Abstract
Suppressor of tumorigenicity 14 (St14) encodes matriptase, a serine protease, which regulates processing of profilaggrin to filaggin in vivo. Here, we report that transgenic mice with 1% of wild-type St14 levels (St14hypo/–) display aberrant processing of profilaggrin and model human ichthyotic skin phenotypes. Scaling of the skin appears at 1 week of age with underlying epidermal acanthosis and orthohyperkeratosis as well as a CD4+ T-cell dermal infiltrate. Upregulation of antimicrobial peptides occurs when challenged by exposure to the postnatal environment. Direct genomic sequencing of bacterial 16S rRNA genes to query microbial diversity identifies a significant shift in both phylogeny and community structure between St14hypo/– mice and control littermates. St14hypo/– mice have a selective shift in resident skin microbiota with a decrease of the dominant genus of skin bacteria, Pseudomonas and an accompanying increase of Corynebacterium and Streptococcus. St14hypo/– mice provide early evidence that the cutaneous microbiome can be specifically altered by genetic state, which may play an important role in modulating skin disease.
INTRODUCTION
The skin serves as both a barrier to infection and an intricate habitat for a diverse population of microbiota (bacteria, fungi, and viruses). Proposed beneficial roles of resident commensal microbiota include inhibition of pathogenic species and further processing of skin proteins, free fatty acids, and sebum (Roth and James, 1988). Many lines of evidence also suggest that specific microorganisms are also associated with skin diseases, such as atopic dermatitis (AD; eczema), rosacea, psoriasis, and acne (Fredricks, 2001). Resident commensal microbiota may become pathogenic, sometimes in response to an impaired skin barrier (Roth and James, 1988).
Knowledge of the skin microbiota was initially limited to culture-dependent assays, although it is estimated that less then 1% of bacterial species can be cultivated (Staley and Konopka, 1985).
Genomic technology has revolutionized our ability to characterize the microbiota that resides in soil, ocean water, and sites in and on the human body (Turnbaugh et al., 2007). The 16S small subunit ribosomal (rRNA) genes are universal among prokaryotes. Analysis of bacterial phylogeny is based on the survey-based aggregate sequence of the prokaryotic 16S rRNA gene, which serves as a molecular clock of bacterial evolution. Broad-range PCR primers anneal to highly conserved regions and allow amplification from the majority of known bacteria.
These 16S rRNA primers flank species-specific variable regions of the gene that are used to infer phylogenetic relationships (Hugenholtz and Pace 1996; Pace 1997). Bacterial species are traditionally defined as having 16S rRNA sequences ≥97% identical to each other (Gevers et al., 2005). Sequence analysis of 16S rRNA genes from a single clinical or environmental sample provides a window into the bacterial diversity of culturable, fastidious, and an even un-culturable species inhabiting that particular niche.
Sequence analysis of 16S rRNA genes from human skin of antecubital fossa and volar forearm both revealed much greater diversity than previously appreciated (Gao et al., 2007; Grice et al., 2008). Analysis of mouse ear skin, commonly used for inflammation studies, found that the microbial community membership and structure is very similar to the antecubital fossa (Grice et al., 2008). Here, we utilize high throughput genomic sequencing to examine the skin microbiota of an ichthyotic animal model with a selective alteration in the genetic state.
Mice lacking the serine protease matriptase (St14–/–), also known as MT-SP1, TADG15, and epithin, die perinatally due to severe skin barrier impairment (List et al., 2003). Mutations in the human ST14 locus underlie autosomal recessive ichthyosis with hypotrichosis (ARIH: OMIM 610765) (Basel-Vanagaite et al., 2007; Alef et al., 2008; Desilets et al., 2008). Mice possessing one null and one hypomorphic allele of St14 (St14hypo/–), express ~1% of St14 at birth and survive to adulthood (List et al., 2007). These St14hypo/– mice recapitulate many of the hallmark features of ARIH, including impaired desquamation, hypotrichosis, and tooth defects.
The skin barrier is formed in the exterior layers of the epidermis and is comprised of cornified envelopes (enucleated keratinocytes), held together by a lipid matrix (Segre, 2006a). The skin barrier defect in both mice and humans with mutations in ST14 have been associated with impaired filaggrin (FLG) processing (List et al., 2003; Desilets et al., 2008). FLG is expressed in the granular layer of terminally differentiating epidermal cells and encodes a 400 kDa precursor protein, profilaggrin that undergoes dephosphorylation and proteolysis to yield 37 kDa repeat filaggrin peptides. FLG peptides are thought to contribute to skin barrier function through aggregation of keratin intermediate filaments to form the chemically cross-linked cornified envelope as well as through hydration and acidification of the stratum corneum (Elias et al., 2008; Steinert et al., 1981). Here, we examine the integration of (1) alterations in skin barrier proteins; (2) innate immune response; and (3) selective shift in the microbiota of St14hypo/– mice as an animal model of human ichthyotic phenotypes.
