Abstract
Prostate cancer cells expressing prostate-specific membrane antigen (PSMA) have been targeted with RNA aptamer–small interfering (si)RNA chimeras, but therapeutic efficacy in vivo was demonstrated only with intratumoral injection. Clinical translation of this approach will require chimeras that are effective when administered systemically and are amenable to chemical synthesis. To these ends, we enhanced the silencing activity and specificity of aptamer-siRNA chimeras by incorporating modifications that enable more efficient processing of the siRNA by the cellular machinery. These included adding 2-nucleotide 3´-overhangs and optimizing the thermodynamic profile and structure of the duplex to favor processing of the siRNA guide strand. We also truncated the aptamer portion of the chimeras to facilitate large-scale chemical synthesis. The optimized chimeras resulted in pronounced regression of PSMA-expressing tumors in athymic mice after systemic administration. Anti-tumor activity was further enhanced by appending a polyethylene glycol moiety, which increased the chimeras’ circulating half-life.
Treatment of advanced prostate cancer relies mainly on nonspecific therapies, such as chemotherapies and ionizing radiation, which have low efficacy and are highly toxic to normal tissues1,2. Gene-specific mRNA knockdown with synthetic siRNAs may offer several advantages over these approaches3–5, including target specificity, ease of siRNA production and the possibility of silencing virtually any gene. In addition, recent advances in the understanding of the molecular mechanisms of RNA interference (RNAi) enable rational optimization of the potency, specificity and in vivo activity of siRNAs6–10. However, before siRNAs can be broadly used in the clinic, safe and effective approaches for their targeted delivery in vivo must be developed11.
Most siRNA targeting approaches involve the formation of siRNA-containing complexes that also include charged peptides12,13, proteins14,15 or polymers16–20. Although these reagents silence the targeted genes when administered systemically in experimental animals, their complicated formulation is likely to confound their large-scale production and regulatory approval. The potential toxicity of the materials used poses another challenge. As a result, applications involving the direct local delivery (for example, to the eye and lung) of naked or nuclease-resistant (chemically modified) siRNA duplexes have been the first to be evaluated in clinical trials10,21–24.
We previously developed a simple RNA-only approach for delivering cytotoxic siRNAs targeting prostate cancer–specific pro-survival genes (Plk1 and Bcl2) directly to prostate cancer cells via an RNA aptamer25. The aptamer portion of these chimeras binds PSMA25,26, undergoes cell internalization and delivers its siRNA cargo to the intracellular RNAi machinery. This results in the silencing of the siRNA target gene and pronounced cancer cell death in vitro. When injected intratumorally, the PSMA-targeting chimera substantially decreased tumor volume in a xenograft mouse model of prostate cancer25, inducing apoptosis only in tumors expressing PSMA and having no adverse effects on PSMA-negative tumors or normal cells. Although the first-generation PSMA-binding aptamer/Plk1-siRNA (A10-Plk1) chimera inhibited tumor growth when administered intratumorally25, systemic administration will be necessary for treatment of advanced prostate cancer, thus presenting a variety of additional challenges. In particular, systemic administration requires greater therapeutic doses (leading to higher treatment costs), and carries a greater risk for harmful side effects owing to greater therapeutic exposure of nontargeted tissues. Improvements that would minimize the necessary dose of the chimera would reduce both the cost of treatment and the risk for harmful side effects. Toward this end, we have modified several aspects of the A10-Plk1chimera. The resulting optimized chimeras exhibit potent antitumor activity when administered systemically to mice bearing PSMA-positive prostate cancer tumors.
RESULTS
Second-generation optimized PSMA-Plk1 chimeras
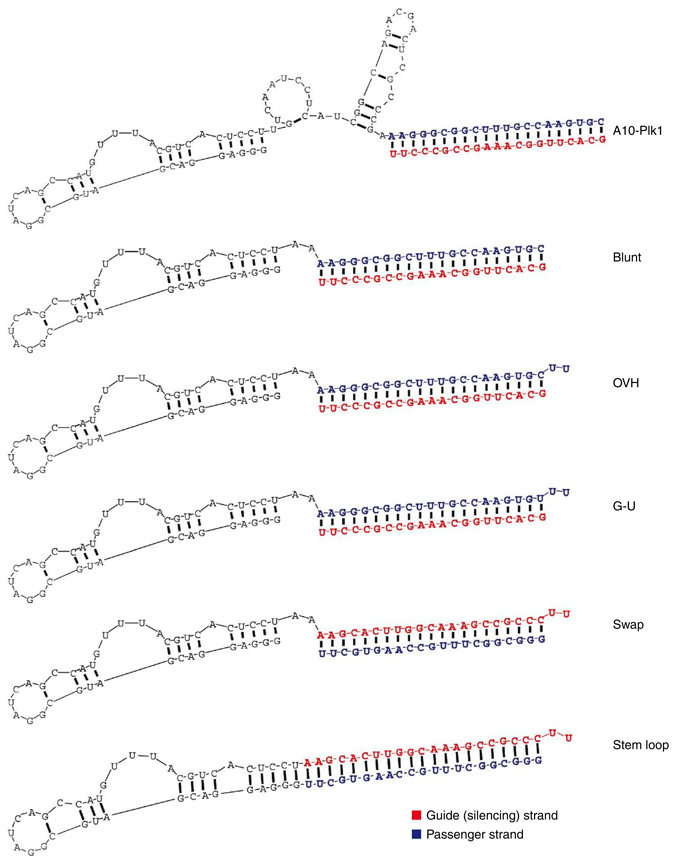
We designed second-generation PSMA-Plk1 chimeras, aiming to (i) facilitate chemical synthesis, (ii) enhance silencing activity and specificity and (iii) enable modifications to optimize in vivo kinetics. Representative second-generation PSMA-Plk1 chimeras developed in this study are shown in Figure 1.
Figure 1.
Optimized PSMA-Plk1 chimeras. Blunt, truncated version of first-generation chimera (A10-Plk1) described previously25. The aptamer portion of the chimera has been truncated from 71+ nt to 39 nt. OVH, overhang chimera similar to blunt, but with 2 nt (UU)-overhangs at the 3′ end of the siRNA duplex. G-U, G-U wobble chimera identical to OVH, but with a wobble base pair at the 5′ end of the antisense siRNA strand (silencing/guide strand). Swap, sense and antisense strands of siRNA duplex are reversed. Stem loop, hairpin chimera where the siRNA duplex (stem) is continuous with the aptamer (loop). Structural predictions were generated using RNAstructure V 4.6.
To facilitate chemical synthesis, we reduced the aptamer portion of the A10-Plk1chimera from 71 (corresponding to original A10 aptamer)26 to 39 nucleotides (nt) (A10-3.2 aptamer)). The longer RNA strand of the second-generation chimeras is modified with 2′-fluoropyrimidines and the shorter RNA strand is unmodified. An exception is the stem loop chimera, which is fully modified. The truncated version of the first-generation chimera (A10-Plk1)25 is referred to as the blunt chimera, as the Plk1 siRNA is a blunted duplex (Fig. 1), identical to the siRNA part of the first-generation chimera.
