do not encode protein; instead, they regulate the level of other proteins by decreasing messenger RNA (mRNA) levels or inhibiting translation by binding the 3′UTR of the target mRNA (8). Surprisingly, non-coding RNA accounts for 98% of all genomic output in humans (11), and it has been proposed that the proportion of non-coding RNA to protein-coding RNA is correlated with developmental complexity (12).
Previous reports implicated miRNAs in the differentiation of osteoclasts and osteoblasts (13–21). However, their importance in the regulation of osteoblast differentiation in vivo, if any, remains to be established. Here we show that one particular miRNA, miR-206, is expressed in the osteoblastic cell lineage and that its expression gradually decreases in parallel with osteoblast differentiation. Interestingly, modulating miR-206 expression in osteoblasts markedly affects their differentiation potential in part by altering the accumulation of connexin 43 (Cx43). Finally, osteoblast-specific expression of miR-206 in vivo leads to severe bone loss due to impairment of osteoblast differentiation. Thus, this study reveals a physiological regulatory mechanism of osteoblast differentiation mediated by miRNA.
Results
Identification of miRNAs Whose Expression Varies During Osteoblast Differentiation.
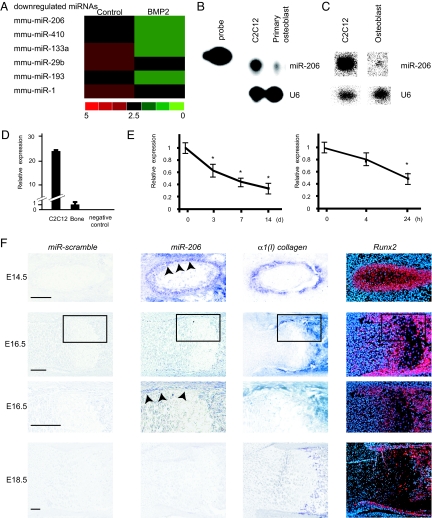
To study the potential involvement of miRNAs in osteoblast differentiation, we first attempted to identify miRNAs that are expressed in the osteoblastic cell lineage, particularly miRNAs whose expression is altered during osteoblast differentiation. To that end, we treated multipotent C2C12 mesenchymal progenitor cells with recombinant BMP-2 for 2 days, an established model for studying osteoblast differentiation (22). We then comprehensively analyzed the expression of miRNAs before and after BMP-2 treatment using a microarray that detects all known miRNAs (23). miR-133a was downregulated by BMP-2 treatment (Fig. 1A and Figs. S1 and S2A) as previously reported (13), suggesting that the experiment was properly conducted. Osteoblasts express many miRNAs, and most of the miRNAs (36%) were downregulated by BMP-2 treatment, while only 4% of them were upregulated (Fig. 1A and Fig. S1). Of these, we were interested in miR-206 because its expression was most significantly downregulated during osteoblast differentiation (Fig. 1A). As miR-206 was originally shown to be expressed exclusively in skeletal muscle and heart (24–28), we first verified its expression in the osteoblastic lineage using primary mouse osteoblasts and bones by four different experiments. First, as shown by an RNase protection assay, miR-206 was clearly expressed in primary osteoblasts (Fig. 1B). Second, by Northern blot analysis, which is almost 10 times less sensitive than an RNase protection assay (29, 30), miR-206 was also shown to be expressed in primary osteoblasts (Fig. 1C). Third, by real-time PCR analysis specific to miR-206, we observed miR-206 expression both in femur and primary osteoblasts (Fig. 1 D and E), and interestingly, miR-206 expression gradually decreased during the course of osteoblast differentiation (Fig. 1E). Fourth and most importantly, to investigate the dynamic pattern of miR-206 expression in bone, we performed in situ hybridization analysis using DIG-labeled probes. At E14.5, miR-206 was expressed in muscle and perichondrium osteoblastic cells, whose identity was verified by the coexpression of α1(I) collagen and Runx2, markers for osteoblasts (Fig. 1F). At E16.5, miR-206 was still expressed in the cells of the bone collar, although the expression was decreased compared to the expression at E14.5 (Fig. 1F). At E18.5, miR-206 expression in bone was close to background level (Fig. 1F). The gradual decrease of miR-206 expression during skeletogenesis in vivo is consistent with its gradual decrease of expression during the course of in vitro osteoblast differentiation. To further confirm in vivo expression of miR-206 in osteoblasts, we also performed double staining for in situ hybridization to detect miR-206 and immunohistochemistry to detect Runx2. High resolution confocal microscopic analysis revealed that miR-206 colocalize with Runx2 in osteoblast (Fig. S3). Taken together, these four independent experiments confirmed that miR-206 is expressed in the osteoblastic cell lineage.
