Abstract
We explored the use of a fiber-optic probe for in vivo fluorescence spectroscopy of breast tissues during percutaneous image-guided breast biopsy. A total of 121 biopsy samples with accompanying histological diagnosis were obtained clinically and investigated in this study. The tissue spectra were analyzed using partial least-squares analysis and represented using a set of principal components (PCs) with dramatically reduced data dimension. For nonmalignant tissue samples, a set of PCs that account for the largest amount of variance in the spectra displayed correlation with the percent tissue composition. For all tissue samples, a set of PCs was identified using a Wilcoxon rank-sum test as showing statistically significant differences between: 1) malignant and fibrous/benign; 2) malignant and adipose; and 3) malignant and nonmalignant breast samples. These PCs were used to distinguish malignant from other nonmalignant tissue types using a binary classification scheme based on both linear and nonlinear support vector machine (SVM) and logistic regression (LR). For the sample set investigated in this study, the SVM classifier provided a cross-validated sensitivity and specificity of up to 81% and 87%, respectively, for discrimination between malignant and fibrous/benign samples, and up to 81% and 81%, respectively, for discriminating between malignant and adipose samples. Classification based on LR was used to generate receiver operator curves with an area under the curve (AUC) of 0.87 for discriminating malignant versus fibrous/benign tissues, and an AUC of 0.84 for discriminating malignant from adipose tissue samples. This study demonstrates the feasibility of performing fluorescence spectroscopy during clinical core needle breast biopsy, and the potential of this technique for identifying breast malignancy in vivo.
Index Terms: Biopsy, breast cancer, fluorescence, spectroscopy
I. Introduction
Percutaneous core needle biopsy plays an important role in the early detection of breast cancer and has been increasingly used as an alternative to open surgical biopsy for the diagnosis of suspect mammographic abnormalities in the breast. It has many advantages over open surgical biopsy. It is less invasive and faster, costs less, causes minimal or no scarring, and requires a short recovery time. Needle biopsy can obviate the need for a surgery in women with benign lesions and reduce the number of surgical procedures performed in women with breast cancer [1]–[9]. However, a major caveat of this minimally invasive procedure is that only a few small pieces of tissue are sampled from the suspicious region, and it is possible that the pathologic area is missed altogether or undersampled [10]. As a result, the frequency of missed cancers in trials ranges from 0.3% to 8.2% [3]–[5], [7], [11] and repeat biopsies are required in 9%–18% of patients [6], [7], [9] primarily due to the discordant mammographic and pathologic findings. Furthermore, statistics showed that approximately 80% of biopsies are benign, according to the American Cancer Society [12], which suggests a large number of unnecessary sampling of nonmalignant tissues.
Given the life time probability that a woman has to undergo a breast biopsy procedure and the physical, emotional, and financial costs to the patient, it is essential to improve the sampling accuracy of this minimally invasive diagnostic procedure. The sampling accuracy highly depends on the appropriate positioning of the biopsy needle. Several imaging modalities are being used to guide needle positioning into the suspicious lesion, such as mammography and ultrasonography. However, such imaging techniques rely on a 2-D image to locate a 3-D target; an error in estimating the location of the target may cause significant offset and the needle may end up missing the pathologic area even though it appears to project into the lesion [5]. Another factor that may affect the sampling accuracy is the skill level of the operator. Findings from previous studies [3], [13] have shown that false negative cases were more frequent among less experienced radiologists, which elucidated the importance of operator experience in performing a successful breast biopsy.
One way to improve the sampling accuracy of core needle biopsy is to incorporate a “sensor” as an adjunct to the biopsy needle, to survey multiple tissue sites and provide nondestructive tissue pathology before the sample is removed. Optical spectroscopy, and in particular, fluorescence and diffuse reflectance spectroscopy, have the potential to serve as an adjunct diagnostic modality to core needle biopsy. The onset of carcinogenesis in tissue can result in structural, physiological, and biochemical changes, which alters the spectroscopic properties of tissue fluorescence and diffuse reflectance [14]. Optical spectroscopy can be deployed through fiber-optic probes, which can be easily adapted for use in the biopsy needle to quickly and nondestructively characterize tissue pathology at the needle tip during a breast biopsy procedure. A positive reading from the optical measurement could potentially indicate that a biopsy is being sampled from a tumor site. If the optical measurement reads negative, then the needle could be repositioned (along the needle track) to a new tissue site. Currently, six to 24 biopsies are taken during a core needle biopsy procedure. If the optical spectroscopy method can maximize sampling from tissue sites that are most likely to be cancerous and minimize unnecessary removal of many normal tissues, it could make the breast biopsy procedure more accurate, less traumatic to the patient, and also reduce the number of biopsies that need to be processed in order to obtain a confirmatory diagnosis.