To increase the silencing activity and specificity of the A10-Plk1 chimera, we engineered several chimeras, four of which are described here with various modifications in the siRNA portion. First, a chimera with a 2-nt (UU)-overhang at the 3′ end of the siRNA duplex (OVH chimera) was designed to favor recognition by the RNase enzyme Dicer6. Second, a wobble base pair was engineered at the 5′ end of the guide (silencing) strand of the OVH chimera by introducing a mutation (C→U) in the passenger strand (G-U wobble chimera). This modification was intended to increase silencing specificity by favoring loading of the guide strand into the RNA-induced silencing complex (RISC)7,8,27. Third, we swapped the passenger and guide (Fig. 1) strands of the siRNA duplex (swap chimera). This configuration was intended to accommodate 5′ terminal modifications of the shorter RNA strand without loss of function28,29. This modification also takes advantage of strand-loading bias introduced by the interaction of the 3′ overhang with the PAZ domains of Argonaute2 (Ago2) and/or Dicer. This configuration favors loading of the guide strand (strand containing 3′ overhang) into RISC30,31. Fourth, a stem loop chimera, where the siRNA duplex (stem) is continuous with the aptamer (loop), was designed to mimic endogenous miRNA precursors.
Binding of optimized chimeras to PSMA-expressing cells
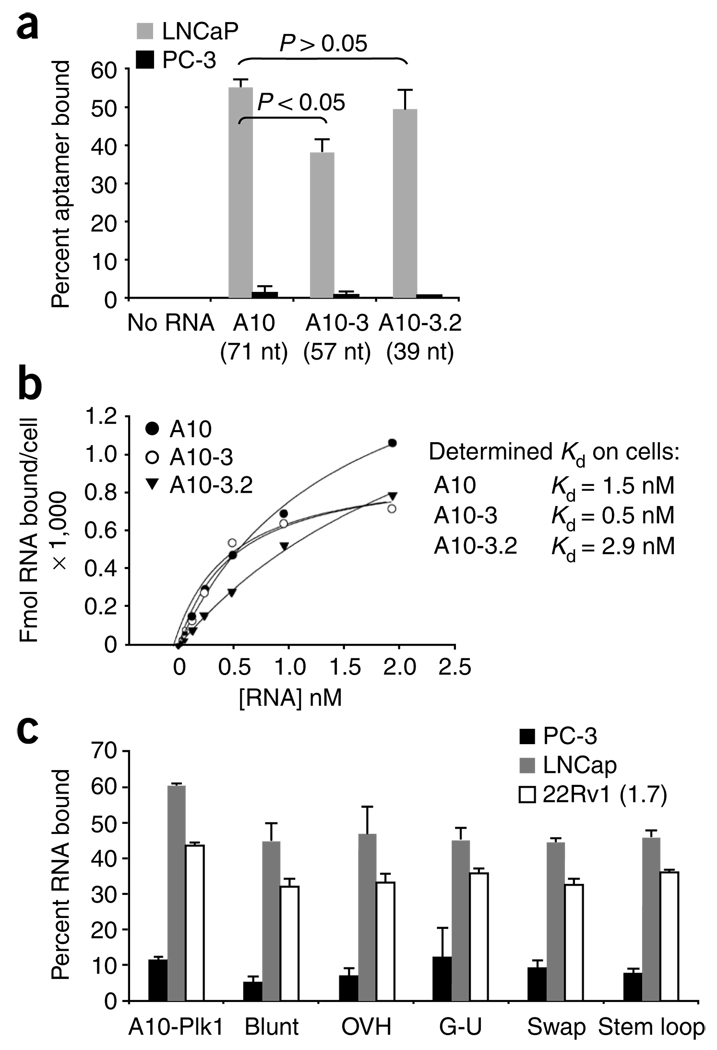
We tested the ability of the truncated PSMA aptamer to bind the surface of prostate cancer cells expressing PSMA (cell lines LNCaP and 22Rv1 clone 1.7). A PSMA-negative prostate cancer cell line (PC-3) was used as a control for specificity. Surface expression of PSMA was verified using flow cytometry (data not shown). To determine whether the truncated aptamer can bind the surface of cells expressing PSMA, we incubated 32P-labeled aptamers A10 (ref. 26), A10-3 (57 nt)26 and A10-3.2 with either LNCaP or PC-3 cells (Fig. 2a). Binding of these aptamers was specific for cells expressing PSMA and was dependent on a region within A10-3.2 as this truncated RNA retained specific binding to PSMA-expressing cells. The A10-3.2 aptamer was found to bind LNCaP cells with comparable affinity to the full-length A10 RNA aptamer (Fig. 2b).
Figure 2.
Binding of truncated versions of PSMA A10 aptamer and optimized chimeras to cells expressing PSMA. RNAs were end-labeled with 32P. (a) LNCaP cells and PC-3 cells were incubated with either the full-length PSMA aptamer A10 (71 nt) or truncated versions of the PSMA aptamer, A10-3 (57 nt) or A10-3.2 (39 nt). 32P-labeled bound/internalized RNAs were determined by liquid scintillation counter (LSC) or filter binding assay (data not shown). (b) Relative affinity of A10 PSMA aptamer and truncated A10 aptamers to cells expressing PSMA. Varying amounts (0–2 nM) of end-labeled A10, A10-3 and A10-3.2 were incubated with fixed LNCaP cells. Bound counts were determined by filter binding assay. (c) First-generation chimera (A10-Plk1) and optimized chimeras were incubated with either PC-3 cells (black bars) or LNCaP and 22Rv1 (1.7). Cells were processed as in a. Bound counts were determined with LSC.
Next we tested the ability of A10-3.2 to bind to PSMA-expressing cells in the context of the modifications to the siRNA part of the chimeras (Fig. 2c). All chimeras retained binding to PSMA-expressing prostate cancer cells. These experiments confirm that modifications made to the first-generation chimera did not affect binding or target specificity.
