Abstract
Metabolomics is an emerging area of research focused on measuring small molecules in biological samples. There are a number of different types of metabolomics, ranging from global profiling of all metabolites in a single sample to measurement of a selected group of analytes. Microfluidics and related technologies have been used in this research area with good success. The aim of this review article is to summarize the use of microfluidics in metabolomics. Direct application of microfluidics to the determination of small molecules is covered first. Next, important sample preparation methods developed for microfluidics and applicable to metabolomics are covered. Finally, a summary of metabolomic work as it relates to analysis of cellular events using microfluidics is covered.
Keywords: Microfluidics, metabolomics, metabolic profiling, review
I INTRODUCTION
The goal of this review is to summarize the use of microfluidics as a novel tool set for metabolomics, metabolic profiling, and other metabolite related biological studies. To the knowledge of the authors this is the first such review generated, and thus we will include articles dating back as far as 2000 to give the reader a comprehensive view of the state of the art. While this is the first review of this topic, a number of important reviews on topics ranging from microfluidic and general analytical methods applicable to metabolomics [1–6] to metabolomic and metabolic profiling [7–12] have been published previously. For specific topics, it is suggested the reader consult these works. These reviews are by no means a comprehensive list of all reviews in this field given that several dozen review articles have been published over the last nine years ranging in scope from very specific to more general.
Before discussing specific applications of microfluidics to metabolomics we will first define metabolomics and the various subsets of this discipline. While many people are familiar with genomics and proteomics, the terminology associated with metabolomics is not as clearly defined. For the sake of this review, we define metabolomics as five separate categories: 1) metabolomics, 2) metabonomics, 3) metabolic fingerprinting, 4) metabolic profiling, and 5) targeted metabolic profiling [13]. Metabolomics is the overall study of metabolite expression at any given point in time. Commonly this is done using qualitative techniques like nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) coupled to a high-resolution separation technique like liquid (LC) or gas chromatography (GC). These techniques provide the ability to detect many species in a single sample. In metabonomics, the goal is to follow changes in the concentrations of large numbers of metabolic markers over time [14]. Again, NMR and MS techniques are common in this form of metabolomics. Often times metabonomic studies make use of radiolabeled nutrients or other inputs to quantitatively compare different profiles. To date, metabolomics and metabonomics make up the largest research segment in metabolomics largely because they can be used to determine biomarkers of disease as well as helping elucidate specific pathways associated with biological changes. The remaining three areas of metabolomics have more specific applications. Metabolic fingerprinting seeks to measure a global profile of metabolites with identification of specific profiles based on pattern recognition. In these applications, non-specific detection methods are used reducing the analysis cost and often increasing method throughput. The major downfall of metabolic fingerprinting is the inability to identify specific biomarkers for a disease state or therapeutic endpoint. Metabolic profiling seeks to measure a specific subset of compounds such as amino acids or carbohydrates. A common area of research in the field of metabolic profiling is lipidomics which seeks to measure lipids present in a sample. Lipids represent an important class of metabolites that are of significant interest because their multitude of roles in biology. Metabolic profiling can use a wide variety of techniques depending on the specific analytes in question. The final area of metabolomics is targeted metabolic profiling. In this area of research, one or two analytes are tracked with time. This area of research has the longest history and is often excluded from the discussion of metabolomics because of the limited number of analytes profiled. It is, however, a very useful tool for understanding biological systems.
As mentioned above, the most common tools in metabolomics are NMR, GC-MS, and LC-MS because they are capable of detecting and identifying many analytes in a single sample. Microfluidics has only recently been added to the metabolomics toolbox but has the potential to a play a significant role in this field. For example, extensive, time-consuming sample preparation techniques consisting of multiple extractions and derivatization reactions are commonly associated with metabolomic studies [15]. These steps could be carried out in a microfluidic format with reduced time and increased efficiency. To date the vast majority of microfluidic systems have focused on coupling selective separation and detection systems for determination of biological samples. In addition, a number of microfluidic systems have been developed that integrate biological systems (cells) with a microfluidic system to perform metabolomic studies. This review will first cover microseparation techniques since this is the most common use of microfluidics in metabolomics. The review then covers sample preparation techniques such as filtration and solid phase extraction that have been developed for microfluidics and are applicable to metabolomic studies. Finally, we will cover integrated microfluidic systems that combine aspects of biology with microanalytical systems. Table 1 summarizes pertinent information for research papers introduced in this review, including type of metabonomics, analyte investigated, detector type, and microfluidic material.
Table 1.
Summary of research articles introduced within this review. Each references is categorized by microfluidic application type, first author name, publication year, type of metabolic analysis, the analyte(s) investigated, material used for microfluidics, and detection mode. Pertinent comments are noted for each reference in the right-most column.
| Methodology | First author name | Reference | Publication year | Type of metabolic analysis |
Type of analyte | Type of microfluidic device |
Detection mode | Other comments |
|---|---|---|---|---|---|---|---|---|
| Microchip-CE | J. Wang | [33] | 2007 | Targeted metabolic profiling |
Lactate and glucose | Glass | Electrochemical | Analysis of lactate and glucose in human serum and blood |
| Microchip-CE | Q.L. Zhang | [30] | 2005 | Targeted metabolic profiling |
Caffeine and theophylline |
PDMS | Electrochemical | 4 µm LOD for caffeine an theophylline |
| Microchip-CE | Q.L. Zhang | [31] | 2007 | Targeted metabolic profiling |
Morphine and codeine |
PDMS | Electrochemical | Monitoring of human urine |
| Microchip-CE | M.A. Schwarz | [27] | 2004 | Targeted metabolic profiling |
Neurotransmitters | PMMA | Electrochemical | 10–100nM LOD |
| Microchip-CE | M. Vlckova | [29] | 2007 | Targeted metabolic profiling |
Neurotransmitters | PMMA | Electrochemical | Analysis of brain homogenates |
| Microchip-CE | T. Miyado | [17] | 2004 | Targeted metabolic profiling |
Nitrate and nitrite | Quartz | UV–vis | Analysis of nitrate and nitrite in human serum and saliva |
| Microchip-CE | T. Miyado | [18] | 2006 | Targeted metabolic profiling |
Nitric oxide | Quartz | UV–vis | 53 and 41 µM LOD for nitrite and nitrate, respectively |
| Microchip-CE | T. Miyado | [19] | 2008 | Targeted metabolic profiling |
Nitric oxide metabolites |
Quartz | UV–vis | Separation of nitrate and nitrite in under 6.5 s |
| Microchip-CE | N. Callewaert | [37] | 2004 | Targeted profiling | N-glycans | Glass | Fluorescence | Dynamic coating, gel based separation |
| Microchip-CE | C.M. Chen | [32] | 2009 | Targeted metabolic profiling |
Mercaptopurine | PMMA | Electrochemical | 100 nM LOD |
| Microchip-CE | E. Guihen | [20] | 2005 | Targeted metabolic profiling |
Phloroglucinols | Quartz | Fluorescence | 15 s separation |
| Microchip-CE | M. Fouad | [21] | 2008 | Plant metabolomics |
Glucosinolates | PMMA | Fluorescence | Does not require offline derivitization |
| Microchip-CE | P.B. Allen | [34] | 2009 | Metabolic profiling | Mitochondria metabolites |
Glass | Fluorescence | Single mitochondria analysis |
| Microchip-CE | R.E. Holcomb | [23] | 2009 | Targeted metabolic profiling |
Catecholamines | PDMS | Electrochemical | Electrode array detector |
| Microchip-CE | B.F. Liu | [36] | 2003 | Targeted metabolic profiling |
Flavins | Glass | Fluorescence | Native fluorescence detection, sub-second separations |
| Microchip-CE | C.D. Garcia | [26] | 2004 | Targeted metabolic profiling |
Creatinine, creatine, uric acid |
PDMS | Electrochemical | Detecting markers of renal function |
| Microchip-CE | C.D. Garcia | [24] | 2004 | Targeted metabolic profiling |
Glucose | PDMS | Electrochemical | 1.2 µM LOD |
| Microchip-CE | R.W. Hompesch | [25] | 2005 | Targeted metabolic profiling |
Catechins | PDMS | Electrochemical | Micellar separation using SDS |
| Cellular Analysis | W. Satoh | [70] | 2009 | Targeted metabolic profiling |
Ammonia | PDMS and glass | Electrochemical | Analysis of hepatocyte cells |
