Abstract
We report the construction of a synthetic flavo-heme protein that incorporates two major physiological activities of flavoproteins: light activation of flavin analogous to DNA photolyase and rapid intramolecular electron transfer between the flavin and heme cofactors as in several oxidoreductases. The functional tetra-α-helix protein comprises two 62-aa helix-loop-helix subunits. Each subunit contains a single cysteine to which flavin (7-acetyl-10-methylisoalloxazine) is covalently attached and two histidines appropriately positioned for bis-his coordination of heme cofactors. Both flavins and hemes are situated within the hydrophobic core of the protein. Intramolecular electron transfer from flavosemiquinone generated by photoreduction from a sacrificial electron donor in solution was examined between protoporphyrin IX and 1-methyl-2-oxomesoheme XIII. Laser pulse-activated electron transfer from flavin to meso heme occurs on a 100-ns time scale, with a favorable free energy of approximately −100 meV. Electron transfer from flavin to the lower potential protoporphyrin IX, with an unfavorable free energy, can be induced after a lag phase under continuous light illumination. Thus, the supporting peptide matrix provides an excellent framework for the positioning of closely juxtaposed redox groups capable of facilitating intramolecular electron transfer and begins to clarify in a simplified and malleable system the natural engineering of flavoproteins.
Nature selects from a battery of redox active cofactors and assembles them within a protein matrix to facilitate essential functions such as substrate binding, electron transfer, energy conversion, and chemical catalysis. The cofactors often are juxtaposed closely within a single structure with the physical chemical properties adjusted by the protein environment to perform the desired function satisfactorily and with high fidelity. Studies aimed at understanding the molecular basis of catalytic fitness in enzymes often are confounded by the sheer complexity of natural proteins. It is our goal to uncover the engineering of natural oxidoreductase protein design by simplifying the problem and synthesizing the minimalistic protein structures that assemble working arrays of cofactors and reproduce native-like function. We call these working structures “molecular maquettes.”
To date there has been much success with this approach. The peptide scaffold of choice is a tetra-α-helix-bundle that assembles and is stable over a wide range of conditions (1, 2). This is exemplified by the remarkable utility of the simple scaffold for successful incorporation of up to four hemes as a maquette for the cytochrome b subunit of the cytochrome bc1 complex (3–8), the covalent attachment of a pendant porphyrin dimer as the fundamental component of a photosynthetic reaction center maquette (9, 10), and the incorporation of an iron–sulfur cluster into the loop region to generate a ferredoxin–heme maquette (11). All of the aforementioned systems exhibit key properties of their natural counterparts with respect to the physical chemical characteristics of the cofactors and the influence that the peptide environment exerts on their properties. However, none of these systems was designed to have any inherent function associated with them. In this paper, we describe a molecular maquette that is capable of supporting one of the most fundamental of all biological processes—light activatable electron transfer. Unique to this maquette is its stable assembly in aqueous solution and the ability to incorporate flavin and heme as native-like cofactors within the protein hydrophobic core, resulting in significant modification of their spectral and redox properties. Ultimately, we anticipate that maquettes will be able to support elementary functions, such as classic “dark” oxidoreduction reactions that reflect the action of many natural enzymes.
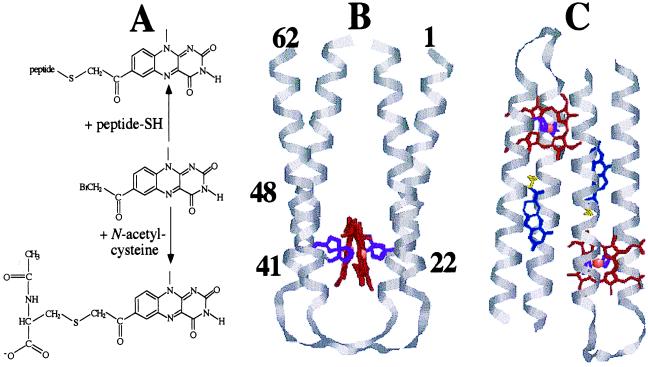
In biology, the most common function of flavin is as a cofactor with versatile redox chemistry in an array of oxidoreductases (12, 13). This is exemplified by flavocytochrome b2, where the flavin is involved in both catalytic dehydrogenation of l-lactate to pyruvate and the subsequent electron transfer to a heme cofactor (14). Flavin also can be used as an efficient photoreceptor and mediator of light-induced electron transfer by DNA photolyase (15) and the plant blue light photoreceptor (16). Irradiation of flavin in solution with blue photons generates the excited triplet state, which is a powerful oxidant. In the presence of a sacrificial electron donor (irreversible reaction) such as EDTA, singly reduced flavosemiquinone is generated, which subsequently can react further with protein-bound cofactors (17–19). Flavin is commonly used as a flash-activatable intermolecular source of electrons for investigating protein-mediated electron transfer (20) and recently has been used as an in situ intraprotein electron donor, where it was covalently attached to a cysteine residue on the surface of cytochrome c to generate semisynthetic flavocytochrome c (21). Thus, we chose to use a flavin chromophore as the light-activatable species in our system. The actual flavin of choice was 7-acetyl-10-methylisoalloxazine (Fig. 1A), which although not naturally occurring, exhibits the same chemistry as natural flavins (22, 23). From a thorough investigation of existing flavoprotein structures, we concluded that engineering a flavin binding site into our existing maquette framework would be premature and would pose considerable design problems. It is evident that flavoproteins, including even the simple flavodoxins, possess a completely different architecture to that offered by tetra-α-helix-bundles (24). However, many flavoproteins contain flavin covalently bound to the polypeptide (25). Therefore, there exists a precedent for localizing the flavin within the helix-loop-helix framework by covalently attaching it to an amino acid side chain situated within the hydrophobic core of the bundle. The maquette design was similar to that of our earlier peptides (2–5), but with the holo maquette containing two covalently bound flavins and two bis-histidine coordinated hemes situated within the tetra-α-helix-bundle (Fig. 1B). Here, we describe the suitability of this minimalistically designed system for supporting light-induced intramolecular electron transfer.