Effect of chimera modifications on RNAi
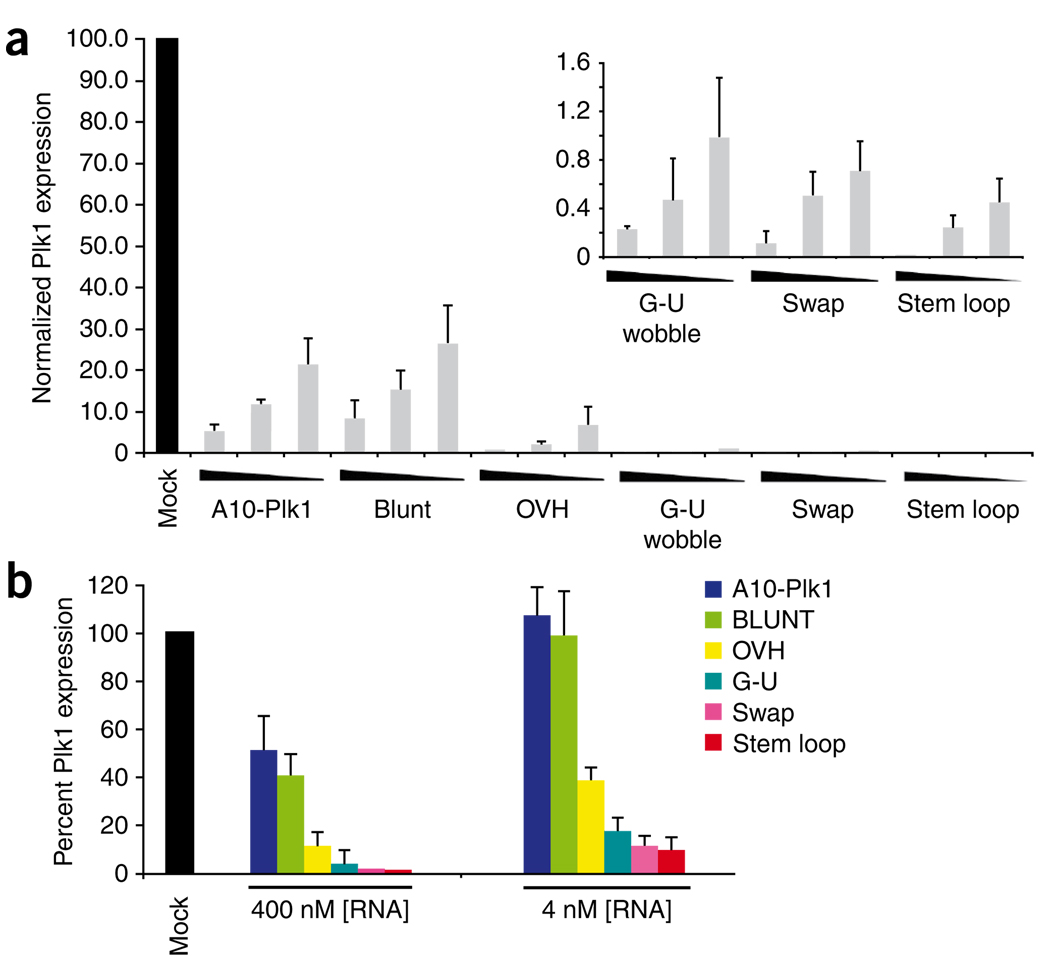
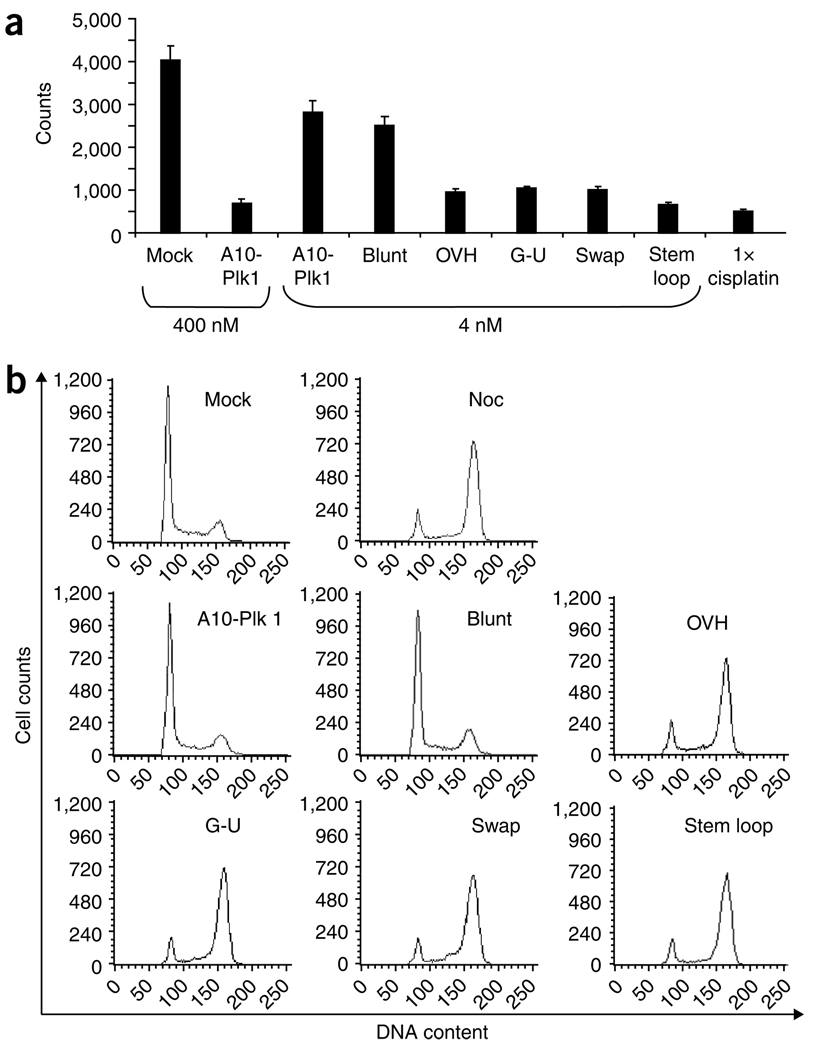
To determine whether the second-generation chimeras can silence target gene expression and whether they have enhanced silencing activity compared to the first-generation chimera, we tested for gene-specific silencing using quantitative real time-PCR (qRT-PCR) (Fig. 3). PSMA-expressing cells (22Rv1 (1.7)) were transfected with increasing amounts (4, 40, 400 nM) of A10-Plk1 or of the second-generation chimeras using a cationic lipid reagent (Fig. 3a). As a control, cells were transfected with a control nonsilencing siRNA (Mock). Elevated expression of Plk1 in 22Rv1 (1.7) cells was confirmed using immunoblotting (Supplementary Fig. 1). This was specific to cancer cells as normal cells (human fibroblasts) have little-to-no Plk1 protein (Supplementary Fig. 1). The modifications introduced within the siRNA portion of the chimera, enhanced Plk1 silencing. The most active of the second-generation chimeras were the swap and the stem loop chimeras, which resulted in >99% silencing at concentrations as low as 4 nM.
Figure 3.
Silencing ability of PSMA chimeras. 22Rv1 (1.7) cells were transfected with 400, 40 or 4 nM of each chimera. Cells were processed for qRT-PCR 24–48 h after transfection. Percent Plk1 expression was normalized to that of mock-transfected (mock) cells. (a) Comparison of silencing efficiencies of the blunt, OVH, G-U Wobble, swap and stem loop chimeras to that of the first-generation chimera (A10-Plk1). (a, inset) Percent Plk1 expression of G-U wobble, swap and stem loop ≤ 1.0 and are depicted on an adjusted y axis. Experiments were performed several times (n = 3). (b) 22Rv1 (1.7) cells were treated with either 400 nM or 4 nM of each of the optimized RNA chimeras in the absence of transfection reagent. Cells were processed for qRT-PCR 4 d after treatments.
Next we verified the ability of the second-generation chimeras to silence target gene expression in the absence of transfection reagent (Fig. 3b). 22Rv1 (1.7) cells were incubated with media containing the various RNA chimeras for 4 d. The modifications made to the siRNA portion of the chimeras substantially enhanced the chimeras’ silencing potential (<10% versus >60% for A10-Plk1 and second-generation chimeras, respectively, at 4 nM concentration of the RNAs,) without affecting binding to PSMA on the cell surface. No effect was observed on PSMA-negative prostate cancer cells (data not shown). These experiments indicate that the second-generation chimeras have silencing activities >50 times stronger than that of the first-generation chimera A10-Plk1.