RESULTS
Icthyotic phenotype of St14hypo/– mice
The skin phenotype of St14hypo/– mice is grossly characterized by body-wide scaling that begins after 1 week of age and persists for several weeks but gradually improves, or is harder to appreciate with the overlying hair. In adult mice, some scaling persists in all body areas with less hair, for example, tail and paws, but it is most prominent on the ears (Figure 1a). Consistent with the body-wide scaling, western blots of back skin from 10-day-old St14hypo/– mice display less processed FLG with an accumulation of larger molecular weight proteins, representing both normal and abnormal processing intermediates, similar to the pattern observed in St14–/– mice (Figure 1b) (List et al., 2003). Adult ear skin in St14hypo/– mice displays a similar defect in FLG processing with less processed FLG in spite of increased expression of Flg mRNA (Figure 1b and data not shown). Adult St14hypo/– ear skin displays an increase of FLG antibody reactive proteins of small molecular weight (<30 kDa), representing further degradation products, which were not seen in St14–/– mice, but have been observed in normal mice under xeric stress conditions (Scott and Harding, 1986). In St14hypo/– mice, upregulation of the Flg gene may indicate the presence of a specific feedback mechanism because by RT-PCR we observe no change in expression of the other tandemly arrayed filaggrin-like genes (filaggrin-2, hornerin, cornulin, repetin, trichohyalin) (data not shown) (Segre, 2006b).
Figure 1. Scaly skin and misprocessing of FLG in St14hypo/– mice.
(a) St14hypo/– mice display body-wide scaling at 10 days of age. In adult mice, the scaly phenotype is most pronounced in the ears. (b) St14hypo/– skin cells display aberrant FLG processing, resulting in visibly reduced processed FLG on western blots of 10-day-old back skin and epidermis from adult ears. Lane 1 and 3 are WT; lane 2 and 5 are St14hypo/– with equal loading; lane 4 is St14hypo/– underloaded for comparison. Keratin 14 (K14) is shown as a control.
St14hypo/– mice display a normal spatiotemporal pattern of barrier acquisition in utero (Figure S1). Transepidermal water loss (TEWL) of affected skin in 10-day-old St14hypo/– mice is normal (data not shown), consistent with the normal TEWL and increased scale demonstrated in grafted skin from TGase–/– mice, another adult animal model of impaired skin barrier (Kuramoto et al., 2002).
Histologic features of St14hypo/– ear skin with immune cell infiltration
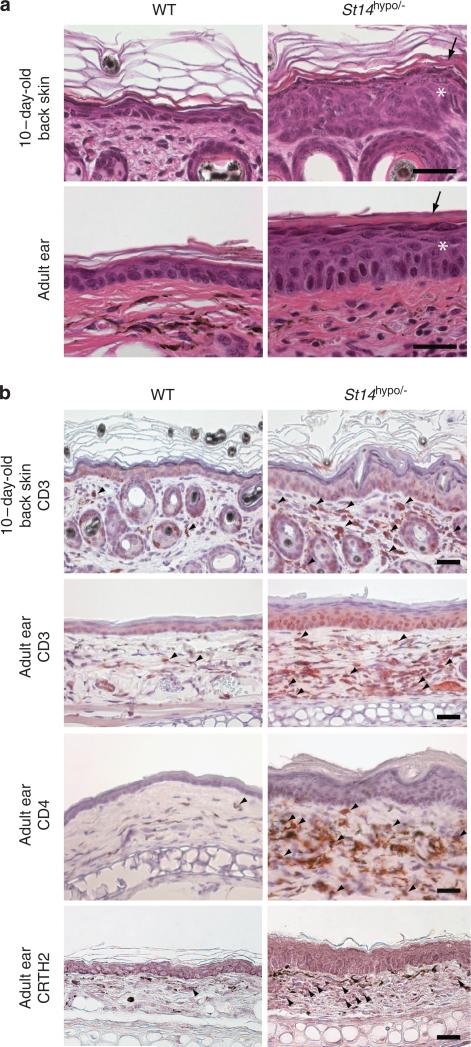
St14hypo/– ear and back skin has a thickened epidermis (acanthosis) and compacted and thickened stratum corneum (orthohyperkeratosis), features common to ichthyotic disorders (Figure 2a). St14hypo/– mice do not show absence of the granular layer as is observed in the most severe cases of ichthyosis vulgaris (Compton et al., 2002). No change in expression of epidermal differentiation markers Keratin1, Keratin14, Involucrin or Loricrin was observed in St14hypo/– skin when analyzed by immunohistochemistry (Figure S2). To investigate whether St14hypo/– skin had an inflammatory phenotype as seen in AD, immunohistochemistry with lymphocytic markers was performed. Increased infiltration by CD3+ T-cells was present in the dermis of both 10-day-old back skin and adult ears of St14hypo/– mice. Further characterization of this inflammation revealed a predominantly CD4+ versus CD8+ T-cell infiltrate, which is consistent with but not specific for AD-like inflammation (Figure 2b and data not shown). No increase in scratching behavior was observed in St14hypo/– mice, consistent with no increase in mast cells, eosinophils, or serum IgE (data not shown). In these tissue samples, mRNA expression of inflammatory cytokines (IL-1, IL-4, IL-5, IL-6, IL-13, IFN-γ, and TNFα) did not reveal a specific T-helper type 1 or 2 profile (data not shown). However, increased dermal staining for the prostaglandin D receptor (CRTH2) was seen in adult St14hypo/– ears, suggesting that this infiltrate has a T-helper type 2 dominant phenotype (Figure 2b) (Man et al., 2007).
Figure 2. Skin from St14hypo/– mice displays histological features of ichthyotic disorders and an inflammatory infiltrate.
(a) Representative images of hematoxylin and eosin staining of 10-day-old back skin and adult ears display significant acanthosis (marked by asterisks) and orthohyperkeratosis (arrows). Bar = 50 μm. (b) CD3 staining reveals dermal lymphocytic infiltrate in both 10-day-old back skin and adult ears. This is a predominantly CD4+ but not CD8+ lymphocytic infiltrate. Positive staining for prostaglandin D receptor (CRTH2) suggests a predominant T-helper type 2 phenotype. Examples of positively stained cells are marked with arrowheads. Bar = 50 μm.
Increased expression of antimicrobial peptides postnatally
Antimicrobial peptides (AMPs), mediators of the innate immune system, can both directly kill bacteria, and trigger an adaptive immune response through cytokine activation (Braff and Gallo, 2006). Although levels of specific AMPs (for example, cathelicidin) are unchanged in AD skin (Ong et al., 2002), increased gene expression of another AMP (for example, β-defensin-2) is observed in the stratum corneum of AD lesional skin (Ong et al., 2002; Asano et al., 2008). AMPs are significantly upregulated in genetically altered or physically disrupted mouse models of barrier dysfunction (de Guzman Strong et al., 2006; Aberg et al., 2008;). Although transcript levels were unchanged in newborn St14hypo/– mouse skin, we observed upregulation of AMPs in St14hypo/– mouse skin at all later time points (P<0.05) (Figure 3). This suggests that the innate immune response of St14hypo/– skin is activated with exposure to the postnatal environment.
Figure 3. Upregulation of antimicrobial peptides, biomarkers for an impaired skin barrier, occurs with exposure to the terrestrial environment in St14hypo/– mice.
Heat map representation of qRT-PCR data shows upregulation of antimicrobial peptides in skin of St14hypo/– mice beginning at 1 week of age. Red indicates increased expression in St14hypo/– vs WT mice with fold increase specified in legend (all P<0.05 by two-tailed Student's t-test).
Longitudinal survey of murine ear microbiota
During the birthing process and subsequent exposure to the postnatal environment, the skin is colonized by a wide array of microbes, many of which are commensal or symbiotic. To identify the bacteria that reside in and on the skin, we utilized high throughput DNA sequencing. Genomic DNA is prepared directly from a tissue sample with a protocol that ensures proper lysis of bacterial cells. From each DNA, metagenomic 16S rRNA genes are amplified with primers flanking the variable regions (Grice et al., 2008) and 200 unique sequences are obtained to survey the bacterial diversity in every sample.