Figure 1.
Construction of a flavo-heme protein maquette. (A) Synthesis of Cys-F1 and covalent attachment of flavin to peptide cysteine. RasMac 2.6.-generated molecular models of: (B) [H:α-l-α]2 with the dimers aligned in a syn topology and (C) [H:Fl-α-l-α]2 with the dimers aligned in an anti topology. Cα backbone, grey; porphyrin macrocycle, red; iron ion, pink; histidine side chains, purple; flavin, blue; cysteine, yellow. The numbers in B refer to relevent amino acid residues in the primary sequence (1 & 62, N- and C-termini; 22 & 41, histidine; 48, cysteine).
MATERIALS AND METHODS
Chemicals and Solvents.
N-acetyl-l-cysteine, bromine, trifluoroethanol, and trifluoroacetic acid were purchased from Aldrich. Ethanedithiol was purchased from Fluka. Guanidine hydrochloride was used as received from Pierce. Fluorenylmethoxycarbonyl-protected amino acid pentafluorophenyl esters were purchased from PerSeptive Biosystems. NovaSyn PR-500 resin was purchased from Calbiochem–Novabiochem. The micro-protein determination kit was purchased from Sigma. All other chemicals and solvents were reagent grade.
Peptide Sequence and Synthesis.
The helix-loop-helix peptide (α-l-α) has the sequence Ac-L⋅KKLREEA⋅LKLLEEF⋅KKLLEEH⋅LKWLEGGGG-GGGGELLKL⋅ HEELLKK⋅CEELLKL⋅AEERLKK⋅L-CONH2, where Ac is the N-terminal acetyl group and the dots emphasize the underlying heptad repeat pattern of the helices. Peptide synthesis was performed on a continuous-flow Milligen 9050 solid phase synthesizer by using standard fluorenylmethoxycarbonyl/tBu methodology. The yield of total peptide cleaved from the resin was ≈95%, and the subsequent yield of reverse-phase HPLC purified α-l-α was 15% of the total peptide. The molecular weights of α-l-α and derivatives were confirmed by laser desorption MS.
Flavin Synthesis and Attachment.
The synthesis of 7-acetyl-10-methylisoalloxazine and the subsequent bromination reaction were performed as described by Levine and Kaiser (22). The bromination reaction typically yielded 70% of the desired 7α-bromoacetyl-10-methylisoalloxazine (Br-Fl), together with 3% of the dibrominated species and 27% unreacted flavin. This material was sufficiently pure to use for subsequent flavination of the peptide. The flavin was covalently attached to the unique cysteine of α-l-α by reacting 5 molar equivalents of Br-Fl in 1:1 dimethylformamide and H20 with 1 equivalent of peptide at room temperature. The reaction progress was monitored by HPLC and, when 95% complete (3 h), was dialyzed against H20 at 4°C to remove dimethylformamide and unreacted Br-Fl. Flavinated peptide, Fl-α-l-α, was purified by HPLC, lyophilized, and stored at −20°C until required. The yield of Fl-α-l-α was 60%. N-acetyl-cysteinyl flavin (Cys-Fl) was synthesized and stored in a similar manner to Fl-α-l-α.
Solution Molecular Mass Determination.
Size-exclusion chromatography was performed on a Beckman System Gold HPLC system equipped with a diode-array detector. A Supelco Sigmachrom GFC-100 column (300 × 7.5 mm) was used, and the sample was eluted with aqueous buffer (50 mM Tris/100 mM NaCl, pH 8.0). The column was calibrated by using aprotinin (6.5 kDa), horse heart cytochrome c (12.1 kDa), chymotrypsinogen (25.0 kDa), ovalbumin (43.0 kDa), and BSA (67.0 kDa).
UV-Vis Spectroscopy.
UV-Vis spectra were recorded on Perkin–Elmer Lambda 2 or Beckman DU-550 diode array spectrophotometers. The α-l-α peptide concentrations were determined by using ɛ280 = 5,600 M−1⋅cm−1 for tryptophan. The Fl-α-l-α peptide concentrations were determined by using ɛ430 = 10,900 M−1⋅cm−1 for oxidized flavin (22). This value was verified to be the same as that for the free flavin (±10%) by assaying the amount of peptide by using a micro-protein determination kit and comparing the values for α-l-α and Fl-α-l-α.