There have been several studies in which reflectance spectroscopy has been performed on breast tissues through biopsy needles. Bigio et al. [15] measured diffuse reflectance spectra from breast tissues using an optical probe through a core biopsy (Trucut) needle. They made single-point measurements in vivo from 59 normal sites and 13 cancer sites. They reported a sensitivity and specificity of 69% and 85% using an artificial neural network (ANN) classification, and 67% and 79% using a hierarchical cluster analysis (HCA) in this pilot study. van Veen et al. [16] performed differential path length spectroscopy (DPS) on 12 patients through a modified biopsy gun (Bard Magnum) and demonstrated the feasibility of DPS combined with large-core needle biopsy to determine the local optical properties of healthy and malignant breast tissues in vivo. In a recent pilot study by our group [17], frequency-domain near-infrared (NIR) diffuse optical spectroscopy was performed through a core biopsy needle on ten normal/benign breast tissues, and the preliminary results showed the feasibility of using a side-firing fiber-optic probe for in vivo NIR spectroscopy during core needle biopsy of the breast. Fluorescence spectroscopy has been extensively investigated for breast cancer diagnosis, as it is sensitive to a number of biological fluorophores in human breast tissues. Previous studies carried out ex vivo or in vitro have demonstrated that there are significant differences in the UV–Vis fluorescence spectra of normal, benign, and malignant breast tissues [18]–[23]. However, to our knowledge, no studies have been carried out to explore the diagnostic potential of this tool for in vivo breast cancer diagnosis.
In this paper, we present a pilot study to address this gap in the scientific literature. In this study, fluorescence spectroscopy of breast tissues was performed on patients undergoing image-guided percutaneous core needle biopsy to assess the feasibility of incorporating in vivo fluorescence spectroscopy into a clinical core needle breast biopsy procedure. The measured tissue spectra were also analyzed to evaluate the potential of diagnosing breast cancer using the spectroscopic contrast contained in the in vivo tissue spectra.
II. Methods
A. Patient Recruitment
The clinical protocol for the optical spectroscopy study was reviewed and approved by the Institutional Review Board (IRB) at the University of Wisconsin. The clinical study was carried out at the University of Wisconsin Hospital and Clinics (UWHC). Patients with a mammographically and/or sonographically visible abnormality and who were scheduled for a core needle biopsy procedure were invited to participate in the study. An informed written consent was obtained from each participating patient before the optical intervention was performed. Patient information including age, menopausal status, number of pregnancies or year of first full-term pregnancy, personal and family history of breast cancer were obtained from a self-reporting patient history datasheet. A radiologist (EB) determined the mammographic breast density from a retrospective review of the patient’s mammography report.
B. Instrument (Fiber-Optic Probe and Optical Spectrometer)
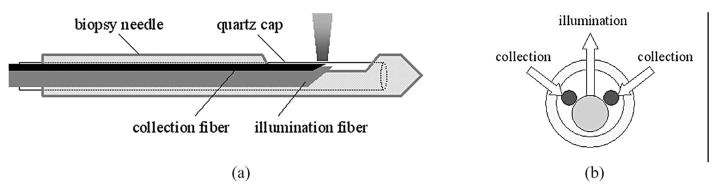
A fiber-optic probe has been designed to work compatibly in a nine-gauge vacuum-assisted core biopsy needle (Suros ATEC, Suros Surgical Systems, IN), which is currently employed for image-guided breast biopsy at the UWHC. The nine-gauge Suros needle has a side-facing aperture that is 20 mm long and 3.7 mm wide at the needle tip. A side-firing probe has been configured to enable the illumination and collection of light through this aperture. Fig. 1(a) shows side view of the side-firing probe inserted in the biopsy needle and Fig 1(b) shows cross section of the probe assembly. The arrows indicate the direction of light illumination and collection. The probe consists of a centrally positioned illumination fiber with a core diameter of 600 μm and a numerical aperture (NA) of 0.22, and two adjacent collection fibers, each with a core diameter of 200 μm and an NA of 0.22. The tips of these fibers are obliquely polished at an angle of 43° such that the light paths are perpendicular to the fiber axis. The illumination and collection fibers are radially oriented and aligned to yield a center-to-center distance of approximately 600 μm between the illumination and collection areas at the probe–tissue interface. The fluorescence sensing depth yielded from this probe geometry was evaluated using a scalable Monte Carlo simulation [24] and ranges from 560 to 750 μm in homogeneous tissue phantoms with absorption coefficients ranging from 0.3 to 20 cm−1 and reduced scattering coefficients ranging from 8.4 to 15 cm−1. These optical properties are representative of that in breast tissues in the UV–Vis spectral range [25]. In the final probe assembly, all the fibers were encased in a transparent quartz cap that transmits light in the UV–Vis spectrum and these fibers relay the optical signals between the tissue and the optical spectrometer via flexible tubing. The entire fiber-optic probe was fabricated according to our design specifications by Polymicro Technologies, LLC (Phoenix, AZ).