Fig. 1.
Expression of miR-206 during osteoblast differentiation. (A) miRNA array expression data from C2C12 cells cultured in growth medium (Control) or in differentiation medium containing BMP-2. Red denotes high expression and green denotes low expression relative to the median; only the representative miRNA nodes that were significantly downregulated in the differentiation medium are shown. (B–D) Expression of miR-206 in osteoblasts: RNase protection assay (B), Northern blot analysis (C), and quantitative RT-PCR analysis (D) to detect miR-206 expression in osteoblasts. Total RNA was isolated from primary mouse osteoblasts, mouse femur or C2C12 cells. U6 RNA was used as a loading control. Note the distinct expression of miR-206 in primary osteoblasts and bone (B–D). (E) Change in miR-206 expression during osteoblast differentiation: quantitative RT-PCR analysis. Mouse primary osteoblasts were treated in differentiation medium (Left) or with the addition of BMP-2 (Right) for each indicated length of time. Note the significant decrease in parallel with the progression of osteoblast differentiation. *, P < 0.05 vs. 0 time point, n = 6. (F) In situ hybridization analysis of miR-206 expression in mouse embryos: Rib (E14.5, Top) and femur (E16.5, middle and E18.5, Bottom) cryosections. Bottom panel of E16.5 embryo shows the higher magnification of the black rectangular region in the top panel. Adjacent sections were hybridized with scramble-miRNA (left), miR-206 (Middle Left), α1(I) collagen (Middle Right), and Runx2 (Right) probes. Note the miR-206 expression in perichondrium osteoblastic cells at E14.5 (arrowheads) and in cells of bone collar at E16.5 (arrowheads). (Scale bar, 100 μm.)
miR-206 Regulates Osteoblast Differentiation.
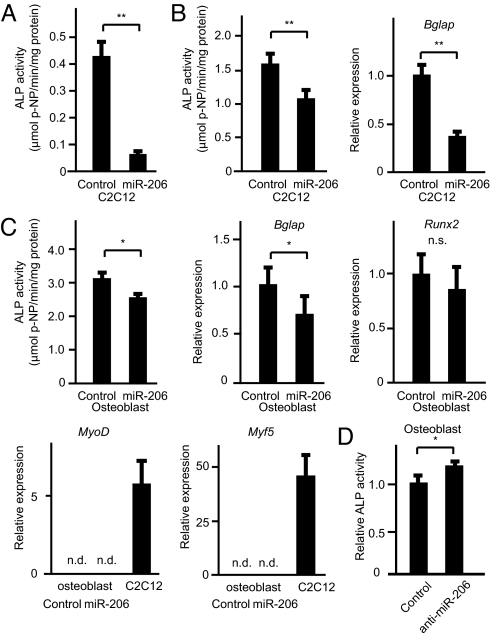
The decrease in the expression of miR-206 during osteoblast differentiation prompted us to test if miR-206 inhibits osteoblast differentiation. To this end, we infected a vector expressing both miR-206 and a blasticidin-resistance gene into C2C12 cells and isolated five stable blasticidin-resistant clones that also express miR-206 to determine if continuous expression of miR-206 affected their ability to differentiate into osteoblasts. As controls, we also infected either an empty vector or a miR-133-expressing vector. C2C12 cells expressing empty vector differentiated normally into the osteoblastic lineage upon BMP-2 treatment. In contrast, osteoblastic differentiation of C2C12 cells expressing miR-133 was significantly impaired (Fig. S2B), as previously reported. Importantly, none of the five clones expressing miR-206 differentiated into the osteoblastic lineage, as shown by the lack of induction of alkaline phosphatase activity (Fig. 2A). To rule out the possibility that stable expression of miR-206 altered the properties of C2C12 cells, we also transiently transfected a miR-206-expressing vector into C2C12 cells. In this transient DNA transfection assay, miR-206 also repressed osteoblastic differentiation (Fig. 2B). To test if miR-206 regulates osteoblast differentiation in a physiological manner, we next used primary mouse osteoblasts because C2C12 is a myogenic cell line. The results showed that continuous expression of miR-206 significantly inhibited osteoblast differentiation as demonstrated by the decrease in alkaline phosphatase activity and bglap expression (Fig. 2C and Fig. S4). Interestingly, miR-206 expression did not affect Runx2 mRNA expression, indicating that miR-206 regulates osteoblast differentiation independently of Runx2 (Fig. 2C).