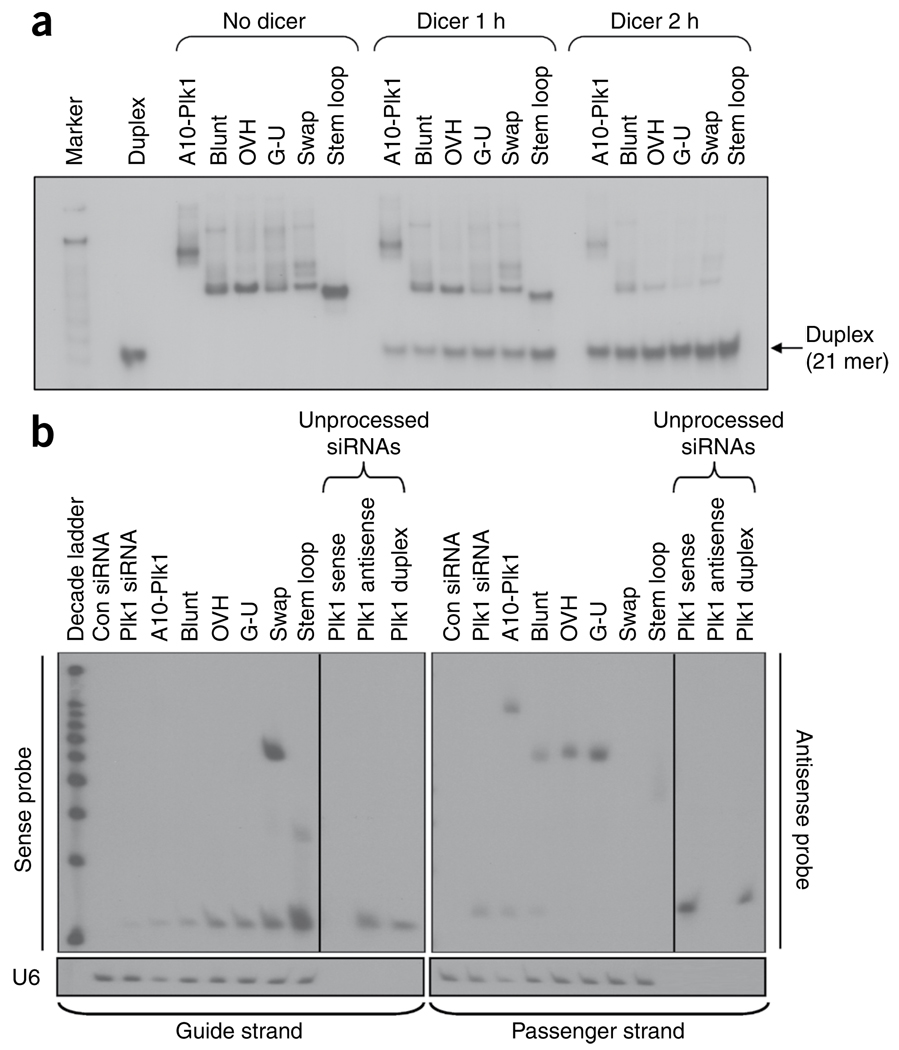
We next assessed whether Dicer can process the siRNA portion of the chimeras in the context of the A10-3.2 PSMA aptamer (Fig. 4a). 32P-labeled chimeras (labeled on the 5′-terminus of the short RNA strand) were incubated with recombinant human Dicer for 1 h and 2 h and the cleavage products were analyzed by nondenaturing gel electrophoresis. Incubation with Dicer resulted in 32P-labeled cleaved products corresponding to the size of the duplex Plk1 siRNA (21 mer). These data suggest that the RNA chimeras are Dicer substrates. The size of the 32P-labeled cleaved products (~21 mer) also indicates from which side Dicer enters the chimera and cleaves32. These results suggest that Dicer enters from the 3′-end of the longer RNA strand and cleaves ~21 nt upstream. Increased levels of processed duplex in the second-generation constructs compared to the first-generation A10-Plk1 chimera suggest these may be better substrates for Dicer.
Figure 4.
Analysis of chimera processing by the RNAi machinery. (a) In vitro Dicer processing. The 32P-labeled PSMA-Plk1 chimeras were incubated with recombinant human Dicer enzyme for either 1 or 2 h. The Dicer cleavage or uncleaved (No Dicer) products were visualized after 15% nondenaturing PAGE. (b) Assessment of strand bias: loading of siRNA silencing strand into RISC. Small fragment northern blot of RNA isolated from 22Rv1 (1.7) cells transfected with 200 pmols of each of the optimized aptamer-siRNA chimera constructs. Loading of the siRNA silencing strand into RISC protects the siRNA strand from degradation (this can be detected with a specific probe using a modified northern blot assay). The strand that is not loaded is rapidly degraded. U6 RNA was used as a loading control. Duplex, Plk1 siRNA duplex; A10-Plk1, first-generation chimera. Blunt, OVH, G-U, swap and stem loop chimeras are described in Figure 1. Probe controls show hybridization efficiencies of the sense and antisense probes. The varying intensities of unprocessed chimeras (upper bands on blots) are due to differential probe binding to these species and do not reflect their amounts (this same trend was observed when equal amounts of each chimera was directly loaded on gel and processed as described here (data not shown)).
Next we tested whether modifications made to the siRNA portion of the second-generation chimeras affect loading of the correct siRNA-silencing strand into RISC (Fig. 4b). Loading of the correct strand into the RNAi machinery generates siRNAs with increased activity and reduced off-target effects10,31,33. Loading into RISC was assessed by small fragment northern (strand-bias assay)34. This assay allows a quantitative measure of the guide strand of the siRNA duplex that is incorporated into RISC and thereby protected from nuclease degradation. The strand that is not incorporated into RISC is rapidly degraded. The assay suggests that modifications made to the first-generation chimera substantially enhanced loading of the correct strand into RISC. Although the addition of the 2-nt (UU)-overhang at the 3′ end of the siRNA duplex alone enhanced loading of the correct strand into RISC, incorporation of a wobble base at the 5′ end of the guide strand had no effect. Notably, swapping the passenger strand with the guide strand resulted in a substantial increase in loading of the guide strand into RISC (Fig. 4b). An even greater effect on strand loading was observed with the stem loop chimera, which has the same passenger guide configuration as the swap chimera (Fig. 4b). Together, these data indicate that the modifications made to the siRNA portion of the PSMA-Plk1 chimeras enhance silencing activity and specificity by promoting optimal RNAi processing.