Fig. 1.
(a) Side view of the side-firing probe inserted in the biopsy needle. (b) Cross section of probe assembly. Arrows indicate the direction of illumination and collection light.
A multiwavelength spectrometer was used for the fluorescence measurements, which consists of a 450 W xenon lamp (FL-1039, J.Y. Horiba), a scanning double excitation monochromator (Gemini 180, J.Y. Horiba), a filter wheel, an imaging spectrograph (Triax 320, J.Y. Horiba), and a charged-couple device (CCD) camera (CCD3000, J.Y. Horiba). Details of the spectrometer can be found in earlier publications [23], [26].
C. In Vivo Optical Spectroscopy During Breast Needle Biopsy
A total of 73 patients, including 43 undergoing stereotactic and 30 undergoing ultrasound guided core needle breast biopsy participated in this study. Out of 73 cases, 51 including 26 from stereotactic and 25 from ultrasound guided biopsy were successful, i.e., yielded good optical measurements and accompanying histological diagnosis, while the remaining 22 failed. A case was considered a failure when the quality of optical measurements was very poor or histological diagnosis of each biopsy core was unavailable. The quality of the spectral measurement was acceptable if the fluorescence peak intensity was at least twice that of the background fluorescence intensity measured in water (which does not fluoresce). Several occurrences can cause the failure, which include but may not be limited to the following:
probe failure due to damaged fibers;
probe was not correctly oriented to enable the light transmission through the open aperture;
the cutting cannula inside the biopsy needle was not retracted to expose the aperture;
the probe was not put in direct contact with tissue;
biopsy cores were mixed together, thus correlating the tissue sample pathology with the corresponding optical measurement was impossible.
During the core biopsy procedure, the biopsy needle was positioned into the lesion with mammographic (stereotactic) or ultrasound guidance. The sterile probe was then inserted into the needle to make fluorescence measurements on breast tissues at the needle tip through the side-facing aperture. A biopsy was taken from the exact site where fluorescence measurements had been made on in order to obtain the corresponding histopathology for the optical measurement. Three to six measurements at clock positions within a 360° rotation were made on each patient. There were two scenarios for measurement and biopsy: for the first 45 (32 successes and 13 failures) study cases, biopsy sampling was done immediately after the optical measurement at each site, and for the next 28 (19 successes and nine failures) study cases, optical measurements on all tissue sites were completed sequentially by rotation of the biopsy needle and then a biopsy sample was obtained retrospectively from each exact site by repositioning the biopsy needle at those sites. The change to the protocol was made to speed up the optical measurements and core needle biopsy sampling procedures.
At each clock position, fluorescence emission spectra were recorded at seven excitation wavelengths from 300 to 420 nm in 20 nm increments. For each excitation wavelength, the fluorescence emission spectrum was measured over a 260 nm wavelength range, with the shortest wavelength red-shifted by 20 nm from the excitation wavelength. This setup allows for the comprehensive characterization of primary endogenous fluorophores present in breast tissues including tryptophan, collagen, nicotinamide adenine dinucleotide hydride (NADH), and retinol content, and indirectly, the affect of absorption and scattering on the fluorescence. The slit width of the imaging spectrograph was set to provide a spectral resolution of 7.9 nm. Each intensity-wavelength point in the emission spectrum was binned over 18 pixels on the CCD chip, resulting in a wavelength increment of 4.7 nm. The integration time for acquiring one fluorescence spectrum was 3 s. For each tissue site, it took about 1 min to complete the fluorescence spectral measurements at all seven excitation wavelengths, and with three to six tissue sites measured, the duration of the optical study was approximately 5–10 min. The patients were advised of the additional time needed for the study when they were consented.