Fig. 2.
Regulation of osteoblast differentiation by miR-206. (A and B) Effect of miR-206 expression on BMP-2-dependent C2C12 cell differentiation: C2C12 cells constitutively expressing miR-206 (A) or transiently transfected with miR-206 (B). Alkaline phosphatase activity (A and B, Left) and bglap gene expression (B, Right) were analyzed. Note the decreased osteoblastic differentiation in miR-206 expressing cells. **, P < 0.01, n = 6–8. (C) Effect of miR-206 continuous expression on primary mouse osteoblast differentiation: primary mouse osteoblasts infected with pAd-miR-206 or control adenovirus were cultured. Subsequently, alkaline phosphatase activity assay (Top Left) and quantative RT-PCR analysis for the indicated genes were analyzed. GAPDH was used as an internal control. n.s., not significant. n.d., not detected. *, P < 0.05, n = 6–8. (D) Effect of miR-206 knockdown on osteoblast differentiation: osteoblasts were transfected with a miR-206 inhibitor or control. Subsequently, alkaline phosphatase activity was analyzed. Note the significant increase by the miR-206 inhibitor. *, P < 0.05, n = 6.
Because it has been shown that expression of miR-206 induces myogenic differentiation (24, 27, 28), we asked whether miR-206 expression induces myogenic transdifferentiation of osteoblasts; however, no expression of myogenic genes such as MyoD or Myf5 (31) was detected, indicating that miR-206 does not induce myogenic differentiation (Fig. 2C)
Since overexpression of miR-206 inhibits osteoblast differentiation, we next asked whether decreasing miR-206 expression would accelerate their differentiation. Indeed, knockdown of miR-206 significantly induced osteoblast differentiation (Fig. 2D). Taken together, these results demonstrate that miR-206, which is expressed in the osteoblastic cell lineage, physiologically regulates osteoblast differentiation.
Connexin 43 Is One Molecular Target of miR-206 in Osteoblasts.
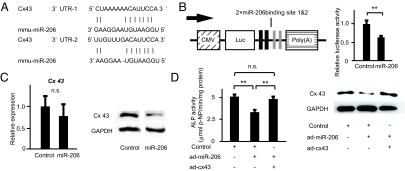
We next studied the molecular mechanism by which miR-206 inhibits osteoblast differentiation. To identify target genes of miR-206, we relied on a computational approach using two different established databases (32, 33). Among the many genes that were predicted to be potential targets by both databases, we focused on Cx43, a gap junction protein expressed in osteoblasts that plays a major role in osteoblast differentiation and function; indeed, Cx43-deficient mice display low bone mass due to osteoblast dysfunction (34, 35). Two putative target sequences for miR-206 were found in the 3′UTR region of Cx43 (Fig. 3A). At first, we tested if miR-206 regulated Cx43 expression using a reporter plasmid in which the two putative binding sites of the Cx43 3′UTR were cloned into the 3′UTR of the luciferase gene (Fig. 3B). As expected from in silico analysis, ectopic expression of miR-206 significantly decreased luciferase activity (Fig. 3B); furthermore, ectopic expression of miR-206 downregulated endogenous Cx43 protein expression without affecting Cx43 mRNA expression (Fig. 3C). Taken together, these results identify Cx43 as a bona fide target of miR-206 in vivo.
Fig. 3.