Effect of chimeras on prostate cancer cell growth and survival
Depletion of Plk1 in cancer cells leads to a G2/M arrest that decreases cell proliferation and subsequent cancer cell death due to mitotic catastrophe (crisis), a type of cell death which results from mitotic DNA damage35. To determine whether treatment with the various PSMA chimeras results in reduced cellular proliferation, 22Rv1 (1.7) cells were transfected with each of the chimeric RNAs using a cationic lipid reagent and cell proliferation was measured by 3H-thymidine incorporation (Fig. 5a). Mock-treated cells (treated with control nonsilencing siRNA) were used to determine the normal rate of cellular proliferation before treatment. Cisplatin (positive control) was used to inhibit cell proliferation and induce cell death. As previously observed, silencing of Plk1 by the A10-Plk1 chimera (at 400 nM) substantially inhibited cell proliferation25. Lowering the concentration of A10-Plk1 to 4 nM reduced the effect on cellular proliferation by ~sixfold, whereas the second-generation chimeras still inhibited cell proliferation at such low concentrations (Fig. 5a). This correlated with cells arrested in the G2/M phase of the cell cycle, as measured by propidium iodide (PI) staining of DNA content and flow cytometry (Fig. 5b). Nocodazole, a microtubule-depolymerizing drug, was used as a positive control to arrest cells in G2/M.
Figure 5.
Effect of PSMA-Plk1 chimeras on prostate cancer cell growth. (a) 22Rv1 (1.7) cells were transfected (or treated in the absence of transfection reagent (data not shown)) with either 400 nM or 4 nM of A10-Plk1 or 4 nM of each of the optimized chimeras. 3H-thymidine was added to the media 24 h after transfection and cells were incubated in the presence of 3H-thymidine for another 24 h. The next day cells were lysed with 0.5 N NaOH and incorporated counts determined by liquid scintillation counter. Cisplatin was used as a positive control for this assay. (b) Cell cycle profile of 22Rv1 (1.7) cells transfected with 4 nM of each of the optimized chimeras. DNA content of treated cells was determined by flow cytometry 48 h after transfection after staining cells with PI. Nocodazole (Noc) treatment was used as a positive control for this assay to arrest cells in mitosis.
Next, we determined whether the second-generation chimeras induced apoptosis in treated cells and whether modifications of the siRNA moiety increased their apoptotic activity. 22Rv1 (1.7) cells were treated with 4 nM of the various chimeras in the absence of transfection reagent. Cisplatin was used as a positive control for induction of apoptosis, which was assessed by measuring production of active caspase 3 (Casp3) by flow cytometry (Table 1 and Supplementary Fig. 2). As expected, the modifications introduced within the Plk1 siRNA sequence greatly enhanced cell death from 22% (for blunt chimera) to 75 and 85% (for swap and stem loop chimeras, respectively) at a concentration of 4 nM. The swap and stem loop chimeras efficiently induced apoptosis at concentrations 100-fold lower than was necessary for the A10-Plk1 chimera (Table 1). Together these data suggest that the modifications made to the second-generation chimeras greatly enhance silencing as well as Plk1-mediated mitotic catastrophe and subsequent cell death.
Table 1.
Effect of PSMA-Plk1 chimeras on prostate cancer cell viability
| Treatment | Concentration | Percent caspase 3 positive cells (average of three experiments) |
|---|---|---|
| Untreated | – | 17 |
| Cisplatin | 2 nM | 90 |
| A10-Plk1 | 400 nM | 52 |
| A10-Plk1 | 4 nM | 27 |
| Blunt | 4 nM | 22 |
| OVH | 4 nM | 64 |
| G-U | 4 nM | 72 |
| Swap | 4 nM | 75 |
| Stem loop | 4 nM | 85 |
| Plk1 siRNA only | 4 nM | 13 |
22Rv1(1.7) PSMA-positive prostate cancer cells were incubated with either 400 nM or 4 nM of A10-Plk1 chimera or 4 nM of each optimized chimera in the absence of transfection reagents. Media containing fresh RNAs was replaced every other day for the course of the experiment. Cells were collected on day 6, stained with an antibody specific for active caspase 3 and processed for flow cytometry. Cisplatin was used as a positive control for apoptosis in this assay. Data were averaged from three independent experiments. (One representative experiment is shown in Supplementary Fig. 2).
In vivo efficacy of optimized PSMA-Plk1 chimeras
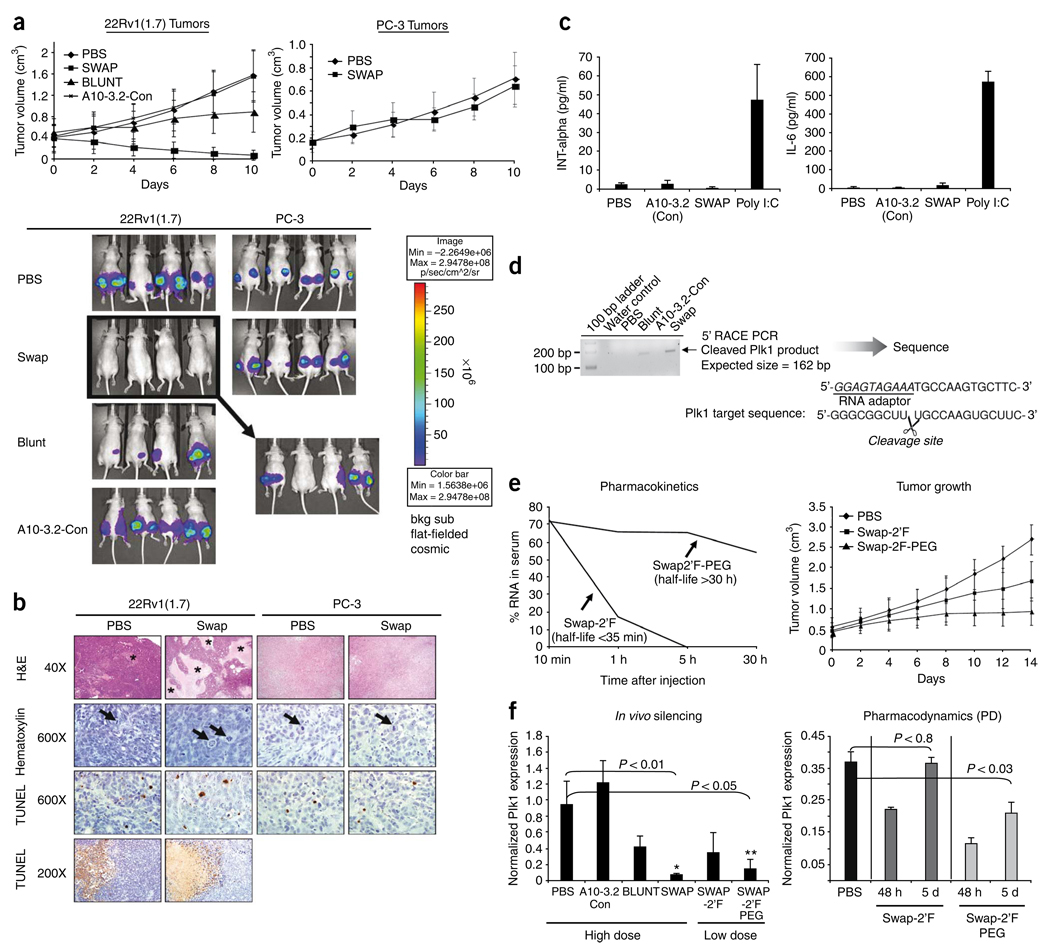
We next assessed the ability of the second-generation chimeras to limit tumor growth in athymic mice bearing tumors derived from either 22Rv1 (1.7) or PC-3 cells (Fig. 6a). PSMA expression in tumors was confirmed by immunoblot analysis (Supplementary Fig. 3a). For the in vivo experiment, we compared the cytotoxic effects of the swap chimera to those of the blunt chimera. Athymic mice (at least ten mice per treatment group) were subcutaneously injected in the flanks with either 22Rv1 (1.7) or PC-3 cells. Both cancer cell lines express luciferase36, which allows measurement of tumor growth using bioluminescence imaging (Fig. 6a and Supplementary Fig. 3b). Mice with 22RV1 (1.7)-derived tumors were injected intraperitoneally each day for a total of 10 d (starting on day 0) with either PBS or 1 nmol each of the indicated chimeric RNAs. Mice bearing PC-3 tumors were treated only with PBS or the swap chimera. No difference in tumor volume was observed in PC-3 tumors after treatment with either PBS or the swap chimera, indicating that the swap chimera did not have nonspecific cell-killing effects (Fig. 6a). A pronounced reduction in tumor volume was observed for 22Rv1 (1.7) tumors treated with the swap chimera (Fig. 6a). In contrast, an increase in tumor volume was observed for tumors treated with PBS or a nonsilencing PSMA chimera, A10-3.2-Con (Supplementary Fig. 3b). Notably, ~70% of all swap-treated tumors completely regressed by the end of the treatment. Of the remaining, ~30% (Fig. 6a, see inset indicated by arrow for representative tumor-bearing mice), the growth rate of all tumors was slowed by as much as 2.3-fold (Fig. 6a). Although regression of PSMA-positive tumors was most evident in swap-treated mice, tumor growth was significantly slowed in mice treated with the blunt chimera (compare PBS with blunt) (P < 0.001). No morbidity or mortality was observed following the 10-d treatment with the chimeric RNAs, suggesting that these compounds are not toxic.