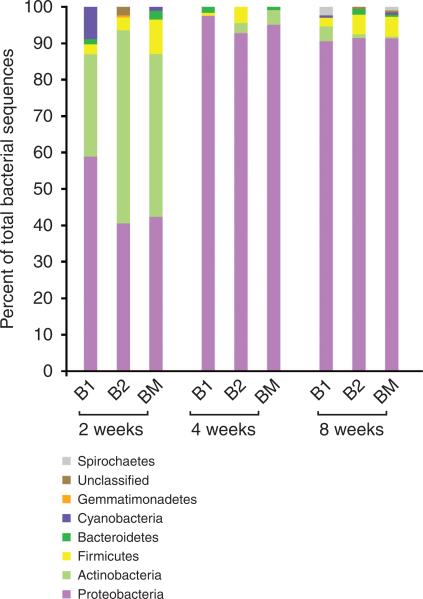
We first assessed the intra-murine and inter-murine fluxes of ear skin microbiota in genetically identical C57BL6/J littermates over the first months of life. At two weeks, the relative abundance of bacteria at the division level ranges from 42–58% Proteobacteria; 28–53% Actinobacteria; and 3–10% Firmicutes and <1–8% Cyanobacteria (Figure 4). The individual microbiota compositions fluctuate for the first month, but by 4 and 8 weeks they begin to normalize and resemble an “adult” skin microbiota profile with very little flux at the division level (Figure 4 and (Grice et al., 2008)).
Figure 4. During the first postnatal month, ear skin microbiota communities vary widely between C57BL6/J littermates.
Direct sequencing of bacterial 16S rRNA genes from 2 C57BL6/J littermates (B1 and B2) surveyed at 2 weeks, 4 weeks and 8 weeks after birth along with their mother (BM). 16S rRNA sequences are grouped into bacterial divisions (phylum).
Selective shift in St14hypo/– skin microbiota
To analyze whether the cutaneous microbiota is altered in St14hypo/– mice, we surveyed the bacterial 16S rRNA genes when the microbiota has stabilized (2 months). Three St14hypo/– mice and three control littermates were housed together at birth and then individually since weaning. To compare the community structure of the bacteria residing on these mice, we measured the pairwise relationship of each mouse and clustered the data to create a dendogram. Specifically, we clustered bacterial sequences into operational taxonomic units (OTUs; “phylotypes”) using the DOTUR program by the furthest neighbor-joining method and a similarity cutoff of 97% (Schloss and Handelsman, 2005); that is, every sequence is ≥97% identical to every other sequence within the OTU. Next, with the SONS program, we calculated θ, an index that measures community structure similarity as the relative abundance of OTUs (Yue and Clayton, 2005; Schloss and Handelsman, 2006). Values of θ can fall between 0 and 1: a value of 1 implies identical community structure and a value of 0 implies dissimilar community structures. θ values for each pairwise comparison (Table S1) are clustered to generate a dendogram, which shows that the bacterial community structure of the 3 St14hypo/– mice is distinct from the bacterial community structure of the control littermates (Figure 5a). As independent confirmation, we employed phylogenetic tree-based methods to compare the skin microbiota communities of St14hypo/– mice and control littermates. The P (phylogeny) test examines the distribution of unique sequences and their co-variation with phylogeny, measured as the number of parsimonious changes required to account for the observed distribution of sequences in the tree (Lozupone et al., 2006). Comparing the St14hypo/– mice with control littermates, we obtained a significant value p<0.01, implying that these 2 classes of mice harbor distinct phylogenetic lineages (Maddison, 1991). These results all suggest that microbiota community composition differs between wt and St14hypo/– mice.
Figure 5. Direct sequencing of bacterial 16S rRNA genes of St14hypo/– mice and WT littermates reveals significant shift in microbiota.
(a) Microbial community structure differs between WT and St14hypo/– mice. Dendogram of pairwise θ values (Table S1) compares the community structure of WT littermate (n = 3) and St14hypo/– mice (n = 3). The length of the scale bar represents a distance of 0.10 (1–θ). (b) Selective shift in abundance of bacterial divisions and genera, comparing WT littermate (n = 3) and St14hypo/– mice (n = 3). 16 s rRNA sequences are grouped into bacterial divisions. Mean values for the abundance of each of the four major divisions are plotted as percent of total sequences for WT littermate and St14hypo/– mice with SD. In St14hypo/– mice abundance of the Proteobacteria division is decreased, whereas that of Actinobacteria and Firmicutes are increased, with P values indicated (two-tailed Student's t-test). Each division bar is further broken down into its component bacterial classes. When a specific bacterial genus dominates the class, its name is noted as Class (Genus) in the legend. indicates the statistically significant increase in abundance of the genera Corynebacterium and Streptococcus for St14hypo/– mice with a decrease in Pseudomonas (close to significance at P = 0.056) and no change in levels of Janthinobacterium.