| Cellular analysis | R. Gomez-Soderberg | [64] | 2005 | Metabonomics | Unknown | Silicon | Impedance | Measuring bacterial metabolic activity |
| Cellular analysis | R. Gomez | [65] | 2002 | Metabonomics | Unknown | Silicon | Impedance | Measuring bacterial metabolic activity |
| Cellular analysis | D.O. Fesenko | [66] | 2005 | Metabonomics | Arsenite | Glass | Fluorescence | Cells immobilized in hydrogels |
| Cellular analysis | W. Cheng | [72] | 2006 | Targeted metabolic profiling |
Lactate, calcium ion |
Glass and SU-8 | Electrochemical | Single cardiomyocyte |
| Cellular analysis | J.Q. Boedicker | [67] | 2008 | Metabonomics | Cefoxitin | PDMS | Fluorescence | Staphylococcus aureus |
| Cellular analysis | J.C. Zguris | [78] | 2005 | Metabonomics | Cytochrome P450 | PEG and PDMS | Fluorescence | Microsomes and hepatocytes |
| Cellular analysis | R. Baudoin | [79] | 2007 | Targeted metabolic profiling |
Glucose, glutamine, ammonia |
PDMS | UV–vis | Madin Darby Canine Kidney (MDCK cells) |
| Cellular analysis | L.H. Wang | [77] | 2008 | Targeted metabolic profiling |
Glutathione | PDMS and glass | Fluorescence | MCF-7 breast cancer cells |
| Cellular analysis | S. Faley | [76] | 2008 | Targeted metabolic profiling |
Calcium ion | PDMS | Fluorescence | T cell capture and analysis |
| Cellular analysis | A.M. Clark | [71] | 2009 | Targeted metabolic profiling |
Glycerol | Glass | Fluorescence | Adipocytes |
| Cellular analysis | R. Davidsson | [69] | 2004 | Targeted metabolic profiling |
Glucose, ethanol | Silicon | Chemiluminescence | Monitoring release from yeast cells |
| Cellular analysis | J.V. Rocheleau | [75] | 2004 | Targeted metabolic profiling |
NAD(P)H, calcium ion |
PDMS | Fluorescence | Glucose stimulation of pancreatic islets |
| Cellular analysis | T.D. Oblak | [81] | 2009 | Targeted metabolic profiling |
Glutathione, c-peptide |
PDMS | Fluorescence | Diagnostic for type 2 diabetes |
| Cellular analysis | N. Klauke | [73] | 2007 | Metabolic profiling | Calcium ion | PDMS | Fluorescence | Intracellular signaling of cardiomyocytes |
| Cellular analysis | H. Zhu | [82] | 2008 | Targeted metabolic profiling |
Cytokines, t cells | PDMS | Fluorescence | Microarray for infectious diseases |
| Cellular analysis | P. Hersen | [74] | 2008 | Targeted metabolic profiling |
Proteins | PDMS | Bandwidth, fluorescence |
Signal processing of single cells |
| Cellular analysis | S.K. Yoo | [80] | 2007 | Targeted metabolic profiling |
Reactive oxygen species |
Glass and silicon | Bioluminescence | Toxicity monitoring with bacterial cells |
| Cellular analysis | A. Carraro | [83] | 2008 | Targeted metabolic profiling |
Albumin | PDMS | UV–vis | Culture of hepatocytes and monitoring liver function |
| Cellular analysis | B. Ma | [56] | 2009 | Targeted metabolic profiling |
Acetaminophen metabolites |
PDMS and quartz | UV–vis, fluorescence |
Drug metabolism and cell cytotoxicity |
| Cellular analysis | J.P. Urbanski | [55] | 2008 | Metabolic profiling | Glucose, pyruvate, lactate |
PDMS | Fluorescence | Integrated device for analyzing embryo viability |
| Microchip-LC | P. Sajonz | [44] | 2007 | Pharmacological profiling |
Dihydrobenzoin enantiomers |
Eksigent Express 800 |
UV–vis | Pharmaceutical analysis of chiral mixtures |
| Microchip-LC | C.E. Hop | [45] | 2006 | Pharmacological profiling |
Metabolites | Silicon | MS | Pharmacokinetics and metabolism |
| Microchip-LC | Y.Q. Lin | [47] | 2008 | Metabolomics | Cyanobacterial metabolites |
Fused silica | MS and NMR | LC and droplet NMR |
| Microchip-LC | E.R. Wickremsinhe | [46] | 2005 | Targeted metabolic profiling |
Reboxetine spiked dog plasma |
Unknown | MS | Metabolism of reboxetine |
| Microchip-LC | L. Wang | [48] | 2008 | Metabolic profiling | Glucose | Microelectromechanical systems |
Thermopile | Continuous monitoring of glucose |
| Microfluidic assay | B.M. Murphy | [58] | 2008 | Targeted metabolic profiling |
Metabolites, proteins |
PDMS and silicon | Fluorescence | Micromosaic immunoassays |
| Microfluidic assay | H.M. Wu | [57] | 2008 | Metabolic profiling | Glucose | PDMS | Radiological | Microfluidic device for blood sampling |
| Sample preparation | K. Jo | [59] | 2007 | Peptide profiling | Neuropeptides | PDMS and silicon | MS | MALDI-MS on-chip |
| Sample preparation | C.D. Mansfield; R.A. Shaw | [49,50] | 2006 | Targeted metabolic profiling |
Creatinine | polymer | IR | Laminar fluid diffusion interface for sample cleanup |
| Sample preparation | H.Y. Tan | [54] | 2008 | Targeted metabolic profiling |
Sarin | PMMA | UV–vis | Sample preparation of whole blood |
| Solid phase extraction | A.M. Tan | [52] | 2003 | Drug metabolism | Antidepressants | Cyclic olefin (zeonor) |
MS | Array of solid phase extraction elements |
| Solid phase extraction | S. Benetton | [51] | 2003 | Drug metabolism | Antidepressants | Cyclic olefin (zeonor) |
MS | Integrated solid phase extraction |
| Solid phase extraction | D.J. Marchiarullo | [53] | 2008 | Targeted metabolic profiling |
Dihydroxybenzoic acids |
Glass | UV–vis | Integrated solid phase extraction |
II MICROSEPARATIONS
As discussed above, many approaches for metabolomic analysis utilize a high resolution separation technique to aid in the identification or measurement of complex mixtures of metabolites. Separations on the microfluidic format are typically performed by electrophoretic means, such as microchip capillary electrophoresis (microchip-CE), or through pressure driven flow, such as miniaturized-LC. The following sections introduce examples of microchip-CE and miniaturized-LC for various applications of metabolic analysis.
A. Microchip Capillary Electrophoresis
Capillary electrophoresis (CE) has been successfully miniaturized to microchip-CE using microfluidics techniques, and numerous reviews have been published describing applications of CE and microfluidics for the determination of biomolecules and metabolites [13, 16]. As with other microfluidic devices, microchip-CE has the advantages of rapid analysis times, small reagent and sample consumption, portability, and low cost as compared to bench-top methods. Solutions and samples can be manipulated throughout channels on-chip by applying voltages to integrated electrodes, and electroosmotic flow can be controlled by altering the surface chemistry of the microchip substrate material and the composition of the buffer. Analytes are separated in microchannels that typically range from 10–75 µm in critical dimensions, and a variety of CE separations modes can be utilized to introduce mobility differences between analytes of interest. Electrochemical and fluorescence detection are the most common detection methods for applications of microchip-CE in the determination of metabolites. Electrochemical methods typically do not require derivatization, and use more portable instrumental components. However, fluorescence methods offer limits of detection several orders of magnitude lower than those of electrochemical detection. The following sections discuss recent reports in the literature using microchip-CE for the determination of metabolites or metabolic activity.
Microchip-CE: UV absorbance
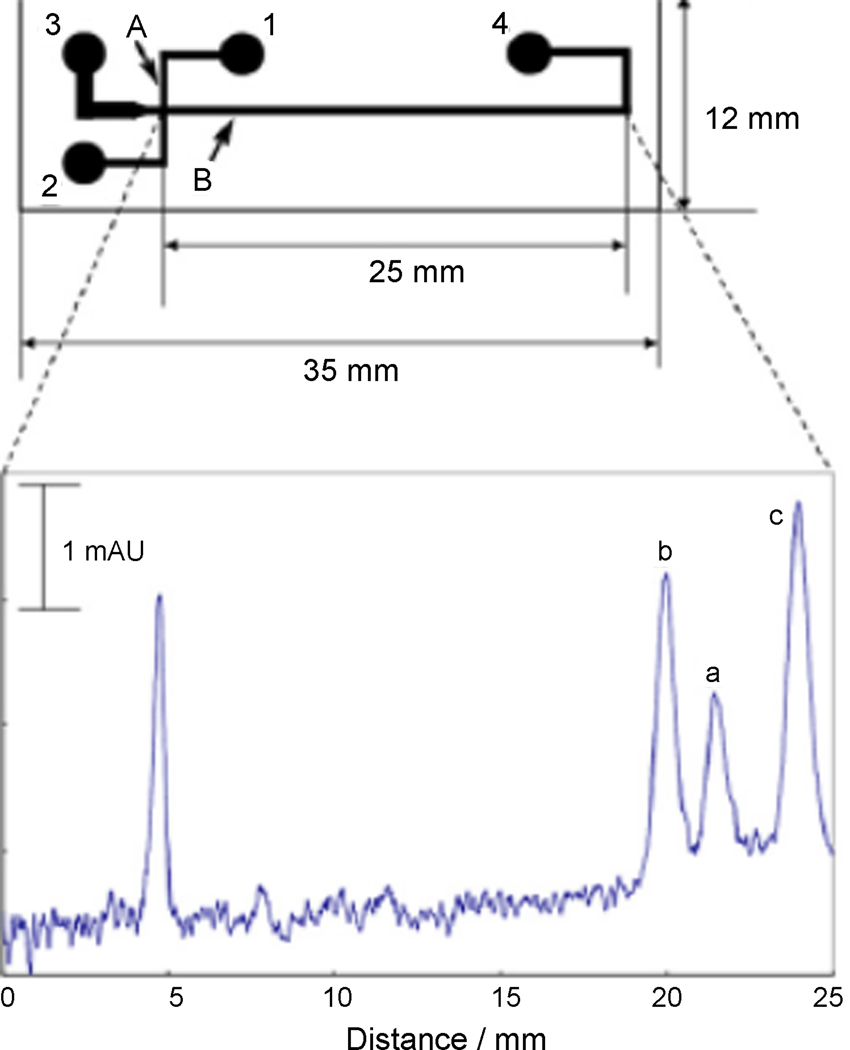
Several reports describe the use of commercially available microchip-CE devices for metabolic analysis. Miyado et al. report the use of a quartz glass microchip-CE device from Shimazdu to separate and detect nitric oxide metabolites in human plasma [17–19]. This high-throughput assay does not require deproteinization, and the zwitterionic surfactant CHES was employed to generate separations in less than 1 minute. Limits of detection of 53 µM for NO2− and 41 µM for NO3− were reported using absorbance detection; however, improvements in sensitivity are necessary to cover the clinical range of NO metabolites in plasma. Figure 1 shows the design of the microchip-CE device used and an example separation of nitrite, nitrate, and iodide in spiked human plasma. In a second report, a Shimadzu MCE 2010 system was used to separate antimicrobial metabolites of Pseudomonas fluorescens, and linear imaging UV detection was employed to quantify resorcinol, monoacetylphloroglucinol (MAPG), and 2,4-diacetylphlororglucinol (2,4-DAPG) [20]. The quartz microchip utilized a 30 µm × 30 µm channel which was 25 mm in length. Different sample introduction methods were tested, and the optimized MCE method resolved MAPG and 2,4-DAPG in less than 15 seconds with a limit of detection of 20 µM. Foaud et al. reported a microchip-CE method for the determination of natural plant products and metabolites [21]. Qualitative determination of glucosinolates was demonstrated on a Hitachi SV1100 microchip electrophoresis instrument based on fluorescent detection of a charge transfer complex (CTC) formed between the glucosinolates and several xanthene dyes. Glucosinolates are important natural products that occur in cruciferous plants, and have been utilized as anticancer agents due to the enzyme modulation behavior of their metabolic degradation products. The microchip material was poly(methylmethacrylate) (PMMA). CTC formation between glucosinolates and phloxine dye was used for indirect quantitative determination of total plant glucosinolate content, and this type of detection system could be applied to assay other types of metabolites. All of the above examples clearly show the ability to use absorbance detection for selected applications where analytes are either at sufficiently high concentrations to allow detection or have large molar extinction coefficients that permit direct detection.