Figure 6.
In vivo efficacy of optimized PSMA chimera in a xenograft model of prostate cancer. (a) 106 luciferase-expressing (PSMA-positive or PSMA-negative) prostate cancer cells were injected into the flanks of nude (nu/nu) mice 2 weeks before treatment with optimized chimeras. Treatment with the optimized chimeras commenced when tumors reached a volume of ~0.4 cm3. 1 nmol of either blunt, swap or A10-3.2-Con was administered intraperitoneally in mice bearing 22Rv1 (1.7) tumors. As a control for specificity, a mouse xenograft model of prostate cancer bearing PSMA-negative prostate cancer cells (PC-3) was also treated with the swap chimera. A total of ten treatments were administered for each treatment group. Treatment occurred every day for 10 consecutive days. Tumors were measured with calipers every other day for the course of the experiment. Saline (PBS) treated animals were used as a control. Animals were euthanized 2–3 d after the last treatment. n ≥ 10 mice per treatment group. Bottom panels: Bioluminescence imaging of 22Rv1 (1.7) and PC-3 prostate tumors was carried out after treatment with optimized chimeras (day 10). Examples show tumor growth in four representative animals from each treatment group. Insert indicated by arrow represents bioluminescence imaging images of ~30% of 22Rv1 (1.7) tumor–bearing mice treated with the swap chimera that still had palpable tumors (17 out of 48 total tumors) by day 10. All sites represent tumor growth ~25 d after injection of tumor cells. Log-scale heat map (right) of photon flux applies to all panels. (b) Histology of 22Rv1 (1.7) and PC-3 tumors treated with the various optimized chimeras. Areas of necrosis (asterisks) were readily detected in swap-treated 22Rv1 (1.7) tumors, but not frequently seen in PBS-treated tumors (H&E, 40×). Mitotic figures (arrows) were often detected in tumors from all treatment groups including occasional large bizarre mitoses in swap-treated 22Rv1 (1.7) tumors (Hematoxylin, 600×). TUNEL staining was detected in scattered cells throughout the tumor section of each group (TUNEL staining, 600×) and at the interface of viable tissue and necrotic foci (TUNEL staining, 200×). Representative sections from the PBS and swap treatment groups are shown. (c) Assessment of potential chimera-dependent immunostimulatory effects. Serum from mice treated with either saline (PBS), A10-3.2-Con, swap or poly I:C was screened for levels of cytokines INT-a and IL-6 using ELISA. (d) 5′-Rapid amplification of cDNA ends (5′-RACE) PCR analysis to assess siRNA mediated cleavage of Plk1 mRNA in tumors treated with the various PSMA-Plk1 chimeras. (e) Pharmacokinetic profile and efficacy of the swap chimera with polyethylene glycol (PEG). (f) In vivo silencing assessed by quantitative RT-PCR. Plk1 mRNA levels in treated tumors were normalized to GAPDH mRNA levels. Panel on left shows Plk1 levels of tumors (9 tumors/group for this experiment) from animals processed for experiments shown in a and e.
As assessed with gross inspection or histological analysis, tumors from mice treated with swap (but not PBS) had liquefactive necrosis–like material that exuded from the tumor mass during gross sectioning. This coincided with large areas of necrosis commonly detected in tumors from the SWAP mice, but substantially less so in those from the PBS-treated mice (Fig. 6b). Mitotic cells were detected in all groups including some occasional large and unusual mitotic phenotypes in swap-treated mice. TUNEL staining was seen throughout the tissues as brown staining of random individual cells and along the interface of necrotic and viable tumor tissue. No substantial difference in tumor histology was noted for PC-3 tumors treated with the swap chimera versus the PBS control, suggesting that no nonspecific uptake and subsequent processing of this chimera occurred after systemic administration.
Within each tumor type (22Rv1 (1.7) or PC-3), there were no detectable differences in cellular inflammation between treatments (for example, PBS versus swap). Moreover, cellular inflammation was uncommon and mild. Immune cells were detected only along the peripheral border of the tumor and comprised mainly scattered neutrophils with few mononuclear cells. This suggests that tumor regression is not dependent on an immune response. As an additional measure of immune responsiveness, serum from treated mice was screened for levels of interferon-α (INT-α) and interleukin-6 (IL-6) using enzyme-linked immunosorbent assays (ELISA) (Fig. 6c). No differences were seen in cytokine levels of mice treated with either PBS or the A10-3.2-Con or swap chimeras. This was in contrast to mice treated with polyinosinic:polycytidylic acid (poly I:C), an established immune stimulator. These data suggest that the chimeras do not trigger an innate immune response and may be safe for in vivo applications.
To determine whether the siRNAs released from the chimeras were indeed triggering RNAi in vivo, we performed a modified 5′ rapid amplification of cDNA ends (RACE)-PCR (as previously described25) on mRNA from tumors of animals treated with the indicated chimeras (Fig. 6d). Sequencing of the 5′RACE-PCR products generated with Plk1-specific primers demonstrate that Ago2-mediated cleavage occurs between bases 10 and 11 relative to the 5′-end of the guide Plk1 siRNA strand. PCR products were not observed in samples from control-treated tumors. This result confirms specific siRNA-mediated cleavage products of Plk1 mRNA in treated tumors in vivo.