We identified a statistically significant shift among St14hypo/– mice in the proportions of three of the four dominant skin divisions (Figure 5b). Specifically, the relative abundance of Proteobacteria was decreased in St14hypo/– mice (P = 0.001), whereas that of Actinobacteria and Firmicutes were increased (P<0.001 and P = 0.018, respectively). At the genus level of classification, Corynebacterium (division: Actinobacteria) and Streptococcus (division: Firmicutes) were over-represented on St14hypo/– skin, 13.6 vs 0.1% (P<0.0001) and 6.3 vs 0.5% (P<0.02), respectively (Figure 5b). The Staphylococcus genus (division: Firmicutes) was also more prevalent on St14hypo/– as compared with WT ears, 6 vs 3 sequences, but this difference did not approach significance in this study. No sequences matching S. aureus bacteria were detected, which could be because of the power of the study or efforts to eliminate pathogens from the animal facility in which these mice were housed. Although the relative abundance of Pseudomonas decreased on St14hypo/– ears, (33.0 vs 47.2%), this was only marginally significant in this population size (P = 0.056). Janthinobacterium, the second most prevalent genus, remained constant between St14hypo/– and littermates (30.0 vs 34.3%). The intra- and inter-murine temporal variation observed among genetically homogenous C57BL6/J littermates in the first month confounds our ability to pinpoint the initiating event in the shift of both AMPs and microbiota. Nevertheless, in adult mice, we observe that Corynebacterium and Streptococcus are selectively filling the niche previously occupied by Pseudomonas.
DISCUSSION
Together these results support the conclusion that St14hypo/– mice represent a physiologically relevant animal model of ST14 deficiency and manifest some but not all features of human ichthyotic skin disorders. St14hypo/– mice may have defects that extend beyond the FLG deficiency and contribute to the ichthyotic skin phenotype. The recent discovery that null mutations in the filaggrin (FLG) gene, which encodes an important epidermal structural protein, are strongly associated as semi-dominant traits with both Ichthyosis vulgaris and AD underscores the primary role of barrier impairment in the pathogenesis of these skin diseases (Williams, 1992; Ogawa and Yoshiike, 1993). The incidence of atopic disorders (AD, asthma, and allergic rhinitis) has risen significantly in the last several decades, suggesting that undetected gene–environment interactions might underlie this increase.
Microbes have the potential to alter both the skin barrier by producing proteases and the immune response by producing specific antigens. Staphylococcus aureus (S. aureus) skin infections are more common in AD patients (Ogawa et al., 1994) and are often associated with disease flares, that is, episodic exacerbation (Baker, 2006). Most recent studies focus on the role of barrier dysfunction and innate immunity defects underlying AD onset and severity (De Benedetto et al., 2009). Although colonization with S. aureus has long been associated with AD, genomic sequence technology and analysis can now expand the understanding of the microbial component of AD beyond what is cultivatable in a microbiology laboratory. Alterations in microbiota, innate and adaptive immune responses and epidermal barrier all have the potential to influence each other.
The NIH Roadmap for Medical Research recently launched the Human Microbiome Project (HMP) with the mission to comprehensively characterize human microbiota and analyze its role in human health and disease state (http://nihroadmap.nih.gov/hmp/). Although the goals of this project will take years to complete, surveys of the diversity of commensal microbiota inhabiting the gut, oral cavity, skin, nares, and urogenital track are underway. Genomic sequencing of clinically important or abundant microbiota is another component of the project.
To take the St14hypo/– mice as an example, our future goal is to perform full genome sequencing of these Corynebacterium and Streptococcus strains that may identify previously unreported and potentially pathogenic antigens as well as of the Pseudomonas strains to identify a beneficial metabolic function, such as breaking down sebum to produce a natural moisturizing factor. These genomic sequence advances rely on the need to culture diverse fastidious skin microbiota by utilizing the 16S rRNA sequences to design specialized media (for example, lipophilic) and tailoring the growth conditions to skin (appropriate temperature, pH, acidity). These microbial growth studies will also enable an investigation into the specificity of antimicrobial peptides to commensal and pathogenic skin microbiota. As microbes have the potential to alter their genomes more rapidly than humans, genetic changes at the microbial level are one unexplored cause for the increasing prevalence of atopic conditions such as AD and asthma. One example of horizontal gene transfer is the acquisition of genetic elements by the USA300 strain of methicilin resistant S. aureus to promote both growth and survival on human skin (Diep et al., 2006). Future research addressing the pathophysiology of AD and asthma should incorporate this concept of microbe–host interaction, and employ genomic tools to characterize the nature of that relationship.