Identification of miR-206 target genes in osteoblast differentiation. (A) Alignment of miR-206 showing complementary pairing to the Cx43 3′ UTR. (B) Schematic presentation of the reporter plasmid used to analyze the effect of the Cx43 3′ UTR on luciferase activity (Left). Effect of miR-206 expression on a luciferase reporter plasmid carrying the Cx43 3′ UTR was analyzed (Right). Cells were transfected with either the miR-206 expression plasmid or a control. The ratio of reporter (firefly) to control (renilla) was plotted. CMV: cytomegalovirus promoter, Luc: luciferase. **, P < 0.01, n = 6. (C) miR-206 targets Cx43 and regulates its expression. Cells were transfected with the miR-206 expression plasmid or a control. Quantitative RT-PCR analysis (Left) and Western blot analysis for Cx43 (Right) were subsequently performed. GAPDH was used as an internal control. n.s., not significant; n = 6–8. (D) Cx43 rescues the inhibitory effect of miR-206 on osteoblast differentiation. Primary osteoblasts expressing miR-206 were infected with either the Cx43-expressing virus (pAd-Cx43) or control adenovirus. Subsequently, alkaline phosphatase activity was analyzed. Note that the inhibitory effect of miR-206 on osteoblast differentiation was reversed by co-expression of Cx43. (Left, alkaline phosphatase activity; Right, Western blot) **, P < 0.01, n = 6.
To address if Cx43 is a physiologically important target of miR-206 during osteoblast differentiation, we asked whether restoring Cx43 expression rescued the impairment of osteoblast differentiation caused by continuous miR-206 expression. Continuous expression of miR-206 in osteoblasts decreased osteoblast differentiation and Cx43 protein expression (Fig. 3C). However, when we co-expressed Cx43 together with miR-206, the inhibitory effect of miR-206 on osteoblast differentiation was markedly rescued (Fig. 3D). Importantly, the expression level of Cx43 protein was similar between control cells and cells co-expressing both miR-206 and Cx43, which indicates that restoration of Cx43 protein expression is sufficient to obtain normal osteoblast differentiation in miR-206-expressing osteoblasts (Fig. 3D).
miR-206 Is a Regulator of Bone Formation in Vivo.
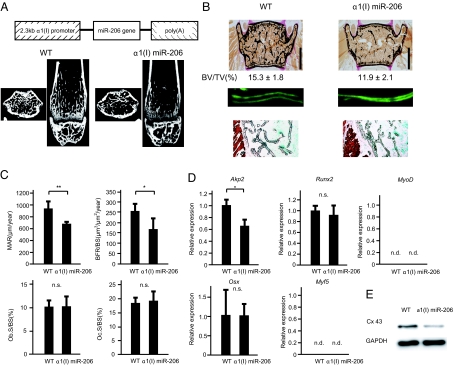
Lastly, to address the in vivo role of miR-206 in bone formation, we generated transgenic (tg) mice specifically expressing miR-206 in osteoblasts using the α1(I) collagen promoter (Fig. 4A and Fig. S5). α1(I) miR-206 tg mice displayed a low bone mass phenotype both in trabecular and cortical bones by μCT analysis and histology (Fig. 4 A and B). Furthermore, bone histomorphometric analysis revealed that the bone formation rate, an indicator of osteoblast function, was significantly decreased in α1(I) miR-206 mice (Fig. 4 B and C). In contrast, osteoclast surface, a marker of bone resorption, was similar in wild-type mice and α1(I) miR-206 tg mice, indicating that osteoclastic bone resorption was not affected (Fig. 4C). The expression of osteoblastic marker genes such as Akp2 was significantly downregulated in α1(I) miR-206 tg mice (Fig. 4D). Importantly, Cx43 protein expression was significantly reduced in α1(I) miR-206 tg mice, while the expression of Runx2 and Osterix was not affected in these mice (Fig. 4 D and E), further indicating that miR-206 regulates osteoblast differentiation through Cx43 independently of Runx2 and Osterix. Of note, the lack of expression of myogenic marker genes in real-time PCR and the absence of Troponin I protein in immunohistochemistry (36) clearly demonstrated that there was no ectopic muscle differentiation in α1(I) miR-206 tg mice (Fig. 4 B and D). Collectively, overexpression of miR-206 in osteoblasts inhibited osteoblast differentiation, which led to low bone mass in vivo.
Fig. 4.