CD Spectropolarimetry.
CD spectra were acquired on an Aviv 62DS spectropolarimeter at 25°C. The buffer used was 10 mM phosphate and 100 mM NaCl (pH 7.0). Peptide denaturation profiles were fit to a dimer folded to monomer unfolded equilibrium by using a nonlinear least-squares routine as described (2).
Redox Potentiometry.
The heme redox midpoint potentials were determined spectrophotometrically at room temperature by using chemical titrations under anaerobic conditions as described (26). Titrations were performed in 50 mM Tris and 100 mM NaCl (pH 8.0). The midpoint potential of the two-electron oxidized to reduced couple (Flox/Flred) for free and peptide-bound flavin were determined by two independent methods. The first used chemical titrations, as described for the heme potentiometry; however, no mediators were used because most absorb light in the same wavelength region as that of the flavin and confuse analysis. We relied entirely on the weak interaction of flavin with the electrode, and the system was left for much longer than usual to establish equilibrium among the electrodes, the solution, and the flavin species after each potential adjustment (30 min). This proved possible because the overall system was reversible and without background oxidative or reductive drift. The second method used the technique introduced by Massey (27). This involves equilibration with an indicator dye whose midpoint potential is known and is within 30 mV of the Flox/Flred couple to be measured. Reduction was initiated and maintained by use of the xanthine/xanthine oxidase system. The dyes that satisfied the selective requirement were 5–5′-indigodisulfonate (Em7 = −140 mV) for free flavin and 5, 5–7′-indigotrisulfonate (Em7 = −90 mV) for Fl-α-l-α.
Fluorescence Spectroscopy.
Fluorescence spectra were acquired on an ISS-2 fluorimeter. The quantum yields of flavin fluorescence were determined by comparing the integral of the emission spectra with that obtained for flavin mononucleotide under identical excitation conditions. Spectra were acquired at room temperature in 50 mM Tris and 100 mM NaCl (pH 8.0) buffer. Quenching constants of flavin and [Fl-α-l-α]2 fluorescence by sodium iodide were determined from the Stern–Volmer equation (28): FO/F = (1 + KSV[I]) exp([I]V), where FO and F are the intensities in the absence and presence of quencher, respectively. KSV (M−1) is the quenching constant, [I] quencher concentration and V is the volume of the quenching sphere of action (M−1).
Photoreduction.
Continuous light photoreduction data were acquired on a Beckman DU-550 diode array spectrophotometer. A Cuda 1–150 light generator was used as a white light source for photoexcitation, and the light was directed onto the sample with a fiber optic light guide. The sample was placed in a sealed glass cuvette and rendered anaerobic by flushing with argon gas while stirring. The buffer in all of the photoreduction experiments was 50 mM phosphate, 100 mM NaCl, and 10 mM EDTA (pH 8.0). Laser flash-induced transient absorption difference spectra in the nanosecond to millisecond time scale were performed with a Quantum Ray DCR-11 YAG laser. The exciting laser pulse was at 355-nm, 20-mJ, 8-ns pulse width, operating at 10 Hz. Detection by a photodiode array was gated at selected intervals following the laser flash and was mediated by a fiber optically guided white light source directed into the sample normal to the incident laser light. The sample was maintained under anaerobic conditions by flushing with argon and was contained in a sealed quartz flow cell connected to a reservoir, such that the sample exposed to laser irradiation was replenished continually. The transient difference spectra were generated by simultaneously subtracting the aquired sample (laser exposed) and reference (no incident laser light). All experiments were performed at room temperature.
RESULTS AND DISCUSSION
Design of [α-l-α]2.
The design was based on our prototype series of heme binding protein maquettes (2–5). The histidines in each 62-aa helix-loop-helix subunit (α-l-α) of the tetra-α-helix-bundle [α-l-α]2 are at positions 22 and 41, appropriate for bis-histidine coordination of bound heme (Fig. 1B). The unique cysteine at position 48 is one complete heptad repeat above that of histidine 41, chosen to place the flavin attachment site at a hydrophobic a position of the helix and is expected to direct the flavin toward the interior of the bundle (4), sterically close to the hemes (Fig. 1C). The flavin of choice was 7-acetyl-10-methylisoalloxazine because it is straightforward to synthesize and has a methyl group in place of the bulky ribose, ribophosphate, or ribophosphate adenine dinucleotide present in naturally occurring flavins (22). Two types of iron protoporphyrin were used in this investigation: protoheme IX (H) and 1-methyl-2-oxomesoheme XIII (OMH) (7). The oxomeso substituents of the porphyrin macrocycle elevate the intrinsic midpoint potential of OMH compared with H and allows the effect of altering the driving force for electron transfer to be probed.
Maquette Architecture and Cofactor Environment.
Size exclusion chromatography confirmed that the apo maquette, [α-l-α]2, and variants eluted with retention times consistent with tetra-α-helix-bundle aggregation states at micromolar concentrations. This was based on column calibration with globular proteins and comparison with the chromatographic behavior of previously characterized tetra-α-helix-bundles (2, 3).