In this study, we observed significant changes in murine skin microbiota during the first month of life. A recently published study of human infant intestinal microbiota demonstrated that the composition and temporal patterns of the microbial communities varied widely from infant to infant until the end of the first year of life, when these idiosyncratic variations in microbial ecosystems converged toward the profile characteristic of the adult intestine (Palmer et al., 2007). It is fascinating to speculate that fluxes in human skin microbiota might also exist in the first year(s) of life and contribute to AD flares.
The skin is an ideal system for pioneering microbial analyses based on its accessibility. Skin possesses many characteristics that will enable us to answer fundamental questions about the role of microbial communities in both health and disease. An initial study of the role of microbiota associated with the common skin condition, psoriasis, has also been published recently (Gao et al., 2008). In coming years, our understanding of other common dermatologic diseases (for example, acne vulgaris and rosacea, psoriasis and folliculitis) and those of countless other organ systems would hopefully benefit from the application of these powerful tools of microbial genomics.
MATERIALS AND METHODS
Mice
Mice were genotyped according to methods published earlier (List et al., 2006) and housed according to NIH Animal Care and Use Committee approved protocols. Matriptase null and hypomorphic alleles were back-crossed onto the C57BL/6J background for >10 generations prior to initiating these studies.
Protein isolation and western blot analysis
Epidermis from adult ears was isolated and proteins purified as described earlier (List et al., 2003). Whole skin was used for analysis of 10-day-old back skin because eruption of hair at this stage prevents epidermal isolation. Western blot analysis was performed using a chicken antibody raised against the filaggrin monomer (1:5,000 dilution, Segre 5562) and a rabbit anti-mouse keratin 1 antibody (1:5,000 dilution, Covance), followed by peroxidase-labeled goat anti-chicken (1:10,000 dilution, Aves Labs, Tigard, OR) and goat anti-rabbit (1:5,000 dilution, GE Healthcare, Piscataway, NJ) secondary antibodies, respectively.
Histology and immunohistochemistry
Ear and back skin samples were stored in 10% neutral-buffered formalin until paraffin embedding and H&E staining. CD3 staining with a primary rabbit anti-human antibody (1:20 dilution, Dako, Carpinteria, CA) was performed on paraffin-embedded tissue sections. CD4 and CD8 staining with primary rabbit anti-mouse antibodies (1:20 dilution, BD Pharmingen, San Jose, CA) were performed on frozen tissue sections fixed in Neg-50 (Richard-Allan Scientific, Kalamazoo, MI). In all cases, a biotin-labeled goat anti-rabbit secondary antibody was used (1:500 dilution, Dako) and staining was visualized using an AEC Substrate kit (Vector Labs, Burlingame, CA). CRTH2 staining with a primary rabbit anti-mouse antibody (1:200 dilution, Cayman, Ann Arbor, MI) was performed on paraffin sections using a biotin-labeled goat anti-rabbit secondary (1:200 dilution, Vector Labs). Staining was visualized with Vector VIP substrate kit (Vector Labs). Images were captured using Openlab 4.0.3 software on a Zeiss AxioSkop microscope equipped with a Zeiss AxioCam (Dublin, CA).
Quantitative real-time PCR
RNA was isolated from murine skin and ears and transcribed into cDNA using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) as described earlier with previously published primers spanning intron–exon boundaries (de Guzman Strong et al., 2006). Samples were normalized to the control gene β-2-microglobulin and validated by comparing with β-actin. Three mice were included for each group at each time point and fold change was statistically validated using a 2-tailed Student's t-test (P<0.05).