Fig. 1.
Design of microchip-CE device and example separation of nitrite and nitrate in spiked human serum. Reprinted from [19] with permission from Elsevier © 2008.
Microchip-CE: Electrochemical Detection
Electrochemical detection (ECD) is an attractive detection method for microfluidics and microchip-CE as miniaturization is possible with minimal loss in detection performance. ECD generally requires analytes of interest to be electroactive which eliminates the need for derivatization and reduces the complexity of microchip-CE analysis. Furthermore, ECD does not require the use of bulky excitation sources and optical detection elements, increasing the portability of the device. This advantage is evidenced by the extensive use of ECD in sensing and biomonitoring applications such as hand-held glucometers. A recent review discussed nanotechnology-based electrochemical sensors used in biomonitoring applications and several examples of electrochemical sensors for metabolite analysis [22].
Microchip-CE devices using ECD detection have been used in several targeted metabolic profiling studies. Holcomb et al. introduced a microchip-CE device with an electrode array detector for monitoring small molecule metabolites and xenobiotics [23]. Eight individually addressable gold working electrodes were used to selectively detect analytes using potential step amperometry, and the system can be used to electrochemically resolve co-migrating species. Other papers from the Henry group have utilized microchip-CE devices for measuring glucose [24], catechins [25], and metabolic markers of renal function [26]. In these examples, the ability of ECD to selectively determine a broad range of metabolites is shown. For a number of these analytes such as glucose and the renal function markers, detection by other modes is much more challenging owing to their low molar extinction coefficients and lack of readily available functional groups for simple derivatization with fluorophores.
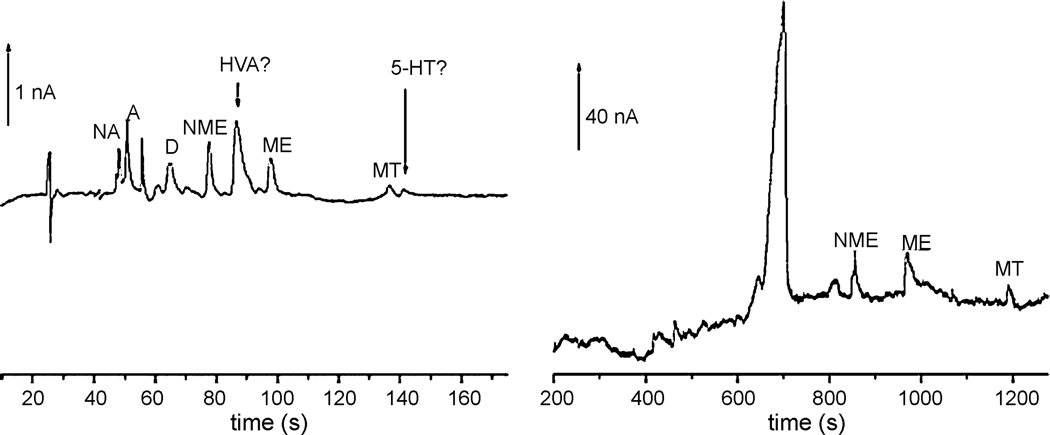
ECD has long been an optimal method for analyzing neurotransmitters and their derivatives. Schwarz et al. describe a microchip-CE system with electrochemical detection used for measuring catecholamines and their O-methoxylated metabolites [27, 28]. The glass chip contained a carbon nanotube-modified working electrode which generated submicromolar limits of detection. An optimized buffer system including a pseudostationary phase containing dendrimers was developed to fully resolve a set of six catecholamines and cationic metabolites. The system was successfully applied to measure multiple metabolic components from both a brain homogenate sample and human urine as shown in Figure 2 [29]. Earlier reports from this group focused on improving the amperometric detection sensitivity of similar microchip-CE systems through the use of enzyme catalyzed reactants such as reduced nicotinamide adenine dinucleotide (NADH). Determination of catecholamines remains as one of the most important applications of microchip CE-ECD as exemplified by these reports due to the excellent selectivity and sensitivity that can be obtained.
Fig. 2.
Microchip-CE separations of neurotransmitters and metabolites from brain homogenate and human urine using enhanced electrochemical detection. Reprinted from [28] with permission from Elsevier © 2007.
Zhang et al. describe a poly(dimethylsiloxane) (PDMS) microchip-CE device for targeted metabolic profiling using electrochemical detection to analyze caffeine and its major metabolite theophylline [30]. Results obtained measuring these two analytes in rat serum and urine were determined to be consistent with that measured by high performance liquid chromatography (HPLC). In a separate work, this group also used a similar microfluidic system to quantify morphine and codeine in human urine samples [31]. In a report from Chen et al., a poly(methylmethacrylate) (PMMA) microchip having gold nanoelectrode ensemble (GNEE) working and decoupling electrodes for ECD was used for determination of mercaptopurine (MP), an important drug for inflammatory bowel disease and acute lymphoblastic leukemia treatment [32]. Monitoring patients’ blood levels for metabolism of MP is crucial for determining treatment efficacy as degradation of MP decreases clinical activity. The limit of detection was 100 nM for MP, and a linear dynamic range (R2 = 0.989) was found from 10 mM to 100 nM, which is more than sufficient for analyzing µM concentrations of MP typically measured in the human body. A second example of microchip-CE-ECD was reported for targeted metabolic profiling of blood samples. Electrophoretic separations and biological enzymatic assays were coupled in a microchip-CE device using ECD to measure levels of glucose and lactate in human blood [33]. The CE separation was performed prior to enzymatic mixing, which reduced interference from oxidizable constituents, and thus increases the number of metabolites the device could potentially analyze in a clinical setting.
Microchip-CE: Fluorescence and Chemiluminescence Detection
Fluorescence detection is the most commonly employed detection method for microchip-CE, in part due to the low nM limits of detection that are possible using fluorescent reporter molecules or dyes. The Chiu group introduced a microchip-CE device for the determination of fluorescently labeled amine contents of a single mitochondria and single 100 nm vesicles [34]. By raising intra-mitochondrial pH using benzylethanolamine (BEA), a membrane permeable dye is used to label mitochondrial metabolites such as amino acids. Individual mitochondria are then photolysed and their contents are rapidly analyzed by microchip-CE. Variations in the fluorescent intensities of the labeled constituents of individual mitochondria suggest heterogeneity in mitochondrial content. Schulze et al. used native fluorescence detection on a microchip-CE device with a deep UV excitation laser and a fluorescent microscope [35]. Fused silica microchips purchased from Micronit were used with the native fluorescence detector to separate and measure aromatic small molecules and standard proteins without the need for derivatization. Liu et al. successfully separated the flavin metabolites riboflavin (RF), flavin mononucleotide (FMN), and flavin-adenine dinucleotide (FAD) using a microchip-CE device and laser induced fluorescence detection [36]. Because the structure of cellular flavins can be excited to fluoresce using a 488 nm laser, derivatization was not needed, and limits of detection of 34 nM, 201 nM, and 127 nM were reported for RF, FAD, and FMN, respectively. The authors optimized parameters for channel length, separation field strength, and pinched injection voltages in order to maximize metabolite resolution and achieve separation in less than one second. Callewaert et al. profiled asparagine-linked glycan (N-linked glycans) in serum proteins from patients having chronic hepatitis and cirrhosis of the liver using a microchip-CE platform [37]. Glycans were labeled with 8-aminopyrene-1,3,6-trisulfonic acid (APTS) and analyzed on an alumina-silicate glass microchip having an 11.5 cm separation channel. The resulting separation profiles were used to determine a diagnostic indicator peak for liver cirrhosis and were compared to separations using ABI 377 gel based DNA sequencer. While this work analyzes glycans rather than small molecule metabolites, it nonetheless demonstrates the ability of microchip-CE to profile chemically relevant biomarkers and provide a complementary method for clinical diagnostics. In total, these examples provide clear evidence of the ability of fluorescence detection to profile small molecules in biological samples with low detection limits and reasonable selectivity.
An alternative to fluorescent detection via derivatization is chemiluminescent or bioluminescent detection where an enzymatic reaction yields a photon emitting product that can be measured to quantitate the presences of metabolites such as reactive oxygen species. A PDMS microchip-CE device using bioluminescent detection was used to measure adenosine triphospate (ATP) and ATP conjugated metabolites from Escherichia coli cells and human urine [38]. The bioluminescence detector utilized the firefly luciferin-luciferase reagents, and the electroosmotic flow of the separation was controlled using the cationic surfactant didodecyldimethylammonium bromide (DDAB). The limit of detection for ATP was determined to be 200 nM, and the linear dynamic range was 50 µM to 200 nM. Separations of ATP were collected in less than 30 seconds. The integrated device was used to quantitate ATP content in E. coli cells that were lysed on chip as well as to determine the levels of ATP-conjugated galactose in human urine.
B. Microchip-LC
LC is the most commonly used separation technique in analytical chemistry as it offers highly effective separations based on a wide variety of chemical interactions and is compatible with a wide variety of sample types. Recently, use of LC in microfluidic devices has attracted increasing interest for fast sample preparation, separation, and subsequent sample analysis using MS [39–43]. The fast analysis times (typically in the seconds to few minutes) of microchip-LC relative to conventional LC significantly increases sample throughput and reduces analysis cost. Thus far, the majority of applications of microchip-LC have focused on proteomic analysis rather than metabolomic analysis. However, the low cost and disposable nature of these devices make them attractive for metabolomic applications as well. Furthermore, with developments in MS instrumentation it is envisioned large scale population screenings could be conducted using microchip-LC-MS technologies. Here, we present some representative applications of microchip-LC-MS for the determination of small molecules and metabolites to demonstrate the potential applicability of these chips for future high-throughput metabolomic studies.