All of the maquettes possess ≈70% α-helical secondary structure. The Θ222/Θ208 ratio for [α-l-α]2 in aqueous buffer is close to 0.95, as opposed to 0.85 in 50% trifluoroethanol. A Θ222/Θ208 ratio of 1.0 is considered diagnostic of a coiled-coil structure (29, 30). The decrease in the Θ222/Θ208 ratio in high concentrations of trifluoroethanol (>4 M) known to disrupt hydrophobic core packing but maintain secondary α-helical structure is representative of the formation of extended or monomeric helices (9, 31, 32).
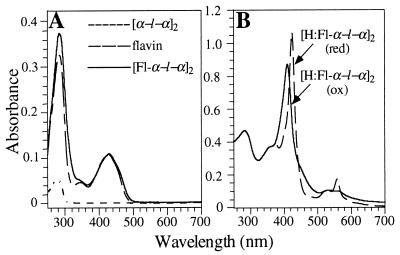
Fig. 2A shows the absorption spectra of flavin compared with that of [Fl-α-l-α]2 in aqueous buffer. Attachment of flavin to [α-l-α]2 causes a shift of the blue absorption peak from 427 to 430 nm, but otherwise the spectra are qualitatively similar. Fig. 2B shows that addition of two hemes per tetra-α-helix-bundle to generate [H:Fl-α-l-α]2 produces a typical bis-histidine ligated ferric b-heme spectrum superimposed on the flavin absorption. On reduction with sodium dithionite, the typical ferrous b-heme spectrum is obtained.
Figure 2.
UV-visible spectra of maquettes in various states of construction: (A) free flavin, [α-l-α]2 and [Fl-α-l-α]2; (B) oxidized [H:Fl-α-l-α]2 (γmax at 411 and 535 nm, corresponding to the Soret and unresolved α/β bands, respectively), and reduced [H:Fl-α-l-α]2 (γmax at 423, 560, and 531 nm, corresponding to the Soret α and β bands, respectively). All concentrations were 10 μM, the buffer used was 50 mM Tris and 100 mM NaCl (pH 8.0), and the spectra were acquired at room temperature.
The thermodynamic stability of [α-l-α]2 and the effect of introducing the flavin and heme cofactors on bundle stability was probed by the use of chemical denaturants. Addition of guanidine hydrochloride (Gdn.HCl) to [α-l-α]2 resulted in a loss of elipticity at Θ222 nm in the CD spectrum, consistent with denaturation to a disordered structure with a [Gdn.HCl]1/2 value of 5.5 M. The free energy for unfolding ΔGH20 was 18.1 ± 0.8 kcal⋅mol−1, and the cosolvation term m was 2.4 ± 0.2 kcal⋅mol−1⋅M−1. Placing flavin in the hydrophobic core of [α-l-α]2 to form [Fl-α-l-α]2 causes the [Gdn.HCl]1/2 value to decrease to 3.8 M, indicating that attachment of flavin destabilizes the bundle (ΔΔG = 6.3 kcal⋅mol−1). The cooperativity of the unfolding transition is also lowered in [Fl-α-l-α]2 (m = 1.5 ± 0.2 kcal⋅mol−1⋅M−1) as compared with [α-l-α]2, probably because of weakened packing of the hydrophobic core due to the presence of a bulky flavin chromophore. Despite this, the [Fl-α-l-α]2 still forms a stable tetra-α-helix-bundle (ΔGH20 = 11.8 ± 0.9 kcal⋅mol−1). Binding of two hemes to both [α-l-α]2 and [Fl-α-l-α]2 shifts the midpoint of the denaturation profile to higher Gdn.HCl concentration, but renders the process less cooperative. No attempt was made to fit the denaturation profile of [H:α-l-α]2 or [H:Fl-α-l-α]2 as the process was not fully reversible because of heme aggregation on unfolding.
Comparison of the flavin and [Fl-α-l-α]2 fluoresence emission spectra revealed that the intensity is quenched 6-fold on attachment to the peptide (quantum yield Φf, flavin = 0.30, and [Fl-α-l-α]2 = 0.05), which is a general property of sulfur-linked flavinyl peptides (22, 33). Stepwise addition of iodide, a dynamic fluorescence quencher, was used to provide an indication for the degree of shielding that the peptide environment affords the flavin in [Fl-α-l-α]2 from bulk solvent. The data were fit to the Stern–Volmer equation to obtain values for dynamic (KSV) and static (V) quenching. The values for KSV and V were 30.4 ± 1.5 M−1 and 2.3 ± 0.4 M−1 for flavin and 7.7 ± 0.3 M−1 and 0.2 ± 0.1 M−1 for [Fl-α-l-α]2, respectively. Thus, the quenching constants are 4-fold (KSV) and 13-fold (V) lower for [Fl-α-l-α]2 compared with flavin in solution. The difference in the magnitude of KSV is comparable to values reported in the literature for the degree of shielding conferred by the protein environment in natural flavoproteins (28). This implies that the peptide matrix significantly shields the flavin from quenching agents in bulk solution.