Bacterial 16S rDNA sequencing and analysis
Comparably sized tissue samples (~1 × 3 mm) were collected from the ears of littermates on the same day using sterile instruments. DNA was extracted with the DNAeasy kit (Qiagen, Valencia, CA), following the modified protocol for Gram-positive bacteria and an additional bead-beating step. For each mouse, two replicate 50-μl 23-cycle PCRs were performed using 8F and 1391R universal primers and products of the two reactions were pooled (Ley et al., 2005). The resulting 1.3-kb amplicons were gel-purified using the QiaQuick gel extraction kit (Qiagen) and subcloned into TOPO TA pCR2.1 (Invitrogen). NIH Intramural Sequencing Center (NISC) sequenced 192 clones per sample bi-directionally. Sequences were trimmed and overlaps assembled with the PHRED and PHRAP software packages. BLAST in GenBank was used to identify and discard mouse or vector sequences. Here, 1113 out of 1152 sequences were retained following these criteria. Bellerophon version 3 (divergence ratio >1.1) was used to check for chimeras (none detected) and sequences were aligned to a core set of sequences using the NAST alignment tool, both available as a suite of programs at Greengenes (http://greengenes.lbl.gov/cgi-bin/nph-Index.cgi) (DeSantis et al., 2006). 1049 sequences successfully aligned and were inserted into reference phylogenetic trees by the neighbor-joining method and the Olsen correction using the ARB software package (www.arb-home.de) (Ludwig et al., 2004). Species identification was performed using the Ribosomal Database Project (RDP-II) release 9 (http://rdp.cme.msu.edu) (Cole et al., 2007). P-test analysis was performed using the UniFrac online suite of tools to compare phylogeny of bacterial communities(Lozupone et al., 2006). Then, 100 permutations were performed to obtain significance values. The DOTUR program, using furthest neighbor settings and a similarity cutoff of 97%, assigned sequences to OTUs (Schloss and Handelsman, 2005). θ similarity index was calculated using the SONS program and an OTU cutoff of 97% (Yue and Clayton, 2005; Schloss and Handelsman, 2006). Differences in prevalence of bacteria by both division and genus were statistically validated using a 2-tailed Student's t-test.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NHGRI and NIDCR Research Programs and by a grant from the Department of Defense (DAMD-17-02-1-0693) to Thomas Bugge. All studies were appropriately reviewed by an NIH Animal Care and Use Committee. We thank Cherry Yang for genotyping, Dr Maria Turner for assisting with histological interpretation, Drs Pamela Schwartzberg and Jennifer Cannons for sharing immunologic expertise, Martha Kirby and Stacie Anderson for FACs analysis, Dr Peter Elias for intellectual discussions on skin barrier analysis and the CRTH2 antibody, Histoserv Inc. for immunohistochemistry services, Alice Young, Robert Blakesly and Gerard Bouffard from NISC for their sequencing expertise and services, Julia Fekecs for assistance with figures, and Drs Christina Herrick and Heidi Kong for reviewing the article. We also thank members of the lab for their underlying contributions.
Abbreviations
- AD
atopic dermatitis
- AMP
antimicrobial peptide
- FLG
filaggrin
- St14
suppressor of tumorigenicity 14
Footnotes
The sequence data from this study have been submitted to GenBank under accession nos. FJ892750–FJ895041 and as NCBI Entrez Genome Project 30121.
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
REFERENCES
- Aberg KM, Man MQ, Gallo RL, Ganz T, Crumrine D, Brown BE, et al. Co-regulation and interdependence of the mammalian epidermal permeability and antimicrobial barriers. J Invest Dermatol. 2008;128:917–25. doi: 10.1038/sj.jid.5701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alef T, Torres S, Hausser I, Metze D, Tursen U, Lestringant GG, et al. Ichthyosis, follicular atrophoderma, and hypotrichosis caused by mutations in ST14 is associated with impaired profilaggrin processing. J Invest Dermatol. 2008;129:862–9. doi: 10.1038/jid.2008.311. [DOI] [PubMed] [Google Scholar]
- Asano S, Ichikawa Y, Kumagai T, Kawashima M, Imokawa G. Microanalysis of an antimicrobial peptide, beta-defensin-2, in the stratum corneum from patients with atopic dermatitis. Br J Dermatol. 2008;159:97–104. doi: 10.1111/j.1365-2133.2008.08613.x. [DOI] [PubMed] [Google Scholar]
- Baker BS. The role of microorganisms in atopic dermatitis. Clin Exp Immunol. 2006;144:1–9. doi: 10.1111/j.1365-2249.2005.02980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Attia R, Ishida-Yamamoto A, Rainshtein L, Ben Amitai D, Lurie R, et al. Autosomal recessive ichthyosis with hypotrichosis caused by a mutation in ST14, encoding type II transmembrane serine protease matriptase. Am J Hum Genet. 2007;80:467–77. doi: 10.1086/512487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff MH, Gallo RL. Antimicrobial peptides: an essential component of the skin defensive barrier. Curr Top Microbiol Immunol. 2006;306:91–110. doi: 10.1007/3-540-29916-5_4. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–72. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton JG, DiGiovanna JJ, Johnston KA, Fleckman P, Bale SJ. Mapping of the associated phenotype of an absent granular layer in ichthyosis vulgaris to the epidermal differentiation complex on chromosome 1. Exp Dermatol. 2002;11:518–26. doi: 10.1034/j.1600-0625.2002.110604.x. [DOI] [PubMed] [Google Scholar]