The measured fluorescence spectrum was corrected for: 1) background; 2) the wavelength-dependent intensity variations of the system; and 3) the throughput of the system. The background spectra were measured with the probe immersed in distilled water in a dark container using the same experimental setup and under the same lighting conditions for fluorescence spectroscopy of breast tissues. The background spectrum was subtracted point-by-point from each tissue spectrum prior to further calibration. The fluorescence emission spectral intensities at each excitation wavelength were divided by the excitation calibration factor, which is the output power of excitation light measured at the probe tip using an optical power meter, to account for the wavelength-dependent variation of the excitation light intensity. The fluorescence spectrum was then corrected for wavelength-dependent response of the collection fibers, imaging spectrograph, and CCD camera, by multiplying it point-by-point by the correction factors measured using a National Institute of Standards Technology (NIST) tungsten calibration lamp. Finally, each fluorescence spectrum was divided by the peak fluorescence intensity (excitation–emission wavelength pair of 460–580 nm) measured with the probe immersed in a Rhodamine B (100 mg/L) solution using the same integration time, to account for the day-to-day changes in the throughput of the instrument.
D. Histopathology of Biopsy Cores
Each biopsy sample was labeled in the sequence of A–E and placed in formalin in a separate container and submitted routinely for histopathology. Several hemotoxylin and eosin (H&E) stained sections were cut from each biopsy core for microscopic evaluation. For the purpose of the optical study, the region of interest on each sample was defined as the 5 mm segment at the center of biopsy core, which is where the fiber probe tip is positioned within the side-facing aperture of the biopsy needle. Histopathologic diagnoses of the region of interest as well as that of the entire biopsy core were performed by a board-certified pathologist (JH). The diagnosis of the optical sample was assigned as the histology of the region of interest on the biopsy core. Some biopsy cores broke into multiple pieces during the biopsy procedure and caused uncertainty in determining the region of interest. In this case, the histology of the entire core was used to describe the diagnosis of the optical sample. The histological composition of each tissue sample was recorded as percentage adipose, fibrous, ducts/lobules, benign tissue content, and cancer. Benign tissue content was further categorized as adenosis, fibroadenoma, cyst, fibrocystic change, and reparative changes, and cancerous tissue content was divided into infiltrating ductal carcinoma (IDC), infiltrating lobular carcinoma (ILC), ductal carcinoma in situ (DCIS), or lobular carcinoma in situ (LCIS). LCIS, while not considered malignant in clinical practice, is a high-risk marker that represents a similar clonal proliferation of neoplastic cells as malignancy. Therefore, we included this pathologic diagnosis in our malignant sample population. In addition, blood content was also recorded as blood clots due to hemorrhage presented in some of the biopsy cores. For the purpose of data analysis in this study, each tissue sample was broadly classified, based on the histologic composition, as benign (samples with >0% of benign content), normal fibrous/fibro glandular (normal samples with >60% of fibrous and ducts/lobular content), normal adipose (normal samples with >60% of adipose content), mixture of fibrous/fibro glandular and adipose (normal samples composed of comparable amount, i.e., 40%–60% of fibrous/ducts/lobules and adipose content), and malignant (samples with >0% of cancer content).
A total of 121 biopsy samples with accompanying histopathologic diagnosis were analyzed in this study, among which 62 were obtained from stereotactically guided biopsy and 59 were obtained from ultrasound-guided biopsy. In all, five out of 62 samples collected from three stereotactic patients, and 22 out of 59 samples collected from 11 ultrasound patients had malignancy. Table I shows the histological breakdown of the biopsy samples and the sample distribution in the two types of biopsy procedure.
TABLE I.
Histological Breakdown of Biopsy Samples Analyzed in this Study
| # of samples |
|||
|---|---|---|---|
| Type and Sub-type | Stereo | US* | Combined |
| Malignant | 5 | 22 | 27 |
| Invasive ductal carcinoma (IDC) | 0 | 9 | 9 |
| Invasive lobular carcinoma (ILC) | 0 | 8 | 8 |
| Ductal carcinoma in situ (DCIS) | 4 | 4 | 8 |
| Lobular carcinoma in situ (LCIS) | 0 | 1 | 1 |
| IDC + DCIS | 1 | 0 | 1 |
| Non-malignant | 57 | 37 | 94 |
| Benign | 14 | 8 | 22 |
| Fibrous/ducts/lobulus | 15 | 10 | 25 |
| Adipose | 22 | 15 | 37 |
| Mix of fibrous/adipose | 6 | 4 | 10 |
| Total | 62 | 59 | 121 |
Note: US-Ultrasound.