The dissociation constants (KD) of the two hemes (H) bound to [α-l-α]2 are 0.035 and 0.135 μM (SD ±20%), similar to reported values for related maquettes (3–5). The KD values for H from [Fl-α-l-α]2 are 0.13 and 1.5 μM (±20%), approximately one order of magnitude weaker than the corresponding values for [α-l-α]2, implying that the presence of the flavin adjacent to the heme hinders binding. The KD values of OMH bound to [α-l-α]2 (0.05 and 0.17 μM) and [Fl-α-l-α]2 (0.15 and 1.2 μM) are similar to H.
Cofactor Potentiometry and Maquette Topology.
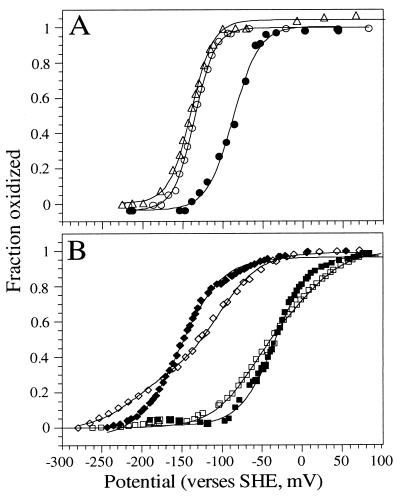
The potentiometric properties of the flavin were examined at pH 8.0 for alteration induced by the protein environment. Fig. 3 shows Nernst plots of redox titrations for the two-electron couple of free flavin, Cys-Fl and [Fl-α-l-α]2, along with the one-electron heme couples for [H:α-l-α]2, [H:Fl-α-l-α]2, [OMH:α-l-α]2, and [OMH:Fl-α-l-α]2. Table 2 summarizes these data. The 50-mV rise in midpoint potential for [Fl-α-l-α]2 compared with free flavin is not simply due to covalent attachment of the flavin to the peptide because the midpoint potential of the Flox/Flred couple is unaltered either when attached to cysteine (Cys-Fl) or to a surface exposed cysteine on cytochrome c (21). This indicates that the shift in the flavin midpoint potential in [Fl-α-l-α]2 is caused by the peptide environment, which stabilizes the fully reduced bound flavin by 1.15 kcal⋅mol−1 relative to the oxidized form. In natural flavoproteins, the two-electron Flox/Flred couple is also typically within 100 mV of that for free flavin. Some natural flavoproteins are engineered to stabilize the single reduced flavosemiquinone (Flsq) (34, 35); however, it is not clear from Fig. 3A whether the tetra-α-helix protein interior stabilizes Flsq relative to solution. The best Nernst fit to the redox data has an n value of 1.6 and could reflect either such stabilization or a small amount of heterogeneity in the flavin environment.
Figure 3.
Oxidation-reduction potentials of flavin and hemes in the maquettes. Nernst plots illustrating Flox/Flred (n = 2) midpoint potentials of: (A) flavin (○), Cys-Fl (▵), and [Fl-α-l-α]2 (•); (B) Hox/Hred (n = 1) midpoint potentials of [H:α-l-α]2 (◊), [H:Fl-α-l-α]2 (♦), [OMH:α-l-α]2 (□), and [OMH:Fl-α-l-α]2 (■). The midpoint potentials are reported in Table 2.
The Em8 values of the two hemes in [H:α-l-α]2 are −220 and −105 mV, respectively, and are the same within experimental error as previously determined values for heme incorporated into related maquettes (3–5). These midpoint potentials are raised in comparison with free bis-imidazole heme: Em8.5 = −235 mV (J. Schifman and P.L.D., unpublished observations), illustrating the relative destabilization of the charged ferric-heme with respect to the neutral ferrous form within the low dielectric protein interior (7). Moreover, the 100-mV splitting of the heme potentials is consistent with negative heme–heme electrochemical cooperativity brought about by the close proximity of two charged ferric hemes in the protein interior (3). This splitting of the potentials has been used to indicate that the (H:α-l-α) subunits in [H:α-l-α]2 have adopted a syn topology (Fig. 1B) (A. M. Grosset & P.L.D., unpublished observations). However, for the flavin containing holo maquette [H:Fl-α-l-α]2, there is no evidence for splitting of the heme potentials; both are at −153 mV. This effect has been shown to be the result of the subunits adopting an overall anti topology, which would place the hemes at diametrically opposite ends of [H:Fl-α-l-α]2 (Fig. 1C). The anti topology greatly diminishes the electrostatic effects attributed to the splitting of the midpoint potentials and gives rise to the same well defined midpoint potential for both hemes. The reason that [H:Fl-α-l-α]2 adopts an anti and not a syn topology as in [H:α-l-α]2 is probably due to the presence of the flavin sterically driving the assembly equilibrium to this conformation, as observed for a related maquette with incorporated spin label probes (2).
The midpoint potentials of OMH in [OMH:α-1-l-α]2 and [OMH:Fl-α-l-α]2 are ≈120 mV more positive than that of the analogous maquettes containing H, due to the electron withdrawing substituents on the OMH porphyrin macrocycle stabilizing the reduced state. The patterns of the redox behavior for OMH and H when bound to [α-l-α]2 or [Fl-α-l-α]2 were identical (Fig. 3B), implying that the adopted maquette topologies are influenced by the same factors.