In one such report, Sajonz et al. investigated use of the Eksigent Express 800 microfluidic eight channel HPLC instrument for multiparallel, normal-phase chiral analysis in high-throughput pharmaceutical process research [44]. Determination of test mixtures containing the two enantiomers of benzoin and the closely related (R, S)-dihydrobenzoin was carried out in a 96-well microplate. This afforded rapid (<2 h) and accurate assessment of enantiopurity. Use of the instrument to support high-throughput catalyst screening of the asymmetric hydrogenation of a prochiral unsaturated ester was also presented. For this work, method development and analysis of a 96-well microplate were carried out in a single working day. Combining multi-parallel chiral HPLC in a microfluidic format dramatically reduces the time needed for high-throughput enantiopurity analysis compared to conventional instrumentation in which analysis usually takes several days to complete.
Advancing the application of microfluidics in LC-MS in part depends on development of an interface to the ionization source. In one report, Hop developed a silicon, chip-based nanoelectrospray device which demonstrated improved practicality and sensitivity advantages over pulled capillary interfaces [45]. Pharmacokinetics and metabolite identification studies were conducted using this device. Another chip-based nanoelectrospray ionization device for coupling to tandem mass spectrometry (nanoESI-MS/MS) has been reported by Wickremsinhe et al. [46]. Reboxetine was extracted from dog plasma samples by liquid-liquid extraction, and the supernatant was concentrated to dryness and redissolved in 90% ACN/water for nanoESI-MS/MS. Determination of reboxetine standards showed improved sensitivity of the nanoESI-MS/MS system compared to LC coupled with tandem mass spectrometry (LC-MS/MS). It is also noteworthy the amount of sample infused during nanoESI-MS/MS was approximately 80-fold less compared to the amount of sample injected during LC-MS/MS. The absence of carryover due to the lack of a common fluid path in the nanoESI technique increased the linear dynamic range of the assay by 500000-fold. This technology offers the possibility for increased throughput for studies supporting drug development and metabolite identification and profiling.
Two more innovations in microscale-LC analysis, the nanoSplitter LC-MS and microdroplet NMR, have been introduced by Lin and coworkers [47]. The LC-MS-NMR platform, which utilizes microfluidics, facilitated the identification of unknown compounds found at low concentrations in complex sample matrixes frequently encountered in metabolomics or natural products discovery. With a flow of 200 nL/min into the MS, the nanoSplitter provided electrospray for high sensitivity MS while allowing 98% of the HPLC effluent to be collected and concentrated by microdroplet-NMR. Microdroplet NMR is a microfluidic NMR loading method which provides several-fold higher sample efficiency than conventional flow injection methods. Interpretable 1D NMR spectra were obtained from analytes at the 200-ng level using automated NMR data acquisition. When applied to a cyanobacterial extract showing antibacterial activity, the platform recognized several previously known metabolites from a single 30-µg injection, down to the 1% level, and prioritized one unknown for further study.
In a final report related to microfluidic-LC, Wang et al. introduced a microelectromechanical system (MEMS) differential thermal biosensor containing integrated microfluidic structures [48]. Channels in the PDMS structure were used to control movement of solution via flow-injection or continuous perfusion mode. Analytes were delivered to two microfluidic chambers where metabolite measurements were made using specific enzymatic assays. Metabolic changes were measured as a function of temperature using an integrated thermopile common to both reaction chambers. The device was tested with glucose solutions at physiologically relevant concentrations. In flow-injection mode, the device demonstrated a sensitivity of approximately 2.1 µV/mM and a resolution of about 0.025 mM. In flow-through mode with a perfusion flow rate of 0.5 mL/h the sensitivity and resolution of the device for glucose measurements were determined to be approximately 0.24 µV/mM and 0.4 mM, respectively. These results illustrate the device, when integrated with subcutaneous sampling methods, can potentially allow for continuous monitoring of glucose and other metabolites in medical applications.
III SAMPLE PREPARATION
One of the goals of microfluidics research is to incorporate sample pre-processing and analysis platforms onto the same device. Integrated microfluidic devices such as these are termed lab on a chip (LOC) or icro-total analysis systems (µTAS), and have many potential benefits for metabolic analyses. The rapid analysis times inherent to these systems make them attractive alternatives to currently used analytical methodologies. In addition, autonomous operation of these systems would further expedite the analysis process and make metabolic analyses less labor intensive. Developments in the fields of LOC and µ TAS for sample pre-processing prior to metabolic analysis are discussed below.
Sample Cleanup and Pre-Concentration
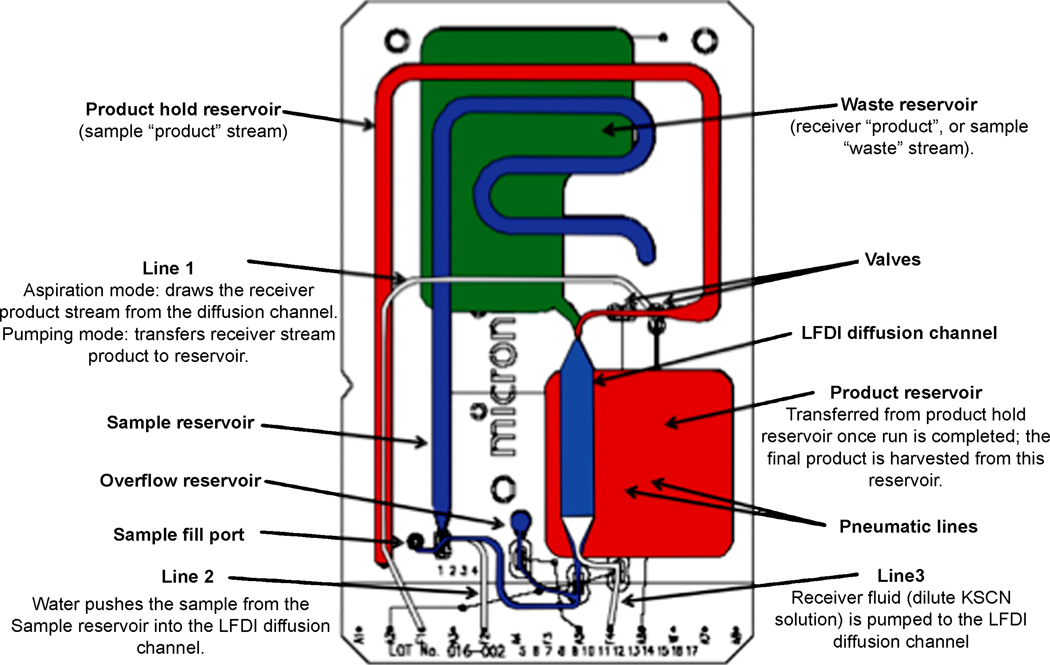
Various methods have been described in the literature for sample cleanup and analyte pre-concentration in microfluidic devices. Mansfield et al. [49] and Shaw et al. [50] described using a microfluidic laminar fluid diffusion interface (LFDI) for sample pre-processing prior to analysis with infrared spectroscopy of dry films (IRDF). The use of LFDI allows low and high molecular weight analytes in biofluids to be separated into separate sample streams prior to analysis. The LFDI accommodates input from both a sample and receiver stream that converge in a common channel. Since the flow regime is laminar the bulk fluid streams do not mix; however, diffusional mixing does occur resulting in low molecular weight components in the receiver stream and high molecular weight components in the original biofluid stream. The two streams can then be split and analyzed using IRDF. The authors demonstrated the utility of this method by profiling creatinine in serum. An illustration of the network of channels utilized in the LFDI device is shown in Figure 3. This work is significant because it provides an example of using the property of microfluidic devices (low Reynolds number flows) to perform an operation that would be very challenging to do in traditional systems.
Fig. 3.
Schematic of LDFI microfluidic device for sample preprocessing prior to IRDF. Reprinted from [50] by permission of the Royal Society of Chemistry.
Three reports described the use of solid phase extraction for sample cleanup and analyte pre-concentration in microfluidic devices. An initial report by Benetton et al. described a monolithic solid phase extraction (SPE) sorbent integrated on-chip for desalting samples prior to MS analysis [51]. The polymeric sorbent was polymerized from butyl methacrylate and ethylene dimethacrylate inside capillaries formed in a thermoplastic (Zeonor). P450 drug metabolism studies were conducted by diffusively mixing human liver microsomes and several antidepressant drugs (imipramine, doxepin, and amitriptyline) on-chip via laminar flow mixing and allowing this mixture to incubate on-chip prior to analysis. The drugs and their metabolites were initially retained on the SPE sorbent following the incubation step to allow for desalting of the sample prior to MS analysis.
In a follow up to this work, Tan et al. described incorporating an array of SPE sorbents into a microfluidic device for sequential sample cleanup and pre-concentration prior to MS analysis [52]. Since SPE elements are typically only reliable for a single use, the authors incorporated an array of eight SPE elements into the microfluidic device to increase device longevity and throughput capabilities. Incorporation of an array of SPE elements allows for more than just a single use of the device. As in the previous report the polymeric sorbents were polymerized from butyl methacrylate and ethylene dimethacrylate inside capillaries molded in a thermoplastic (Zeonor). SPE of a human urine sample as well as a P450 drug metabolism incubation mixture was conducted prior to ESI-MS detection to demonstrate the applicability of this device for handling complex sample mixtures. Similar to the previous study the P450 drug metabolism incubation mixtures consisted of human liver microsomes and imipramine, a common antidepressant drug. Determination of the incubation mixture following SPE resulted in detection of several metabolites of imipramine demonstrating the usefulness of this technique for profiling drug metabolites in complex sample mixtures.
A third SPE application was reported on by the Landers group in which they described a SPE microfluidic device for the determination of metabolic markers of oxidative stress for spaceflight clinical diagnostics [53]. Extreme environments, such as those found in space, can lead to production of reactive oxygen species (ROS) that can damage biomolecules and negatively impact human health. Short-lived hydroxyl radicals are detected using salicylic acid, which forms stable products of 2,3-dihydroxybenzoic acid (DHBA) and 2,5 DHBA when combined with ROS. A microfluidic chip with an integrated SPE sorbent having a hydrophilic-lipophilic copolymer was used to extract and concentrate 2,3-DHBA and 2,5-DHBA from saliva. 5–15 µL of eluent from the microchip was then analyzed by benchtop capillary electrophoresis, and concentration enhancements of up to 80-fold were demonstrated. This work is the first step towards development of a fully integrated device for in-flight space diagnostics that uses SPE and microfluidics for metabolic biomarker analysis
A fully integrated LOC device containing extensive sample pre-processing components for isolating and regenerating sarin in blood samples was reported by Tan et al [54]. Determination of sarin in a matrix such as blood required on-line sample manipulation in the forms of regeneration of the nerve agent, cell lysis, protein precipitation, and filtration. The device was fabricated from PMMA and consisted of five distinct sections. Sarin regeneration was achieved in the first section via chaotic advection with herring-bone micro-mixers. The next section consisted of rectangular pillars for cell lysis and filtration of particulate matter resulting from protein precipitation in the blood sample. This was followed by a filter chamber with microbeads to remove fluoride ions used in the sarin regeneration step. The remaining segments consisted of the detection elements of the device which included enzymatic chromophore generation for indirect optical detection of sarin. This work represents an important level of integration for total analysis of a small molecule from a complex matrix. Examples of this type are lacking in the microfluidics field.