We next determined whether the addition of a 20 kDa polyethylene glycol (PEG) group could extend the circulating half-life of the swap chimera without affecting binding to PSMA (Supplementary Fig. 4a) or Plk1 silencing activity (Supplementary Fig. 4b). A 20 kDa PEG was placed at the 5′ end of a Plk1 siRNA passenger strand by chemical synthesis. This RNA strand included 2′-fluoropyrimidines for decreased nuclease sensitivity. An analogous fully 2′-fluoropyrimidines–modified chimera (swap-2′F) with no terminal PEG was used as a control (Supplementary Fig. 4b). First, we verified that the addition of PEG did not abrogate binding specificity and internalization into PSMA-positive prostate cancer cells (Supplementary Fig. 4a) and confirmed silencing of Plk1 mRNA by the PEG-modified chimera (Supplementary Fig. 4b).
To determine the in vivo half-lives of the swap-2′F and swap-2′F-PEG chimeras, the RNAs were intraperitoneally injected into mice and blood samples were obtained 10 min, 1 h, 5 h and 30 h later. qRT-PCR was used to quantify the amount of RNA present in each blood sample. The in vivo circulating half-life of the swap-2′F chimera was substantially increased (from <35 min to >30 h) by the addition of the 20 kDa PEG (Fig. 6e, left panel). We next addressed whether the PEGylation of the swap chimera leads to increased in vivo efficacy. Mice bearing 22Rv1 (1.7) tumors were intraperitoneally injected with a low dose (250 pmols) of the swap-2′F or swap-2′F-PEG chimeras or with PBS. A total of 5 injections were performed over the course of 10 d. Tumor volume was determined as in Figure 6a. The swap-2′F-PEG chimera inhibited tumor growth at substantially lower doses (Fig. 6a,e, right panel; 10 × 1 nmol versus 5 × 250 pmols). We then determined whether inhibition of tumor growth in treated mice was correlated with silencing of Plk1 gene expression by the PSMA-Plk1 chimeras (Fig. 6f, left panel). Plk1 mRNA expression was significantly reduced in swap- (P < 0.01) and swap-2′F-PEG-treated (P < 0.05) tumors compared to PBS control or to A10-3.2-Con.
To determine whether prolonged silencing of Plk1 gene expression might be responsible for the difference in the in vivo efficacies of the swap-2′F-PEG and swap-2′F chimeras (Fig. 6e), we carried out a pharmacodynamic study to assess silencing over time (Fig. 6f; right panel). In this experiment, PSMA positive tumor–bearing mice were injected with two doses (1 d apart) of 1 nmol each of either swap-2′F or swap-2′F-PEG. Quantitative RT-PCR was then performed on mRNA from tumors to determine the amount of Plk1 mRNA in the treated tumors at the indicated time points. Plk1 mRNA silencing is observed in both swap-2′F- and swap-2′F-PEG-treated tumors at 48 h but only in the swap-2′F-PEG-treated tumors 5 d after the last treatment (Fig. 6f, right panel). These data indicate that the swap-2′F-PEG chimera has greater gene silencing activity in vivo.
DISCUSSION
We developed and characterized PSMA-Plk1 aptamer-siRNA chimeras with enhanced activity, specificity and in vivo kinetics relative to the first-generation PSMA-Plk1 chimera25. These RNA-only chimeras were optimized by incorporating modifications shown to enhance silencing activity and specificity of siRNA6–8,27,30,31,33. The modifications applied to the first-generation chimera include the addition of 2 nt 3′-overhangs and optimization of the thermodynamic profile and structure of the duplex to favor RISC processing of the correct siRNA guide strand6–8,27,30,31. Our targeted approach for treating prostate cancer is effective when delivered systemically and is amenable to chemical synthesis for large-scale production.
As many potential therapeutic applications of chimeras, including cancer therapy, require systemic administration of the therapeutic reagent, it is also necessary to optimize the in vivo kinetics of these chimeras, in addition to enhancing their potency and specificity. Terminal modification of RNAs with PEG increases the half-life and bioavailability of many oligonucleotide-based therapies, including RNA aptamers28,37,38. We found that addition of a 20 kDa PEG to the 5′-terminus of the smaller RNA strand promotes increased retention of the chimera in serum (Fig. 6e) without affecting chimera targeting and silencing (Supplementary Fig. 4). The PEGylated reagent leads to prolonged-silencing in vivo (Fig. 6f, right panel) and to inhibition of tumor growth, at lower doses, in mice bearing PSMA-positive prostate tumors (Fig. 6e, right panel). Although it is possible that the greater degree of silencing induced by the PEGylated chimera at the 5-d time point is a result of prolonged exposure of the tumor to this reagent, an alternative possibility is that this difference is due to the lingering effects of a greater initial knockdown.
As previously described for the first-generation PSMA-Plk1 chimera (A10-Plk1), cellular targeting of the optimized chimeric RNAs was mediated by the interaction of the aptamer portion of the chimeras with PSMA expressed on the surface of prostate cancer cells (Fig. 2c). We found that the first 39 nt of the A10 PSMA aptamer are sufficient for targeting the chimeras to PSMA expressed on the surface of prostate cancer cells. This allowed us to truncate the aptamer portion of the chimeras from 71 nt to 39 nt without loss of function (Fig. 2). Chimeras designed with such short aptamers have a long strand of ≤64 bases, a length that can be efficiently produced with chemical synthesis.
Depletion of Plk1 by the ‘optimized’ chimeras was also specific to PSMA-positive prostate cancer cells (data not shown) and resulted in decreased proliferation and increased apoptosis of the target cells in culture (Fig. 5a, Table 1 and Supplementary Fig. 2). Notably, after modifications to the siRNA portion of the chimera, these effects were observed at concentrations of the reagent >50-fold lower than for the ‘first-generation’ chimera (Fig. 5a and Table 1). In addition, we found that upon depletion of Plk1, the prostate cancer cells undergo a mitotic arrest (Fig. 5b) leading to apoptosis. Coincident with this observation, reducing Plk1 expression has been reported to lead to mitotic catastrophe (crisis) (due to arrest of cancer cells at the G2/M transition of the cell cycle) and death of prostate cancer cells35. Notably, this effect is specific to cancer cells; normal cells resume cell-cycle entry upon restoration of Plk1 expression35,39,40.
An additional measure of specificity was achieved by modifying the siRNA portion of the chimera to enhance loading of the guide strand into RISC (Fig. 4b). Optimal loading of the guide strand into RISC is thought to reduce off-target effects that result from inappropriate incorporation of both siRNA strands into the silencing complex7,8,27,30,31,33. Although we cannot rule out potential off-target effects mediated by the guide strand itself, these effects would likely be restricted to the tumor, as the siRNAs are targeted to PSMA-expressing prostate cancer cells (Fig. 6a).
A major advantage of the PSMA-Plk1 chimera approach as a therapeutic for advanced prostate cancer lies in its target specificity, which is achieved at the level of the aptamer (PSMA-expressing cells are specifically targeted) and at the level of the siRNA (siRNAs are designed against cancer-specific transcripts). Cancer cell-specific targeting could substantially reduce the amount of siRNA needed for effective therapy while reducing systemic cytotoxicity. Most targeted delivery approaches for siRNAs described to date involve the use of complex formulations of synthetic polymers16–20, proteins14,15 or charged peptides12,13. Although such approaches are proving effective in preclinical studies, their multicomponent formulations complicate production and safety assessment41. A one-component system, which involves the direct conjugation of an siRNA to an RNA aptamer, reduces the complexity of the reagent and thus simplifies manufacturing.