Light-Activated Electron Transfer from Flavin to Heme.
Favorable intramolecular electron transfer from Flsq to heme requires a Flox/Flsq couple Em8 more negative than the heme Em8. In solution, Flox/Flsq is ≈30 mV lower than the Flox/Flred midpoint. Thus, if Flsq is not stabilized by the protein relative to solution, electron transfer from Flsq to Hox would be unfavorable by ≈30 mV. However, electron transfer from Flsq to OMHox would be favorable by ≈100 mV. Only if Flsq is stabilized by the protein would Flsq to Hox electron transfer become favorable.
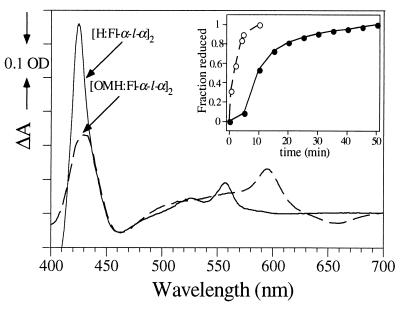
Fig. 4 demonstrates that under continuous illumination, [H:Fl-α-l-α]2 and [OMH:Fl-α-l-α]2 can support photoreduction. The inset to Fig. 4 shows that reduction of OMH is essentially complete after 5 min of irradiation, whereas H reduction takes ≈60 min. Also apparent is a significant 5-min lag phase before the onset of H reduction in [H:Fl-α-l-α]2, presumably caused by a requirement for accumulation of reduced flavin before initiation of thermodynamically unfavorable heme reduction. No such lag phase is evident for [OMH:Fl-α-l-α]2. This demonstrates that the difference in free energy, ΔG°, for intramolecular electron transfer in both systems affects the rate of accumulation of reduced heme. Both H and OMH reduction occur slowly because the reaction is light-limited and the quantum yield of flavin fluoresence in the holo maquettes is low (Φf ≈ 0.01). When the light was switched off, the sample remained fully reduced, illustrating that the reduced state of the system is stable under anaerobic conditions. On aeration of the samples, reoxidation rapidly occurred. In control experiments (not shown), where [H:α-l-α]2 and [OMH:α-l-α]2 (unflavinated) were irradiated with light, no photoreduction occurred, illustrating the absolute requirement of flavin for mediating heme reduction in this system.
Figure 4.
Photoreduction of the flavo-heme maquettes on continuous light illumination. UV-visible reduced minus oxidized difference spectra of [H:Fl-α-l-α]2 and [OMH:Fl-α-l-α]2, illustrating the generation of fully reduced maquettes. The sample concentrations were 10 μM. (Inset) Time course of the reduction process (•) [H:Fl-α-l-α]2 reduction and (○) [OMH:Fl-α-l-α]2 reduction.
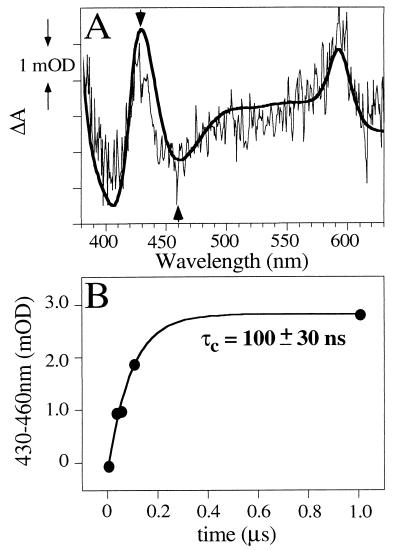
The microscopic time course for accumulation of reduced species in [OMH:Fl-α-l-α]2 was probed by pulsed laser excitation, with subsequent detection of absorbance difference transients at set time intervals after the laser flash. Fig. 5A shows the difference spectrum for [OMH:Fl-α-l-α]2 obtained 10 μs after the laser flash. The spectrum is consistent with the reduced minus oxidized difference spectrum shown in Fig. 4 for [OMH:Fl-α-l-α]2, implying that reduced OMH is generated rapidly in the maquette. It was not possible to observe H reduction in [H:Fl-α-l-α]2 for the laser-activated experiments, consistent with the findings from the continuous light illumination experiments that the ΔG° for Flsq to Hox electron transfer is unfavorable. Fig. 5B depicts the time course for the OMH reduction process, where the data points were obtained for a series of transient absorption spectra gated at set time intervals after the laser flash (spectra not shown). The rate of accumulation of reduced OMH is well described by a single exponential process, with a time constant (τc) of ≈100 ns. Whether τc actually reflects the intramolecular electron transfer from Flsq to OMHox, depends on a number of factors. The scheme below illustrates the photochemical processes and electron transfer events that can occur on light irradiation in the presence of an electron donor (for simplicity, only one subunit is considered): ![]()
Figure 5.