E. Analysis of Tissue Fluorescence Spectra
Prior to spectral analysis, each spectrum was normalized to the integrated spectral intensity over the entire spectrum. This preprocessing removed interpatient variations and possible in-trapatient variations due to variations in probe–tissue contact. Partial least squares (PLS) analysis was then carried out on the normalized fluorescence spectra measured at each of the excitation wavelengths between 300 and 400 nm, one at a time, to extract the principal components (PCs) that not only account for a large amount of the variance in the spectral data, but are also most relevant to the known histological diagnosis (malignant or nonmalignant) of the samples, thus dimensionally reducing the number of variables needed to represent the tissue spectra. The fluorescence spectra at 420 nm excitation were excluded because at this excitation wavelength the spectra had very low SNR. The extracted PCs that accounted for a total of 95% or greater variance in the spectral data were retained for further analysis. Details about the PLS analysis are described in previous publications by our group [23], [26].
For nonmalignant tissue samples, the scores of the primary PCs, which account for the largest amount of variance in the spectra at each excitation wavelength, were evaluated for their correlation with the percent histological composition of the tissue, including percent adipose, percent fibrous, and percent ducts/lobular tissue content. Pearson correlation analysis was used to determine the relationship between the PC scores and the histological variables.
For all tissue samples, a Wilcoxon rank-sum test was carried out on each of the retained PCs (accounting for a total of 95% or greater variance), in order to identify which ones show statistically the most significant differences between malignant and nonmalignant tissue types. Three separate tests were performed on: 1) malignant versus fibrous/benign tissues; 2) malignant versus adipose; and 3) malignant versus all nonmalignant breast tissues. The scores of PCs that discriminated between malignant and nonmalignant tissue types can be used to discriminate breast malignancy. However, some of the diagnostically significant PCs at different excitation wavelengths may have correlated PC scores, as they may represent similar spectral features. To remove data redundancy, only the PC scores that were diagnostically significant as well as least correlated with other diagnostic PCs were input into the classification algorithms to diagnose breast malignancy. Two sets of PC scores were considered un-correlated if: 1) p > 0.01 for the Pearson correlation and/or 2) their corresponding PCs have distinct spectral line shapes (i.e., the spectral line shapes do not overlap), which should be characteristic of different fluorophores. The PC that showed statistically the most significant difference (i.e., lowest p-value) between malignant and nonmalignant tissues was retained as the first diagnostic PC. The second diagnostic PC was then chosen among the PCs that were uncorrelated with the first diagnostic PC and which had the lowest p-value. The “uncorrelated” condition was actually determined by applying the two aforementioned criteria rather than just by the correlation coefficient. The remaining diagnostic PCs were selected sequentially in this way.
F. Discrimination Between Malignant and Nonmalignant Tissues
1) Support Vector Machine Classification
Binary classification based on support vector machine (SVM) learning [27] was performed to discriminate malignant (i.e., class 1, denoted as c1) from nonmalignant tissue samples (i.e., class 0, denoted as c0), which include the following tissue groups: fibrous/benign, adipose, or all nonmalignant tissues. Two SVM classifiers were used in this study, one is a linear SVM and the other is a nonlinear SVM using a radial basis function (RBF) as the kernel function. Candidate classifiers were trained over a large range of penalty parameters and the performance of each SVM classifier was evaluated using a leave-one-out cross-validation scheme, in which a single sample was left out as the testing data and the remaining samples were used as the training data, and this procedure was repeated such that each sample was used once for testing. The optimal classifier was chosen as the one that yielded the best cross-validated classification accuracy. A more detailed description of the SVM classifier and cross-validation is described in our previous publications [22], [23].
2) Logistic Regression Modeling
Logistic regression (LR) modeling [28], [29] was also developed to build the predictive relationship between the tissue category and the scores of PCs that show statistically significant differences between the two tissue categories. The logistic function used to describe the predictive relationship was p(c1 |x) = exp(β′x)/(1 + exp(β′x)), where c1 represents the membership to the malignant category, β represents a vector of regression coefficients βi, i = 0, …, k, and x represents the predictor variables (i.e., score of PCs that showed statistically significant contrast between two tissue categories of interest). Output from this regression model gives the posterior probability of being malignant for each sample. A probabilistic classification scheme was then constructed by varying the threshold: if the output probability for a tissue sample is above the threshold, the tissue was designated as malignant, otherwise nonmalignant. A receiver operating characteristics (ROC) curve was constructed and the area under the ROC curve (AUC) was calculated and used to evaluate the diagnostic performance of the classifier.