Other Integrated Sample Pre-Processing Platforms
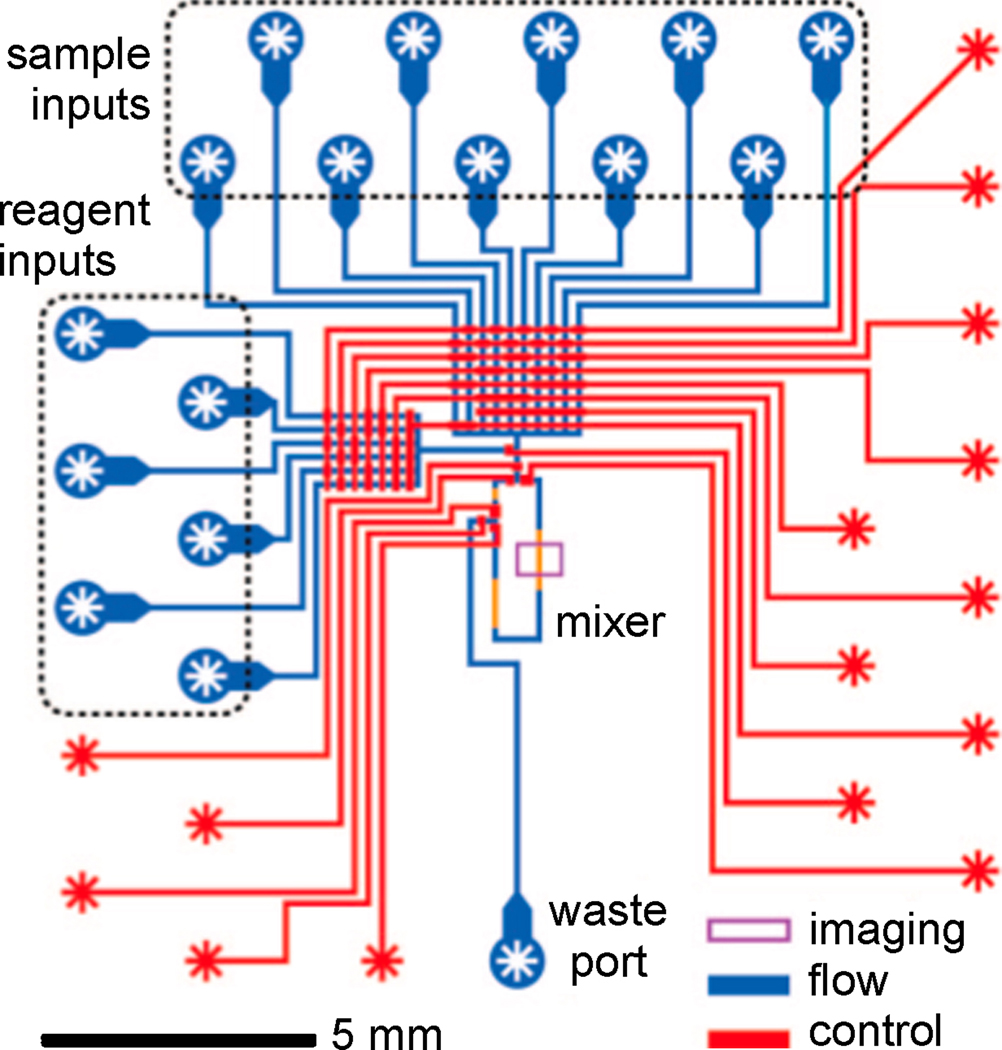
Several reports described the use of other sample pre-processing techniques such as mixing, sample labeling, and metabolite generation on microfluidic devices. One such report was given by Urbanski et al. in which they described determination of embryo metabolism using an integrated microfluidic chip [55]. The chip was constructed from PDMS using multilayer soft lithography techniques and consisted of two layers; an upper flow channel network for loading and mixing sample and reagents, and a bottom control layer for valving and fluidic manipulation of the top layer. After initial sample and reagent loading onto the chip, delivery of sample and reagent aliquots, mixing, data acquisition, and data analysis could be conducted in an autonomous fashion. A schematic of the microfluidic device used for measuring embryo metabolism is shown in Figure 4.
Fig. 4.
Schematic of microfluidic networks for an integrated metabolic profiling microchip. The blue features represent channels in which samples and reagents are delivered and mixed. The red features represent channels used for pneumatic fluid manipulation and valving. The purple box denotes where fluorescent assay imaging occurs. Reprinted with permission from [55]. © 2008 American Chemical Society.
Another report by Ma et al. described integrating sol-gel based bioreactors in a microfluidic device for generation of drug metabolites [56]. Metabolite analysis and cell cytotoxicity assays were also conducted on-chip which required the device be comprised of three layers. A quartz middle layer contained human liver microsome sol-gel bioreactors for drug metabolite generation and CE channels for their subsequent analysis. An upper PDMS layer consisted of solution reservoirs for the middle layer while a bottom PDMS layer contained hepG2 cells positioned below the sol-gel bioreactors for drug and drug metabolite cytotoxicity assays. The effects of acetaminophen and acetaminophen metabolites generated by UDP-glucuronosyltransferase in the sol-gel bioreactors were studied. Metabolite generation was monitored using CE with UV detection while cytotoxicity of the hepG2 cells was monitored using a fluorescent live/dead stain. Integrating drug metabolism bioreactors along with complementary analysis components on a microfluidic platform such as this has applications for rapid drug metabolite profiling.
In a final report by Wu et al. the authors described the implementation of a microfluidic platform for sampling and mixing small volumes of blood to aid in quantitative positron emission tomography (PET) studies of small animals (mice) [57]. The microfluidic chip was fabricated from PDMS and consisted of two layers. The top layer contained microfluidic networks for blood and heparin block solution as well as 18 blood sampling wells. The bottom layer contained channels for pneumatic valving; these pneumatic valves controlled flow of blood and heparin block solution on the upper layer. For blood sampling, fresh blood supplied from an arterial catheter in the mouse was continually flowed through the chip and then transferred into a sample well using a heparin flush. 18F-fluorodeoxyglucose activities in the blood samples were subsequently measured with a well counter and used to analyze glucose metabolism.
Immunoassays and Analyte Immobilization Techniques
Use of selective analysis techniques such as immunoassays negates the need for extensive sample pre-preprocessing and thus allows for the direct determination of biofluids. In one report by Murphy et al. the authors described a competitive immunoassay for the simultaneous detection of proteins and metabolites using micromosaic patterning [58]. For these assays, a BSA conjugate of the target analyte was immobilized on a sensitized silicon nitride substrate by flowing it over the substrate surface in a microfluidic network. The sample containing the analyte of interest and its fluorescently labeled antibody were then patterned in the orthogonal direction and the competitive immunoassay analyzed using fluorescent microscopy. The metabolic biomarkers thyroxine and 3-nitrotyrosine were detected using this technique.
Another report by Jo et al. described immobilizing neuropeptide releasate from Apylsia californica bag cell neurons onto specific regions of a solid substrate using microfluidic networks [59]. The ability to immobilize the analyte of interest onto a solid substrate allows for additional sample cleanup prior to analysis. In this report the authors were able to rinse away non-retained species and spray the solid substrate with matrix after neuropeptide immobilization for direct matrix assisted laser desorption ionization (MALDI)-MS analysis. The microfluidic devices were comprised of a PDMS top layer with reservoirs for the neuronal cells and microfluidic channels for directing fluid flow as well as a bottom silicon layer functionalized with C18 for the capture of neuropeptides. Using this setup the authors were able to profile neuropeptide release pre, during, and post neuronal stimulation. Neuropeptide release from a single neuron was profiled in this manner.
IV MICROFLUIDICS FOR CELLULAR ANALYSIS
Microfluidics technologies are capable of manipulating and mixing small volumes of solutions and reagents using networks of channels and reaction chambers. These features of microfluidic devices also make them well suited for the analysis and/or growth of cells and tissues. There have been extensive applications of microfluidics for miniaturized cell culture, isolation, trapping, concentration, and chemical cellular monitoring after exposure to a stimulus. Other recent review articles have summarized current developments of microfluidic chips for cell sorting applications, single cell analysis, cell culture, and cell biological assays [16, 60–63]. For the scope of this review, attention was given to those microfluidic applications of cellular analysis that measured general metabolic activity or that monitored targeted metabolic products. Research articles in this category are classified as bacterial monitoring, cell stimulus and exposure, toxicity screening, and clinical diagnostics.