Laser-induced rapid accumulation of reduced heme. (A) Laser flash photolysis absorbance difference spectrum of 10 μM [OMH:Fl-α-l-α]2 with the diode array detection gated 10 μs after the laser flash. The spectrum is the mean of 500 acquisitions. Superimposed on this is the steady-state aquired reduced minus oxidized difference spectrum of the same sample, to illustrate that the absorbance difference is due to OMH reduction. (B) Time course for OMH reduction, obtained from a series of transient absorption spectra gated at set intervals after the laser flash (0.03-, 0.05-, 0.07-, 0.1-, and 1.0-μs delay). The absorption difference at 430–460 nm was used to monitor this process (arrows in A). The data are fit to a single exponential, with the indicated time constant (τc) for reduction.
The scheme describes: (1) flavin photoexcitation to the singlet state, with subsequent intersystem crossing to generate the triplet state, (2) abstraction of an electron from EDTA generating Flsq, and (3) intramolecular flavin to heme electron transfer; the scheme also describes that, (4) with constant light irradiation in the presence of excess EDTA, the maquette will become fully reduced (Fig. 4). The superscripts 0, 1, and 2 refer to the redox state of the cofactors: 0, oxidized; 1, Flsq or reduced heme; 2, Flred; 3Fl*, excited triplet state. The second order rate constant for EDTA oxidation by the excited flavin triplet state (3Fl*, reaction 2) is of the order 109 M−1⋅s−1 (17), and under the experimental conditions (0.01 M EDTA), the τc for this process would be anticipated to be ≈100 ns. The lifetime of 3Fl* is ≈10 μs (data not shown) and thus persists for sufficient time to abstract an electron from EDTA in solution. Thus, at this juncture, we can state that the reported τc of 100 ns reflects a pseudo first-order process for electron transfer from EDTA through flavin and thence to OMH (reactions 1–3) because the intramolecular rate may be limited by the generation of Flsq. The yield of OMHred generated by the laser flash in the holo maquette is ≈0.01, which correlates well with the quantum yield for generation of 3Fl*.
The relative simplicity and control conferred by these working maquettes provides the framework for exploring physiologically relevent actions akin to natural photoactivatable flavo-heme proteins (36). The ultimate goal of future studies is to build on this foundation and construct maquettes that are able to support enzymatic catalytic function and thus serve as simple systems to probe the fundamental requirements of catalytic fitness that nature displays in oxidoreductases.
Table 1.
Midpoint potentials of the flavin and heme redox couples
| Redox couple | Em8, mV | n |
|---|---|---|
| Flavin, (Flox/Flred) | −138 ± 5* | 2 |
| Cys-Fl, (Flox/Flred) | −146 ± 5* | 2 |
| [Fl-α-l-α]2, (Flox/Flred) | −95 ± 10 | 1.6 |
| [H:α-l-α]2, (Hox/Hred) | −105 ± 5 (0.68) | 1 |
| −220 ± 10 (0.32) | 1 | |
| [H:Fl-α-l-α]2, (Hox/Hred) | −153 ± 10 | 1 |
| [OMH:α-l-α]2, (OMHox/OMHred) | +14 ± 5 (0.37) | 1 |
| −63 ± 5 (0.63) | 1 | |
| [OMH:FL-α-l-α]2, (OMHox/OMHred) | −30 ± 10 | 1 |
The numbers in parentheses refer to the relative amplitude of the two n = 1 couples contributing to the redox reaction.
These values were obtained from both equilibrium titration and the xanthine/xanthine oxidase system as described in Materials and Methods. In both cases, the values determined were the same within experimental error.
Acknowledgments
MS analyses were performed by the University of Pennsylvania Protein Chemistry Laboratory. We thank Drs. C. Mark Phillips and Sondra Vitols from the Regional Laser and Biotechnology Laboratory, Department of Chemistry, University of Pennsylvania for their invaluable assistance with laser experiments, Dr. Kevin Smith (University of California, Davis) for the gift of OMH, and Dr. Vincent Massey (University of Michigan) for the gift of 5-deaza flavin. This work was supported by the National Institute of Health Grant GM 41048 and in part by the National Science Foundation. R.E.S. and F.R. gratefully acknowledge receipt of postdoctoral fellowships from the North Atlantic Treaty Organization and the European Molecular Biology Organization, respectively.