- De Benedetto A, Agnihothri R, McGirt LY, Bankova LG, Beck LA. Atopic dermatitis: a disease caused by innate immune defects? J Invest Dermatol. 2009;129:14–30. doi: 10.1038/jid.2008.259. [DOI] [PubMed] [Google Scholar]
- de Guzman Strong C, Wertz PW, Wang C, Yang F, Meltzer PS, Andl T, et al. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J Cell Biol. 2006;175:661–70. doi: 10.1083/jcb.200605057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desilets A, Beliveau F, Vandal G, McDuff FO, Lavigne P, Leduc R. Mutation G827R in matriptase causing autosomal recessive ichthyosis with hypotrichosis yields an inactive protease. J Biol Chem. 2008;283:10535–42. doi: 10.1074/jbc.M707012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Elias PM, Hatano Y, Williams ML. Basis for the barrier abnormality in atopic dermatitis: outside-inside-outside pathogenic mechanisms. J Allergy Clin Immunol. 2008;121:1337–43. doi: 10.1016/j.jaci.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN. Microbial ecology of human skin in health and disease. J Investig Dermatol Symp Proc. 2001;6:167–9. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–32. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Opinion: Re-evaluating prokaryotic species. Nat Rev. 2005;3:733–9. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, Young AC, NISC Comparative Sequencing Program. Bouffard GG, Blakesley RW, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–50. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholz P, Pace NR. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14(6):190–7. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- Kuramoto N, Takizawa T, Takizawa T, Matsuki M, Morioka H, Robinson JM, et al. Development of ichthyosiform skin compensates for defective permeability barrier function in mice lacking transglutaminase 1. J Clin Invest. 2002;109:243–50. doi: 10.1172/JCI13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List K, Currie B, Scharschmidt TC, Szabo R, Shireman J, Molinolo A, et al. Autosomal ichthyosis with hypotrichosis syndrome displays low matriptase proteolytic activity and is phenocopied in ST14 hypomorphic mice. J Biol Chem. 2007;282:36714–23. doi: 10.1074/jbc.M705521200. [DOI] [PubMed] [Google Scholar]
- List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am J Pathol. 2006;168:1513–25. doi: 10.2353/ajpath.2006.051071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List K, Szabo R, Wertz PW, Segre J, Haudenschild CC, Kim SY, et al. Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J Cell Biol. 2003;163:901–10. doi: 10.1083/jcb.200304161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WPaS. Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution. 1991;45:1184–97. doi: 10.1111/j.1558-5646.1991.tb04385.x. [DOI] [PubMed] [Google Scholar]
- Man MQ, Hatano Y, Lee SH, Man M, Chang S, Feingold KR, et al. Characterization of a Hapten-induced, murine model with multiple features of atopic dermatitis: structural, immunologic, and biochemical changes following single versus multiple oxazolone challenges. J Invest Dermatol. 2007;128:79–86. doi: 10.1038/sj.jid.5701011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Yoshiike T. A speculative view of atopic dermatitis: barrier dysfunction in pathogenesis. J Dermatol Sci. 1993;5:197–204. doi: 10.1016/0923-1811(93)90767-j. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Katsuoka K, Kawano K, Nishiyama S. Comparative study of staphylococcal flora on the skin surface of atopic dermatitis patients and healthy subjects. J Dermatol. 1994;21:453–60. doi: 10.1111/j.1346-8138.1994.tb01774.x. [DOI] [PubMed] [Google Scholar]
- Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–60. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol. 1988;42:441–64. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–6. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl Environ Microbiol. 2006;72:6773–9. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott IR, Harding CR. Filaggrin breakdown to water binding compounds during development of the rat stratum corneum is controlled by the water activity of the environment. Dev Biol. 1986;115:84–92. doi: 10.1016/0012-1606(86)90230-7. [DOI] [PubMed] [Google Scholar]
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006a;116:1150–8. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA. Epidermal differentiation complex yields a secret: mutations in the cornification protein filaggrin underlie ichthyosis vulgaris. J Invest Dermatol. 2006b;126:1202–4. doi: 10.1038/sj.jid.5700367. [DOI] [PubMed] [Google Scholar]
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–46. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Cantieri JS, Teller DC, Lonsdale-Eccles JD, Dale BA. Characterization of a class of cationic proteins that specifically interact with intermediate filaments. Proc Natl Acad Sci USA. 1981;78:4097–101. doi: 10.1073/pnas.78.7.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ML. Ichthyosis: mechanisms of disease. Pediatr Dermatol. 1992;9:365–8. doi: 10.1111/j.1525-1470.1992.tb00632.x. [DOI] [PubMed] [Google Scholar]
- Yue JC, Clayton MK. A similarity measure based on species proportions. Commun Stat Theor M. 2005;34:2123–31. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.