Bacterial Monitoring
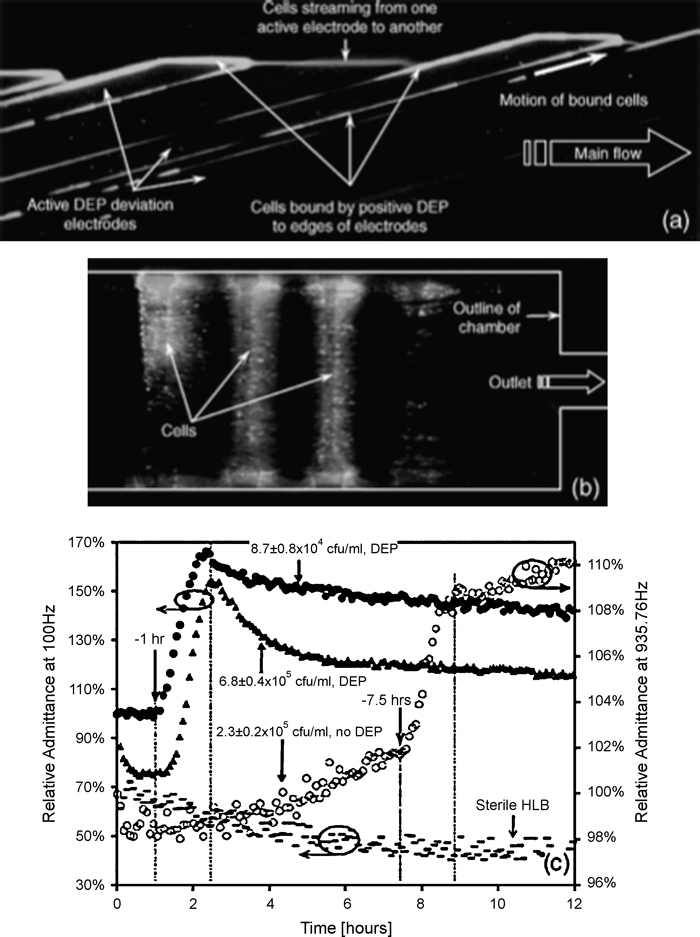
Microfluidic devices that can isolate or immobilize bacterial cells can be used for toxicity measurements, monitoring the presence or absence of bacteria, and investigating metabolic activity of specific bacterial strains. A silicon based microfluidic chip was used to concentrate bacterial cells from dilute samples and measure metabolic activity in small volumes using impedance monitoring techniques [64]. Dielectrophoresis was used to mobilize and concentrate cells into a 400 pL incubation/measurement chamber where interdigitated platinum electrodes were used to measure changes in impedance within the chamber. Metabolic activity from bacterial cell growth releases metabolites into the surrounding solution and in turn changes the measured impedance [65]. The microfluidic system was very sensitive to fluctuations in temperature, and automated temperature control was necessary to reduce deviations in measured impedance. Concentration factors on the order of 104 to 105 were observed for several cultures of bacterial cells, and the microfluidic device significantly reduced the amount of time needed to detect the presence of bacteria. Suspensions of Listeria monocytogenes cells containing as few as approximately 3480 cells were concentrated and analyzed using the microfluidic bioprocessor, and strong metabolic activity was observed after one hour of collection. Figure 5 shows images of fluorescent bacterial cells collected in the incubation chamber and a plot of impendence versus time to show changes in metabolic activity.
Fig. 5.
Images of fluorescently labeled bacteria cells collected within a microfluidic incubator (a, b) and a plot of changes in impedance (c), which correspond to increases or decreases in metabolic activity, for control and normal cells. Reprinted from [64] by permission of the IEEE © 2005.
A hydrogel bacterial microchip was presented for biosensing and monitoring of intracellular metabolism [66]. An array of hydrogel elements, or gel pads, containing microbial cells were fixed to a glass surface and optical measurements obtained using fluorescent reporters or stains. Each gel pad was approximately 0.3–60 nL in volume, contained 104 to 105 bacterial cells, and could be loaded with unique bacterial strains. Growth curve kinetics were measured for strains of E. coli TG-1, and a bioluminescent reporter was used to measure arsenite concentrations (10−5 to 10−6 M). One limitation of the current design is the use of an acrylamide-based hydrogel that can be toxic for some types of bacteria. However, the system has significant promise for cell-based biosensing using E. coli as the sensing system.
A PDMS microfluidic device was presented that isolates single bacterial cells into nanoliter droplets using plug-based microfluidics [67]. An antibiogram, or chart of metabolic activity, was determined for single cells of methicillin-resistant Staphylococcus aureus as well as the minimal inhibitory concentration (MIC) of the antibiotic cefoxitin. Metabolites released by single cells are restricted to the nL sized volume of the microfluidic plug in a process the authors term stochastic confinement. The microfluidic system was used to distinguish between strains of S. aureus, where a higher MIC (8 mg/mL) was observed for the resistant strain than for the sensitive strain (4 mg/mL). The authors determined detection time of bacteria in their device was dependant on plug volume, rather than initial concentration or bacterial growth rate. To show the applicability of the microfluidic system for measuring bacteria samples from human blood were analyzed to differentiate between different drug resistances in order to select an appropriate therapeutic route. In all, the system permits rapid evaluation of the bacterial metabolis as a function of different drugs and has potential for application in both bacteria detection and drug screening applications.
Another example of bacterial monitoring using microfluidics included a PDMS microfluidic bioassay chip containing immobilized bacterial strains for bioluminescent detection. A 5 × 5 well array was used to stimulate bacterial strains using varying concentrations of mitomycin C [68]. Also, as an example of targeted metabolic profiling a microfluidic flow injection system fabricated from silicon was introduced to measure glucose and ethanol secretion from immobilized Saccharomyces cerevisiae cells using chemiluminescence detection [69]. Glucose and ethanol levels were measured for up to 5 days using this biosensor.
Cell Stimulus and Exposure
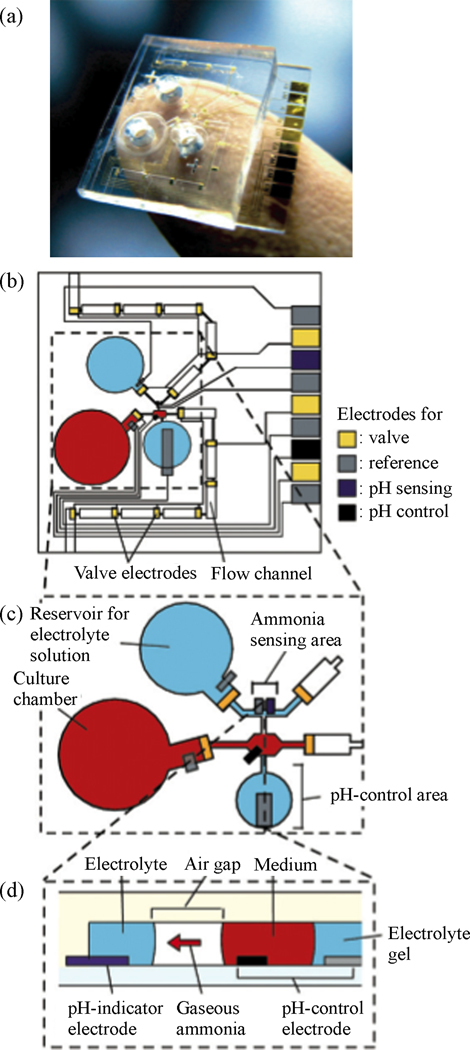
A second class of cellular analysis using microfluidics involves the stimulus or exposure of isolated single cells, tissues, or cultured cell populations. Fully integrated µTAS devices are capable of both exposing or stimulating the cells and measuring the inherent metabolic response. A microfluidic device for monitoring ammonia metabolism from hepatocytes cultured on-chip was presented by Satoh et al. [70]. An iridium oxide pH-indicator electrode and Ag/AgCl reference electrode positioned at an air gap junction within the glass-PDMS microfluidic device served as the ammonia sensor. A secondary gold working electrode and Ag/AgCl reference electrode were positioned at a liquid junction in the culture medium, and were used to control the pH of the media solution. Figure 6 shows an illustration of the detection elements of the microfluidic culture device. By raising the pH of the culture medium ammonia ions were released as gaseous ammonia into the air-gap where they were then dissolved into an electrolyte solution and measured by the ammonia sensing electrode. Ammonia metabolism was measured for cultures of hepatocytes seeded at different cell densities within the microfluidic cell culture device, and the results provided a more detailed description of ammonia levels within the first three hours of sampling compared to conventional absorbance measurement kits.
Fig. 6.
Photo and design of microsystem for monitoring hepatocyte ammonia metabolism. Reprinted from [6] by permission of the Royal Society of Chemistry.
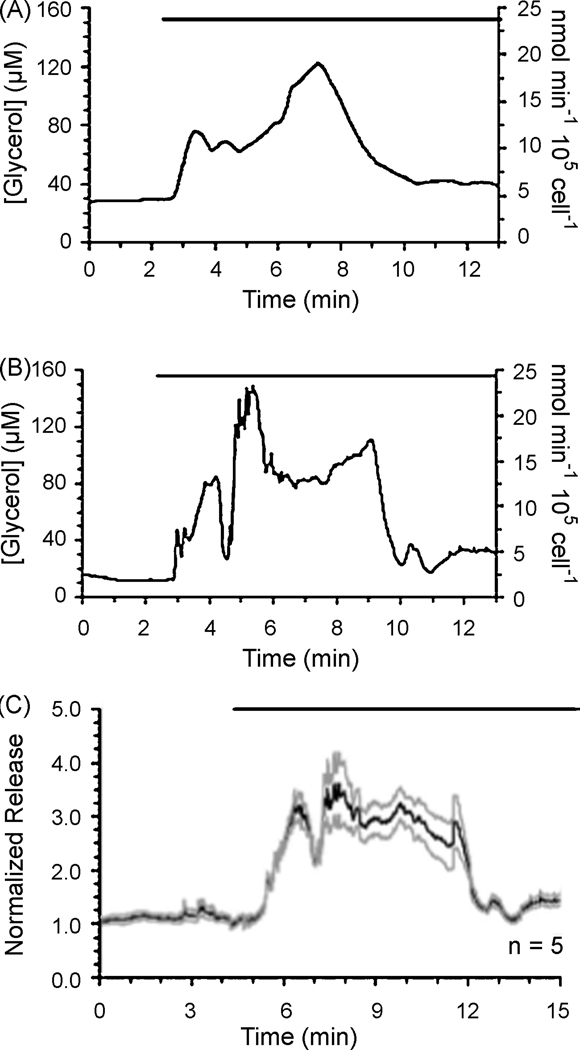
The Kennedy group reports a microfluidic device for monitoring glycerol secretion and metabolism in cultured adipocytes using a continuous flow enzyme assay [71]. The real time assay could detect changes in glycerol concentration every 90 seconds, and had a limit of detection of 4 µM glycerol. Stimulation of the adipocytes with isoproterenol, a B-adrenergic agonist, induced lipolysis and resulted in a 3-fold increase in glyercol secretion. Figure 7 shows changes in glycerol secretion for individual adipocyctes in response to treatment with isoproterenol.
Fig. 7.
Microfluidic monitoring of glycerol secretion from individual adipocytes (a, b) and normalized release from five individual cells (c). Reprinted with permission from [71]. © 2009 American Chemical Society.