ABBREVIATIONS
- Fl
flavin (7-acetyl-10-methylisoalloxazine)
- H
protoheme IX
- OMH
1-methyl-2-oxomesoheme XIII
- α-l-α
helix-loop-helix peptide
- Fl-α-l-α
helix-loop-helix peptide with flavin attached
References
- 1.Bryson J W, Betz S F, Lu H S, Suich D J, Zhou H X, O’Neil K T, DeGrado W F. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 2.Gibney B R, Johansson J J, Rabanal F, Skalicky J J, Wand A J, Dutton P L. Biochemistry. 1997;36:2798–2806. doi: 10.1021/bi9618225. [DOI] [PubMed] [Google Scholar]
- 3.Robertson D E, Farid R S, Moser C C, Urbauer J L, Mulholland S E, Ravindernath P, Lear J D, Wand A J, DeGrado W F, Dutton P L. Nature (London) 1994;368:425–432. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 4.Gibney B R, Rabanal F, Skalicky J J, Wand A J, Dutton P L. J Am Chem Soc. 1997;119:2323–2324. [Google Scholar]
- 5.Gibney B R, Rabanal F, Reddy K S, Dutton P L. Biochemistry. 1998;37:4635–4643. doi: 10.1021/bi971856s. [DOI] [PubMed] [Google Scholar]
- 6.Choma C T, Lear J D, Nelson M J, Dutton P L, Robertson D E, DeGrado W F. J Am Chem Soc. 1994;116:856–865. [Google Scholar]
- 7.Kalsbeck W A, Robertson D E, Pandey R K, Smith K M, Dutton P L, Bocian D F. Biochemistry. 1996;35:3429–3438. doi: 10.1021/bi952662k. [DOI] [PubMed] [Google Scholar]
- 8.Gray K, Daldal F. In: Anoxygenic Photosynthetic Bacteria. Blankership R E, Madigan M T, Brauer C, editors. Dordrecht, The Nertherlands: Kluwer; 1995. pp. 725–774. [Google Scholar]
- 9.Rabanal F, DeGrado W F, Dutton P L. J Am Chem Soc. 1996;118:473–474. [Google Scholar]
- 10.Rabanal F, Gibney B R, DeGrado W F, Moser C C, Dutton P L. Inorg Chim Acta. 1996;243:213–218. [Google Scholar]
- 11.Gibney B R, Mulholland S E, Rabanal F, Dutton P L. Proc Natl Acad Sci USA. 1996;93:15041–15046. doi: 10.1073/pnas.93.26.15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massey V. FASEB J. 1995;9:473–475. doi: 10.1096/fasebj.9.7.7737454. [DOI] [PubMed] [Google Scholar]
- 13.Ghisla S, Massey V. Eur J Biochem. 1989;181:1–14. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 14.Daff S, Ingledew W J, Reid G A, Chapman S K. Biochemistry. 1996;35:6345–6350. doi: 10.1021/bi9522559. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A. Biochemistry. 1994;33:2–7. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- 16.Lin C, Robertson D E, Ahmad M, Raibekas A A, Schuman-Jorns M, Dutton P L, Cashmore A R. Science. 1995;269:68–71. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed I, Cusanovich M A, Tollin G. Proc Natl Acad Sci USA. 1981;78:6724–6728. doi: 10.1073/pnas.78.11.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung J, Tollin G. Biochemistry. 1981;20:5124–5131. doi: 10.1021/bi00521a005. [DOI] [PubMed] [Google Scholar]
- 19.Traber R, Kramer H E A, Hemmerich P. Biochemistry. 1982;21:1687–1693. doi: 10.1021/bi00536a033. [DOI] [PubMed] [Google Scholar]
- 20.Tollin G. J Bioenerg Biomem. 1995;27:303–309. doi: 10.1007/BF02110100. [DOI] [PubMed] [Google Scholar]
- 21.Twitchet M B, Ferrer J C, Siddarth P, Mauk A G. J Am Chem Soc. 1997;119:435–436. [Google Scholar]
- 22.Levine H L, Kaiser E T. J Am Chem Soc. 1978;100:7670–7677. [Google Scholar]
- 23.Kaiser E T, Lawrence D S. Science. 1984;226:505–511. doi: 10.1126/science.6238407. [DOI] [PubMed] [Google Scholar]
- 24.Schulz G E. Curr Opin Struct Biol. 1992;2:61–67. [Google Scholar]
- 25.Mewies M, McIntire W S, Scrutton N S. Prot Sci. 1998;7:7–20. doi: 10.1002/pro.5560070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutton P L. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 27.Massey V. Flavins and Flavoproteins. Berlin: Walter de Gruyer; 1990. pp. 59–66. [Google Scholar]
- 28.Bastiaens P I H, Van-Hoek A, Van-Berkel W J H, de-Kok A, Visser A J W G. Biochemistry. 1992;31:7061–7068. doi: 10.1021/bi00146a006. [DOI] [PubMed] [Google Scholar]
- 29.Zhou N E, Kay C M, Hodges R S. J Biol Chem. 1992;267:2664–2670. [PubMed] [Google Scholar]
- 30.Gradis T J, Myszka D G, Chaiken I M. Biochemistry. 1993;32:12664–126. doi: 10.1021/bi00210a015. [DOI] [PubMed] [Google Scholar]
- 31.Dill K A, Bromberg S, Yue K, Fiebig K M, Yee D P, Thomas P D, Chan H S. Protein Sci. 1995;4:561–565. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau S Y M, Taneja A K, Hodges R S. J Biol Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- 33.Sakamoto S, Aoyagi H, Nakashima N, Mihara H. J Chem Soc Perkin Trans. 1996;2:2319–2326. [Google Scholar]
- 34.Ludwig M L, Pattridge K A, Metzger A L, Dixon M M, Eren M, Feng Y, Swenson R P. Biochemistry. 1997;36:1259–1280. doi: 10.1021/bi962180o. [DOI] [PubMed] [Google Scholar]
- 35.Tegoni M, Janot J-M, Labeyrie F. Eur J Biochem. 1986;140:39–45. doi: 10.1111/j.1432-1033.1984.tb08064.x. [DOI] [PubMed] [Google Scholar]
- 36.Galland P, Senger H. In: Photorecptor Evolution and Function. Holmes M G, editor. London: Academic; 1991. pp. 66–124. [Google Scholar]