Low bone mass in α1(I) miR-206 tg mice due to reduced bone formation. (A) Structure of the construct for osteoblast-specific α1(I) miR-206 tg mice (Top). μCT analysis of the femurs of 6-week-old female wild-type (WT) or α1(I) miR-206 tg mice (Bottom). Axial section (Left) and coronal section (Right). (B) Histological analysis of the vertebrae of 6-week-old female WT or α1(I) miR-206 tg mice. Von Kossa staining (Top). Bone volume per tissue volume (BV/TV). (Scale bars, 1 mm.) The distance between the two calcein labels represents the bone formation rate (middle). Note the significant decrease in bone formation in α1(I) miR-206 tg mice. Immunohistochemical staining for Troponin I (Bottom). Note the absence of Troponin I immunoreactivity (brown) in the vertebrae of α1(I) miR-206 tg mice in contrast to the intense staining in skeletal muscle. (C) Histomorphometric analysis of the vertebrae of 6-week-old female mice. Mineral apposition rate (MAR), bone formation rate over bone surface area (BFR/BS), osteoblast surface area over bone surface area (Ob.S/BS), osteoclast surface area over bone surface area (Oc.S/BS). n.s, not significant. *, P < 0.05, **, P < 0.01, n = 6–7. (D) Gene expression in α1(I) miR-206 tg mice. Quantitative RT-PCR analysis of osteoblastic genes (Akp2, Runx2, and Osx) and myogenic genes (Myf5, MyoD). Primary calvarial osteoblasts were isolated from WT or α1(I) miR-206 tg mice and used for subsequent analyses. n.s.: not significant. *, P < 0.05, n = 6. (E) Western blot analysis of calvaial bones of 2-week-old female mice. Note the decrease in Cx43 protein expression in α1(I) miR-206 tg mice.
Discussion
We demonstrate here an inhibitory role of miR-206 during osteoblast differentiation. First, we showed that miR-206 is expressed in the osteoblastic lineage and that this expression gradually decreases during osteoblast differentiation. Then we demonstrated that modulating the expression of miR-206 in osteoblasts affects osteoblast differentiation and that one of the targets of miR-206 is Cx43. Finally, we showed that miR-206 regulates osteoblast differentiation in vivo. Recent reports suggested the involvement of miRNAs in osteoblast differentiation (13–20). However, these reports were based only on in vitro observations using a cell line. To our knowledge, this study demonstrates an in vivo regulatory role of miRNA in osteoblast differentiation.
It was previously shown that miR-206 induces myogenic differentiation (24, 27, 28). Therefore, we were concerned that the observed inhibitory effect of miR-206 in osteoblast differentiation was attributable only to an increase of differentiation into the myoblastic lineage, or alternatively, that miR-206 transdifferentiates osteoblasts into the myoblastic lineage. However, the experimental evidence argues against these hypotheses. Indeed, primary osteoblasts continuously expressing miR-206 do not express myogenic markers such as MyoD or Myf5. Furthermore, α1(I) miR-206 tg mice do not demonstrate any evidence of ectopic muscle differentiation in bone. Notably, the fact that to overexpress miR-206 we used the α1(I) collagen promoter, which is active only in cells committed to the osteoblastic lineage (37), and that these tg mice developed bone abnormalities strongly suggests that miR-206 directly affects osteoblast differentiation independently from any function it has during myogenic differentiation.
There was also another concern that miR-206 was expressed only in myogenic cells and inhibited osteoblastic genes in them. However, we observed that miR-206 was expressed in osteoblastic cells by four different experimental procedures. Moreover, the fact that the knockdown of miR-206 in osteoblasts accelerated their differentiation demonstrates that miR-206 is expressed in osteoblasts and plays a role in their differentiation.
Muscle-specific miRNAs comprise a well-defined family consisting of miR-1, miR-133, and miR-206. Interestingly, while miR-1 and −133 are expressed in Drosophila and vertebrates, miR-206 is not expressed in Drosophila. Instead, it is only expressed in vertebrates. This suggests that miR-206 evolved at a different period from miR-1 and miR-133 and thus may play a role other than the regulation of myogenic differentiation exhibited by miR-1 and miR-133.