A microfluidic system was developed to determine the amounts of lactate produced from single cardiomyocytes and also monitor changes in extracellular pH and concentrations of Ca2+ [72]. Microelectrodes were incorporated into the device to stimulate the heart cells at specific rates, in an effort to induce the cell to ‘work’ under various metabolic conditions. An enzyme-modified platinum electrode yielded a detection limit of 4.8 fmol of lactate. Electrical stimulation at 75 V cm−1 and 0.5 to 2 Hz caused the heart cell to beat continuously, and indicator dyes were used to measure changes in pH and Ca2+ over the course of 20 minutes. A second microfluidic device made from PDMS was introduced by Klauke et al. to study intracellular cardiomyocyte coupling and signaling [73]. Electrical stimulation was accomplished using microelectrodes, and chemical perturbations of ions or drugs were introduced via microfluidic channels. Intracellular signaling was monitoring using a fluorescent tracer for Ca2+.
A microfluidic device was designed to provide rapid changes in media conditions to the environment of a single cell. Media conditions could be changed as rapidly as twice a second without perturbing cell adhesion. Signaling pathways of Sacchraromyces cerevisiae cells were quantitated by measuring the bandwidth of the cellular response while varying osmolality; the cells behave as low-pass filters with distinct bandwidths [74]. Rocheleau et al. designed a PDMS microchip for the analysis of single pancreatic islets for targeted metabolic profiling. An actuator and two fluid streams were used to stimulate islets with glucose, while NAD(P)H and Ca2+ were measured using fluorescent markers [75].
In an example of high-throughput exposure, a PDMS based microfluidic cell culture device with 440 micro-wells for trapping hematopoietic cells for chemical exposure studies of intracellular signaling was introduced [76]. A fluorescent dye, rhod-2 calcium indicator, was used to monitor calcium release from single primary T cells exposed to ionomycin, antibody coated microspheres, and mature dendridic cells. Wang et al. presented a PDMS microfluidic chip for multiple well cell exposure studies using a laminar flow-based network for generation of a gradient of exposure concentrations [77]. Pharmacological profiling of MCF-7 breast cancer cells was demonstrated by stimulation of 30 individual cell chambers with concentration gradients of As2O3 and N-acetyl-cysteine for glutathione modulation. The dose-dependant impact of induced glutathione activity on chemotherapy sensitivity to adriamycin was investigated using the fluorescent dye naphthalene-2,3-dicarboxadelhyde.
Toxicity Screening
Microfluidic cell culture devices can also be used to investigate the toxicity of drugs and assess the changes in metabolic activity under various conditions. A microfluidic microreactor was introduced for evaluating metabolism and other pharmacological properties of drug candidates by exposing microsomes and hepatocytes entrapped in polyethylene glycol hydrogels [78]. Mass transfer of drug delivery was characterized by calculating the mesh size of the hydrogel, and metabolic activity was measured using a fluorescent reporter for cytochrome P450. Baudoin et al. report a PDMS microchip device for cell culture application and toxicity studies [79]. Cultures of Madin Darby Canine Kidney (MDCK) cells were grown on the PDMS microchip, which was coated with a layer of fibronectin. Metabolic measurements of glucose consumption and production of ammonia via glutamine metabolism were estimated using enzymatic reactions and direct and indirect absorbance measurements, respectively. Furthermore, the effects of flow rate (0 to 25 µL/min) and ammonium chloride concentration (0 to 10 mM) on glucose consumption and ammonia production were determined for the MDCK culture. A bio-MEMS chip was developed for general toxicity monitoring using bioluminescent bacterial cells that respond to the presence of reactive oxygen species [80]. A dose dependant bioluminescent response was observed when exposing the cells to phenol, and exposure to 0.88 mM H2O2 induced a 10 fold increase in bioluminescence.
Clinical Diagnostics
Several examples of the use of microfluidic devices for cell analysis have been applied to distinguish clinical samples. For instance, a parallel channel PDMS device was used to quantitate levels of glutathione and C-peptide in red blood cells and distinguish diabetic samples from control [81]. Also, a microfluidic device for multiplexed isolation of T-cell subsets and cell secreted cytokines from blood was developed for improved HIV diagnosis and monitoring [82]. Carraro et al. describe a novel microfluidic device for hepatocyte culture that contained a microfluidic network capable of providing a low shear stress physiological environment for cell growth and monitoring [83]. Liver function assays were measured for HepG2/C3A and primary rat hepatocytes, and metabolic function was monitored by profiling albumin concentration over time.
V CONCLUSIONS
This review has presented recent publications that utilize microfluidic devices for metabolomics, metabonomics, metabolic fingerprinting, and targeted metabolic profiling applications. While the majority of bioanalyical microfluidics applications are focused towards proteomic and genomic systems, there is a growing trend towards developing microsystems for the determination of small molecules and metabolites. Furthermore, microsystems that can detect and measure metabolic changes in time and in response to a stimulus or chemical exposure will serve as useful tools for biomonitoring, toxicity screening, and clinical diagnostics. The bulk of the microfluidics work has been in the area of microchip electrophoresis as evidenced by the relative length of that section. However, other areas, including sample preparation and cellular analysis are beginning to come to the forefront of the field. The biggest challenge, however, remains in integration of function into microfluidic devices and analysis of true biological samples. The former is challenging on many chemical and engineering levels because of the diversity of operations required. The latter is challenging simply because of the overwhelming complexity of sample matrixes such as blood and tissue. However, the authors anticipate that these areas will be successfully addressed in the near future as the global area of metabolomics continues to garner increasing attention in a variety of medical and toxicological areas of research endeavor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Babu CVS, Song EJ, Babar SM, Wi MH, Yoo YS. Electrophoresis. 2006;27:97. doi: 10.1002/elps.200500511. [DOI] [PubMed] [Google Scholar]
- 2.Whitesides GM. Nature. 2006;442:368. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 3.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 4.Pumera M. Electrophoresis. 2007;28:2113. doi: 10.1002/elps.200600709. [DOI] [PubMed] [Google Scholar]
- 5.Henares TG, Mizutani F, Hisamoto H. Anal. Chim. Acta. 2008;611:17. doi: 10.1016/j.aca.2008.01.064. [DOI] [PubMed] [Google Scholar]
- 6.Ohno K, Tachikawa K, Manz A. Electrophoresis. 2008;29:4443. doi: 10.1002/elps.200800121. [DOI] [PubMed] [Google Scholar]
- 7.Ellis J, Grimm R, Clark JF, Pyne-Gaithman G, Wilbur S, Caruso JA. J. Proteome Res. 2008;7:4736. doi: 10.1021/pr800294r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monton MR, Soga T. J. Chromatogr., A. 2007;1168:237. doi: 10.1016/j.chroma.2007.02.065. [DOI] [PubMed] [Google Scholar]
- 9.Serkova NJ, Spratlin JL, Eckhardt SG. Curr. Opin. Mol. Ther. 2007;9:572. [PubMed] [Google Scholar]
- 10.Clarke CJ, Haselden JN. Toxicol. Pathol. 2008;36:140. doi: 10.1177/0192623307310947. [DOI] [PubMed] [Google Scholar]
- 11.Xiayan L, Legido-Quigley C. Electrophoresis. 2008;29:3724. doi: 10.1002/elps.200700851. [DOI] [PubMed] [Google Scholar]
- 12.Koulman A, Lane GA, Harrison SJ, Volmer DA. Anal. Bioanal. Chem. 2009;394:663. doi: 10.1007/s00216-009-2690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Perez I, Vallejo M, Garcia A, Legido-Quigley C, Barbas C. J. Chromatogr., A. 2008;1204:130. doi: 10.1016/j.chroma.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 14.Lindon JC, Holmes E, Nicholson JK. Curr. Opin. Mol. Ther. 2004;6:265. [PubMed] [Google Scholar]
- 15.Jonsson P, Gullberg J, Nordstrom A, Kusano M, Kowalczyk M, Sjostrom M, Moritz T. Anal. Chem. 2004;76:1738. doi: 10.1021/ac0352427. [DOI] [PubMed] [Google Scholar]
- 16.Huang WH, Ai F, Wang ZL, Cheng JK. J. Chromatogr., B: Analyt. Technol. Biomed. Life Sci. 2008;866:104. doi: 10.1016/j.jchromb.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 17.Miyado T, Tanaka Y, Nagai H, Takeda S, Saito K, Fukushi K, Yoshida Y, Wakida S, Niki E. J. Chromatogr., A. 2004;1051:185. [PubMed] [Google Scholar]