2′-fluoropyrimidines in the PSMA-Plk1 chimera increase in vivo stability and decrease immunotoxicity, and a terminal 20 kDa PEG increase serum retention. Both modifications are well characterized in humans and are reported to be well tolerated with little toxicity10,38. RNA oligonucleotides with similar modifications have already been approved for use in humans (for example, Macugen), with many more quickly moving through the clinical pipeline10,42–47. Although we cannot completely rule out potential intracellular toxicity of 2′ fluoropyrimidine–modified RNAs leading to nonspecific immunostimulation10, based on our findings (Fig. 6c) we do not expect these chimeras to produce problematic toxicity in humans.
In principle, the aptamer-siRNA chimera approach can be applied to develop reagents targeting many different cell types provided that a cell-type-specific receptor exists and that an aptamer against the receptor can be selected. The development of an aptamer-siRNA chimera that targets HIV-infected cells supports this notion32. The same types of modifications that enabled the in vivo efficacy of the PSMA-Plk1 chimera may thus prove useful in developing siRNA-based therapeutics for a wide variety of diseases.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Henry (Department of Molecular Physiology and Biophysics, University of Iowa) for supplying PC-3 and 22Rv1 (1.7) luciferase-positive cells, A. Klingelhutz (Department of Microbiology, University of Iowa) for providing immortalized human fibroblasts, J. Houtman (Department of Immunology, University of Iowa) for help with flow cytometry, and R. Sousa (Department of Biochemistry, University of Texas Health Science Center) for providing a plasmid encoding a mutant T7 RNA polymerase. We are also thankful to M. Behlke for help with the manuscript. J.P.D. is supported by an National Institutes of Health (NIH) graduate student training grant to the Molecular and Cellular Biology Program (University of Iowa), K.W.T. is supported by a Ladies Auxiliary to the Veterans of Foreign Wars postdoctoral fellowship, J.O.M. and A.P.M. are supported by the NIH. This research was supported by an American Cancer Society Institutional Research Grant and a Lymphoma SPORE Developmental Research Award to P.H.G.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
J.P.D., X.-y.L., G.S.T., R.M.W., K.W.T., K.R.S. performed research; D.K.M. provided expertise and analyzed data; A.P.M. provided expertise and useful discussions; J.O.M. designed research, wrote the manuscript and provided useful discussions; P.H.G. designed, coordinated and performed research, analyzed data and wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Mendenhall WM, Henderson RH, Mendenhall NP. Definitive radiotherapy for prostate cancer. Am. J. Clin. Oncol. 2008;31:496–503. doi: 10.1097/COC.0b013e31816d1ca6. [DOI] [PubMed] [Google Scholar]
- 2.Potvin K, Winquist E. Hormone-refractory prostate cancer: a primer for the primary care physician. Can. J. Urol. 2008;15 Suppl 1:14–20. [PubMed] [Google Scholar]
- 3.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 8.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 9.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 10.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 11.Blow N. Small RNAs: delivering the future. Nature. 2007;450:1117–1120. doi: 10.1038/4501117a. [DOI] [PubMed] [Google Scholar]
- 12.Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 13.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv. Drug Deliv. Rev. 2007;59:134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 15.Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc. Natl. Acad. Sci. USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozema DB, et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. USA. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 18.Heidel JD, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc. Natl. Acad. Sci. USA. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard KA, et al. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol. Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Pillé JY, et al. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Hum. Gene Ther. 2006;17:1019–1026. doi: 10.1089/hum.2006.17.1019. [DOI] [PubMed] [Google Scholar]
- 21.Reich SJ, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol. Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- 22.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 23.Li BJ, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeVincenzo J, et al. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral Res. 2008;77:225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 25.McNamara JO, II, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 26.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 27.Keck K, et al. Rational design leads to more potent RNA interference against hepatitis B virus: factors effecting silencing efficiency. Mol. Ther. 2009;17:538–547. doi: 10.1038/mt.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czauderna F, et al. Structural variations and stabilizing modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boomer RM, et al. Conjugation to polyethylene glycol polymer promotes aptamer biodistribution to healthy and inflamed tissues. Oligonucleotides. 2005;15:183–195. doi: 10.1089/oli.2005.15.183. [DOI] [PubMed] [Google Scholar]
- 30.Rose SD, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sano M, et al. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res. 2008;36:5812–5821. doi: 10.1093/nar/gkn584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds A, et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 34.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protocols. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 35.Reagan-Shaw S, Ahmad N. Silencing of polo-like kinase (Plk) 1 via siRNA causes induction of apoptosis and impairment of mitosis machinery in human prostate cancer cells: implications for the treatment of prostate cancer. FASEB J. 2005;19:611–613. doi: 10.1096/fj.04-2910fje. [DOI] [PubMed] [Google Scholar]
- 36.Drake JM, Gabriel CL, Henry MD. Assessing tumor growth and distribution in a model of prostate cancer metastasis using bioluminescence imaging. Clin. Exp. Metastasis. 2005;22:674–684. doi: 10.1007/s10585-006-9011-4. [DOI] [PubMed] [Google Scholar]
- 37.Bozza M, Sheardy RD, Dilone E, Scypinski S, Galazka M. Characterization of the secondary structure and stability of an RNA aptamer that binds vascular endothelial growth factor. Biochemistry. 2006;45:7639–7643. doi: 10.1021/bi0521442. [DOI] [PubMed] [Google Scholar]
- 38.Veronese FM, Mero A. The impact of PEGylation on biological therapies. BioDrugs. 2008;22:315–329. doi: 10.2165/00063030-200822050-00004. [DOI] [PubMed] [Google Scholar]
- 39.Reagan-Shaw S, Ahmad N. Polo-like kinase (Plk) 1 as a target for prostate cancer management. IUBMB Life. 2005;57:677–682. doi: 10.1080/15216540500305910. [DOI] [PubMed] [Google Scholar]
- 40.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 41.Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum. Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 42.Dyke CK, et al. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490–2497. doi: 10.1161/CIRCULATIONAHA.106.668434. [DOI] [PubMed] [Google Scholar]
- 43.Chan MY, et al. Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation. 2008;117:2865–2874. doi: 10.1161/CIRCULATIONAHA.107.745687. [DOI] [PubMed] [Google Scholar]
- 44.Katz B, Goldbaum M. Macugen (pegaptanib sodium), a novel ocular therapeutic that targets vascular endothelial growth factor (VEGF) Int. Ophthalmol. Clin. 2006;46:141–154. doi: 10.1097/01.iio.0000212130.91136.31. [DOI] [PubMed] [Google Scholar]
- 45.Gilbert JC, et al. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation. 2007;116:2678–2686. doi: 10.1161/CIRCULATIONAHA.107.724864. [DOI] [PubMed] [Google Scholar]
- 46.Girvan AC, et al. AGRO100 inhibits activation of nuclear factor-kappaB (NF kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 47.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.