Our observation that the inhibitory effect of miR-206 on osteoblast differentiation was rescued by the restoration of Cx43 suggests that Cx43 is a bona fide target of miR-206 in osteoblast differentiation. Indeed, while miR-206-expressing osteoblasts have a defect in osteoblast differentiation, they do not show any proliferative abnormality. This result is consistent with the normal proliferation of Cx43-deficient osteoblasts (34, 35). However, given that a miRNA can regulate multiple target genes (8), the effect of miR-206 may not depend solely on Cx43. Indeed, although osteoblast-specific Cx43-deficient mice have a normal mineral apposition rate (34), α1(I) miR-206 tg mice have a 30% decrease in the same parameter, suggesting the involvement of other molecules in miR-206-mediated bone formation defects.
Interestingly, parathyroid hormone (PTH), a well-known regulator of osteoblast differentiation, has been shown to regulate Cx43 expression through a posttranscriptional modification of Cx43 mRNA (38), and the anabolic response of PTH is attenuated in Cx43-deficient mice (34). Because miR-206 regulates both osteoblast differentiation and Cx43 mRNA stability, we tested whether PTH regulates Cx43 through a modification of miR-206 expression. However, PTH did not affect miR-206 expression.
It is interesting that miR-206 is strongly expressed in perichondrium, whereas its expression in trabecular bone is less (Fig. 1F). Considering that osteoblasts are derived from immature osteoprogenitor cells located in the perichondrium (39), strong expression of miR-206 in the perichondrium suggests that miR-206 may work to keep osteoblast immature and decrease of miR-206 expression is important for proper osteoblastic differentiation, which is in agreement with our in vitro observations. Currently, the molecular mechanism accounting for the downregulation of miR-206 expression during osteoblast differentiation is unknown. Because miR-206 is expressed in myogenic (24–28), adipocytic (40), and osteoblastic cells, all of mesenchymal origin, and miR-206 regulates both myogenic and osteoblastic differentiation, it is tempting to hypothesize that transcription factors involved in the differentiation of mesenchymal stem cells into specific cell lineages also regulate miR-206 expression. In this context, myogenic factors were shown to bind the upstream sequences of miR-206 (26). Therefore, it is possible that essential transcription factors for osteoblast differentiation, such as Runx2 and Osterix, also regulate miR-206 expression. Indeed, there are many putative binding sites for these factors in the sequence upstream of miR-206.
In conclusion, we demonstrated a regulatory role of miRNA in osteoblast differentiation in vivo. From a clinical point of view, inhibiting miRNA expression (41) may lead to therapies for bone degenerative diseases such as osteoporosis.
Materials and Methods
Cell Culture, Microarrays, Alkaline Phosphatase Assay, and Dual-Luciferase Reporter Assay.
Primary osteoblast and C2C12 cells were cultured and alkaline phosphatase activity (Wako, LabAssay ALP) was measured as previously described (42). Microarray analysis was performed as previously described (23). The activities of luciferase were determined by the dual-luciferase reporter assay (Promega). Further details are provided in the SI Text.
Cloning and Gene Expression.
Genomic fragments of miR-206 precursors were amplified by PCR. miRNA expression was detected by an RNase protection assay using a mirVANA miRNA kit (Ambion) or quantitative RT-PCR with Mx3000P (Stratagene). Northern blot analysis was performed as previously reported (43). 3′-UTR of Cx43 was subcloned into downstream of the luciferase gene for Cx43–3′-UTR reporter construction.
Western Blot Analysis, Immunohistochemistry, and in Situ Hybridization.
Western blot analysis and immunohistochemistry were performed according to a standard protocol (42). In situ hybridization was performed using DIG labeled probe [miR-206, miR-scramble and α1(I) collagen] and 35S-labeled riboprobe (Runx2) as reported in ref. 44 with modifications. Further details are provided in the SI Text.
Transgenic Mice, Histology, and Histomorphometry.
The genomic fragment of the miR-206 precursor was cloned into a plasmid containing a 2.3-kb α1(I) collagen promoter and microinjected as described in ref. 37. We performed histomorphometric analysis using the Osteomeasure System (Osteometrics) as described in ref. 42. Further details are provided in the SI Text.
Statistics.