- 18.Miyado T, Tanaka Y, Nagai H, Takeda S, Saito K, Fukushi K, Yoshida Y, Wakida S, Niki E. J. Chromatogr., A. 2006;1109:174. doi: 10.1016/j.chroma.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Miyado T, Wakida S, Aizawa H, Shibutani Y, Kanie T, Katayama M, Nose K, Shimouchi A. J. Chromatogr., A. 2008;1206:41. doi: 10.1016/j.chroma.2008.07.065. [DOI] [PubMed] [Google Scholar]
- 20.Guihen E, Glennon JD. J. Chromatogr., A. 2005;1071:223. doi: 10.1016/j.chroma.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Fouad M, Jabasini M, Kaji N, Terasaka K, Tokeshi M, Mizukami H, Baba Y. Electrophoresis. 2008;29:2280. doi: 10.1002/elps.200700635. [DOI] [PubMed] [Google Scholar]
- 22.Barry RC, Lin YH, Wang J, Liu GD, Timchalk CA. J. Expo. Sci. Environ. Epidemiol. 2009;19:1. doi: 10.1038/jes.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holcomb RE, Kraly JR, Henry CS. Analyst. 2009;134:486. doi: 10.1039/b816289a. [DOI] [PubMed] [Google Scholar]
- 24.Garcia CD, Henry CS. Anal. Chim. Acta. 2004;508:1. [Google Scholar]
- 25.Hompesch RW, Garcia CD, Weiss DJ, Vivanco JM, Henry CS. Analyst. 2005;130:694. doi: 10.1039/b418368a. [DOI] [PubMed] [Google Scholar]
- 26.Garcia CD, Henry CS. Analyst. 2004;129:579. doi: 10.1039/b403529a. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz MA. Electrophoresis. 2004;25:1916. doi: 10.1002/elps.200305816. [DOI] [PubMed] [Google Scholar]
- 28.Vlckova M, Schwarz MA. Electrophoresis. 2005;26:2701. doi: 10.1002/elps.200410396. [DOI] [PubMed] [Google Scholar]
- 29.Vlckova M, Schwarz MA. J. Chromatogr., A. 2007;1142:214. doi: 10.1016/j.chroma.2006.12.040. [DOI] [PubMed] [Google Scholar]
- 30.Zhang QL, Lian HZ, Wang WH, Chen HY. J. Chromatogr., A. 2005;1098:172. doi: 10.1016/j.chroma.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 31.Zhang QL, Xu JJ, Li XY, Lian HZ, Chen HY. J. Pharm. Biomed. Anal. 2007;43:237. doi: 10.1016/j.jpba.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Chen CM, Ho YH, Wu SM, Chang GL, Lin CH. J. Nanosci. Nanotechnol. 2009;9:718. doi: 10.1166/jnn.2009.c010. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Chatrathi MP, Collins GE. Anal. Chim. Acta. 2007;585:11. doi: 10.1016/j.aca.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Allen PB, Doepker BR, Chiu DT. Anal. Chem. 2009;81:3784. doi: 10.1021/ac900099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulze P, Ludwig M, Kohler F, Belder D. Anal. Chem. 2005;77:1325. doi: 10.1021/ac048596m. [DOI] [PubMed] [Google Scholar]
- 36.Liu BF, Hisamoto H, Terabe S. J. Chromatogr., A. 2003;1021:201. doi: 10.1016/j.chroma.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Callewaert N, Contreras R, Mitnik-Gankin L, Carey L, Matsudaira P, Ehrlich D. Electrophoresis. 2004;25:3128. doi: 10.1002/elps.200406020. [DOI] [PubMed] [Google Scholar]
- 38.Liu BF, Ozaki M, Hisamoto H, Luo QM, Utsumi Y, Hattori T, Terabe S. Anal. Chem. 2005;77:573. doi: 10.1021/ac0490447. [DOI] [PubMed] [Google Scholar]
- 39.de Boer AR, Bruyneel B, Krabbe JG, Lingeman H, Niessen WM, Irth H. Lab Chip. 2005;5:1286. doi: 10.1039/b506559c. [DOI] [PubMed] [Google Scholar]
- 40.Figeys D, Gygi SP, McKinnon G, Aebersold R. Anal. Chem. 1998;70:3728. doi: 10.1021/ac980320p. [DOI] [PubMed] [Google Scholar]
- 41.Fortier MH, Bonneil E, Goodley P, Thibault P. Anal. Chem. 2005;77:1631. doi: 10.1021/ac048506d. [DOI] [PubMed] [Google Scholar]
- 42.Xiang F, Lin Y, Wen J, Matson DW, Smith RD. Anal. Chem. 1999;71:1485. doi: 10.1021/ac981400w. [DOI] [PubMed] [Google Scholar]
- 43.Xue Q, Foret F, Dunayevskiy YM, Zavracky PM, McGruer NE, Karger BL. Anal. Chem. 1997;69:426. doi: 10.1021/ac9607119. [DOI] [PubMed] [Google Scholar]
- 44.Sajonz P, Schafer W, Gong X, Shultz S, Rosner T, Welch CJ. J. Chromatogr., A. 2007;1145:149. doi: 10.1016/j.chroma.2007.01.067. [DOI] [PubMed] [Google Scholar]
- 45.Hop CE. Curr. Drug. Metab. 2006;7:557. doi: 10.2174/138920006777697909. [DOI] [PubMed] [Google Scholar]
- 46.Wickremsinhe ER, Ackermann BL, Chaudhary AK. Rapid Commun. Mass Spectrom. 2005;19:47. doi: 10.1002/rcm.1747. [DOI] [PubMed] [Google Scholar]
- 47.Lin YQ, Schiavo S, Orjala J, Vouros P, Kautz R. Anal. Chem. 2008;80:8045. doi: 10.1021/ac801049k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Sipe DM, Xu Y, Lin Q. J. MEMS. 2008;17:318. [Google Scholar]
- 49.Mansfield CD, Man A, Shaw RA. IEE Proc. Nanobiotechnol. 2006;153:74. doi: 10.1049/ip-nbt:20050028. [DOI] [PubMed] [Google Scholar]
- 50.Shaw RA, Rigatto C, Reslerova M, Ying SL, Man A, Schattka B, Battrell CF, Matthewson J, Mansfield C. Analyst. 2009;134:1224. doi: 10.1039/b821442e. [DOI] [PubMed] [Google Scholar]
- 51.Benetton S, Kameoka J, Tan AM, Wachs T, Craighead H, Henion JD. Anal. Chem. 2003;75:6430. doi: 10.1021/ac030249+. [DOI] [PubMed] [Google Scholar]
- 52.Tan AM, Benetton S, Henion JD. Anal. Chem. 2003;75:5504. doi: 10.1021/ac030196w. [DOI] [PubMed] [Google Scholar]
- 53.Marchiarullo DJ, Lim JY, Vaksman Z, Ferrance JP, Putcha L, Landers JP. J. Chromatogr., A. 2008;1200:198. doi: 10.1016/j.chroma.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 54.Tan HY, Loke WK, Tan YT, Nguyen NT. Lab Chip. 2008;8:885. doi: 10.1039/b800438b. [DOI] [PubMed] [Google Scholar]
- 55.Urbanski JP, Johnson MT, Craig DD, Potter DL, Gardner DK, Thorsen T. Anal. Chem. 2008;80:6500. doi: 10.1021/ac8010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma B, Zhang GH, Qin JH, Lin BC. Lab Chip. 2009;9:232. doi: 10.1039/b809117j. [DOI] [PubMed] [Google Scholar]
- 57.Wu HM, Sui G, Lee CC, Prins ML, Ladno W, Lin HD, Yu AS, Phelps ME, Huang SC. J. Nucl. Med. 2007;48:837. doi: 10.2967/jnumed.106.038182. [DOI] [PubMed] [Google Scholar]
- 58.Murphy BM, He XY, Dandy D, Henry CS. Anal. Chem. 2008;80:444. doi: 10.1021/ac7019046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jo K, Heien ML, Thompson LB, Zhong M, Nuzzo RG, Sweedler JV. Lab Chip. 2007;7:1454. doi: 10.1039/b706940e. [DOI] [PubMed] [Google Scholar]
- 60.Yan H, Zhang B, Wu H. Electrophoresis. 2008;29:1775. doi: 10.1002/elps.200700561. [DOI] [PubMed] [Google Scholar]
- 61.Chen P, Feng X, Du W, Liu BF. Front. Biosci. 2008;13:2464. doi: 10.2741/2859. [DOI] [PubMed] [Google Scholar]
- 62.Bao N, Wang J, Lu C. Anal. Bioanal. Chem. 2008;391:933. doi: 10.1007/s00216-008-1899-x. [DOI] [PubMed] [Google Scholar]
- 63.Paguirigan AL, Beebe DJ. Bioessays. 2008;30:811. doi: 10.1002/bies.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gomez-Sjoberg R, Morisette DT, Bashir R. J. MEMS. 2005;14:829. [Google Scholar]
- 65.Gomez R, Bashir R, Bhunia AK. Sensor. Actuat. B-Chem. 2002;86:198. [Google Scholar]
- 66.Fesenko DO, Nasedkina TV, Prokopenko DV, Mirzabekov AD. Biosens. Bioelectron. 2005;20:1860. doi: 10.1016/j.bios.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Boedicker JQ, Li L, Kline TR, Ismagilov RF. Lab Chip. 2008;8:1265. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tani H, Maehana K, Kamidate T. Methods Mol. Biol. 2007;385:37. doi: 10.1007/978-1-59745-426-1_4. [DOI] [PubMed] [Google Scholar]
- 69.Davidsson R, Johansson B, Passoth V, Bengtsson M, Laurell T, Emneus J. Lab Chip. 2004;4:488. doi: 10.1039/b400900b. [DOI] [PubMed] [Google Scholar]
- 70.Satoh W, Takahashi S, Sassa F, Fukuda J, Suzuki H. Lab Chip. 2009;9:35. doi: 10.1039/b810961c. [DOI] [PubMed] [Google Scholar]
- 71.Clark AM, Sousa KM, Jennings C, MacDougald OA, Kennedy RT. Anal. Chem. 2009;81:2350. doi: 10.1021/ac8026965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng W, Klauke N, Sedgwick H, Smith GL, Cooper JM. Lab Chip. 2006;6:1424. doi: 10.1039/b608202e. [DOI] [PubMed] [Google Scholar]
- 73.Klauke N, Smith G, Cooper JM. Lab Chip. 2007;7:731. doi: 10.1039/b706175g. [DOI] [PubMed] [Google Scholar]
- 74.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7165. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rocheleau JV, Walker GM, Head WS, McGuinness OP, Piston DW. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12899. doi: 10.1073/pnas.0405149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Faley S, Seale K, Hughey J, Schaffer DK, VanCompernolle S, McKinney B, Baudenbacher F, Unutmaz D, Wikswo JP. Lab Chip. 2008;8:1700. doi: 10.1039/b719799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang LH, Liu DY, Bo W, Jie S, Li LH. Chin. J. Anal. Chem. 2008;36:143. [Google Scholar]
- 78.Zguris JC, Itle LJ, Hayes D, Pishko MV. Biomed. Microdevices. 2005;7:117. doi: 10.1007/s10544-005-1589-9. [DOI] [PubMed] [Google Scholar]
- 79.Baudoin R, Corlu A, Griscom L, Legallais C, Leclerc E. Toxicol. in Vitro. 2007;21:535. doi: 10.1016/j.tiv.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 80.Yoo SK, Lee JH, Yun SS, Gu MB, Lee JH. Biosens. Bioelectron. 2007;22:1586. doi: 10.1016/j.bios.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 81.Oblak TD, Meyer JA, Spence DM. Analyst. 2009;134:188. doi: 10.1039/b816740k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu H, Stybayeva G, Macal M, Ramanculov E, George MD, Dandekar S, Revzin A. Lab Chip. 2008;8:2197. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- 83.Carraro A, Hsu WM, Kulig KM, Cheung WS, Miller ML, Weinberg EJ, Swart EF, Kaazempur-Mofrad M, Borenstein JT, Vacanti JP, Neville C. Biomed. Microdevices. 2008;10:795. doi: 10.1007/s10544-008-9194-3. [DOI] [PubMed] [Google Scholar]