All data are presented as means ± SE. (n ≥ 6). We performed statistical analysis by Student's t test, and P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments.
We thank Dr. Gerard Karsenty for discussion and Takako Usami for technical assistance. This work was supported by the Japan Society for the Promotion of Science grants (to Y.A., K.S., and S.T.) and Uehara Memorial Foundation grant (to S.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909311106/DCSupplemental.
References
- 1.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009 doi: 10.1146/annurev.cellbio.042308.113308. in press. [DOI] [PubMed] [Google Scholar]
- 2.Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99:1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 3.Khosla S, Westendorf JJ, Oursler MJ. Building bone to reverse osteoporosis and repair fractures. J Clin Invest. 2008;118:421–428. doi: 10.1172/JCI33612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen CJ. Clinical practice. Postmenopausal osteoporosis. N Engl J Med. 2005;353:595–603. doi: 10.1056/NEJMcp043801. [DOI] [PubMed] [Google Scholar]
- 5.Sambrook P, Cooper C. Osteoporosis. Lancet. 2006;367:2010–2018. doi: 10.1016/S0140-6736(06)68891-0. [DOI] [PubMed] [Google Scholar]
- 6.Franceschi RT, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Ann N Y Acad Sci. 2007;1116:196–207. doi: 10.1196/annals.1402.081. [DOI] [PubMed] [Google Scholar]
- 7.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 8.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 9.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 11.Mattick JS. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luzi E, et al. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 15.Mizuno Y, et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun. 2008;368:267–272. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105:1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuddenham L, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 19.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe T, et al. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 21.Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 2007;101:996–999. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 22.Katagiri T, et al. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohjoh H, Fukushima T. Marked change in microRNA expression during neuronal differentiation of human teratocarcinoma NTera2D1 and mouse embryonal carcinoma P19 cells. Biochem Biophys Res Commun. 2007;362:360–367. doi: 10.1016/j.bbrc.2007.07.189. [DOI] [PubMed] [Google Scholar]
- 24.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci USA. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweetman D, et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206, and miR-133. Dev Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. pp. 7.2–7.87. [Google Scholar]
- 30.Reue K. mRNA quantitation techniques: Considerations for experimental design and application. J Nutr. 1998;128:2038–2044. doi: 10.1093/jn/128.11.2038. [DOI] [PubMed] [Google Scholar]
- 31.McKinsey TA, Zhang CL, Olson EN. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 32.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 33.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 34.Chung DJ, et al. Low peak bone mass and attenuated anabolic response to parathyroid hormone in mice with an osteoblast-specific deletion of connexin43. J Cell Sci. 2006;119:4187–4198. doi: 10.1242/jcs.03162. [DOI] [PubMed] [Google Scholar]
- 35.Lecanda F, et al. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J Cell Biol. 2000;151:931–944. doi: 10.1083/jcb.151.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiso N, et al. Fine mapping of five human skeletal muscle genes: alpha-tropomyosin, beta-tropomyosin, troponin-I slow-twitch, troponin-I fast-twitch, and troponin-C fast. Biochem Biophys Res Commun. 1997;230:347–350. doi: 10.1006/bbrc.1996.5958. [DOI] [PubMed] [Google Scholar]
- 37.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell JA, Ou C, Chen Z, Nishimura T, Lye SJ. Parathyroid hormone-induced up-regulation of connexin-43 messenger ribonucleic acid (mRNA) is mediated by sequences within both the promoter and the 3′untranslated region of the mRNA. Endocrinology. 2001;142:907–915. doi: 10.1210/endo.142.2.7930. [DOI] [PubMed] [Google Scholar]
- 39.Caplan AI, Pechak DG. In: Bone and Mineral Research. Peck WA, editor. New York: Elsevier; 1987. pp. 117–183. [Google Scholar]
- 40.Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific MicroRNAs (myomirs) in brown adipocytes. J Cell Physiol. 2009;218:444–449. doi: 10.1002/jcp.21621. [DOI] [PubMed] [Google Scholar]
- 41.Care A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 42.Sato S, et al. Central control of bone remodeling by neuromedin U. Nat Med. 2007;13:1234–1240. doi: 10.1038/nm1640. [DOI] [PubMed] [Google Scholar]
- 43.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.