III. Results
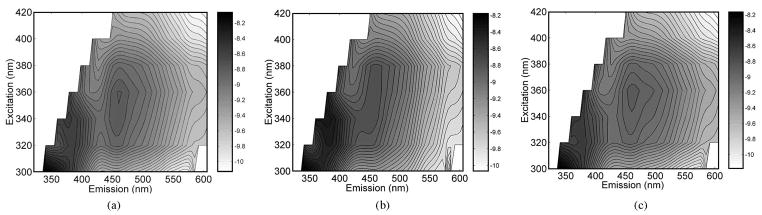
Fig. 2 displays the average in vivo fluorescence excitation–emission matrices (EEMs) of: 1) malignant (n = 27); 2) fibrous (n = 25); and 3) adipose breast tissues (n = 37) measured with the side-firing fiber-optic probe during core needle breast biopsy. All figures are plotted on a log scale. The average EEM of malignant tissues displays three visible fluorescence peaks, at excitation–emission wavelength pairs of (300, 340), (340, 385), and (340/360, 460) nm. The average EEM of normal fibrous tissues has similar fluorescence peaks as the malignant tissue EEM. The average adipose tissue EEM differs from those of malignant and normal fibrous tissues, with a weak presence of the fluorescence peak at (340, 385) nm and a shoulder adjacent to fluorescence peak at (340/360, 460) nm extending up to (340/360, 510) nm. These fluorescence features are very similar to what we have observed in average fluorescence EEMs of ex vivo breast tissues [22], [23].
Fig. 2.
Average in vivo fluorescence EEMs of (a) malignant (n = 27), (b) fibrous (n = 25), and (c) adipose (n = 37) breast tissue samples.
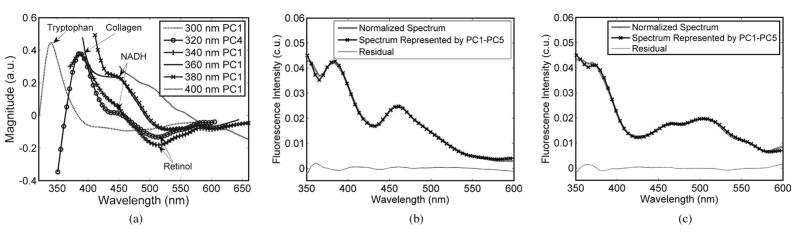
Fig. 3(a) shows the primary PCs obtained from PLS analysis, each of which accounts for the largest amount of variance in the spectra measured at a single excitation wavelength each between 300 and 400 nm, and the fluorophores they likely correspond to. Fig. 3(b) and (c) shows the normalized measured spectra of a representative malignant sample (b) and a representative normal sample (c) at an excitation wavelength of 320 nm, their corresponding PLS fit using a linear combination of five PCs PC1–PC5, and the residual of the PLS fit, as an example. The set of PCs (PC1–PC5) accounted for a total of >95% of the variance of the normalized spectra. The residual was calculated by subtracting the PLS fit point by point from the corresponding normalized spectrum. The particular excitation wavelength of 320 nm is chosen to show here as an example because most fluorophores present in breast tissues can be excited at this wavelength.
Fig. 3.
(a) Primary PCs obtained from PLS analysis, each of which accounts for the largest amount of variance in the spectra measured at a single excitation wavelength each between 300 and 400 nm, and the fluorophores they likely correspond to. The normalized measured spectra of (b) representative malignant sample and (c) representative normal sample at an excitation of 320 nm, their corresponding PLS fit using a linear combination of five PCs PC1–PC5, and the residual of the PLS fit. The set of PCs (PC1–PC5) accounted for a total of >95% of the variance in the normalized spectra.
In Fig. 3(a), the 300 nm PC1 displays a peak at 340 nm, which is similar to the fluorescence spectrum of tryptophan. The 320 nm PC4 and 340 nm PC1 displayed similar spectral profiles, with a peak appearing at 390 nm, which likely corresponds to the emission maximum of collagen, and a valley at 515 nm, which most likely corresponds to the emission maximum of retinol. The 360 nm PC1 and 380 nm PC1 show similar spectral profiles, with shoulders appearing at around 450 nm, which are likely related to NADH fluorescence. The 400 nm PC1 decreases monotonously with increasing wavelength and likely captures the tail end of the NADH fluorescence. In Fig. 3(b) and (c), the most notable difference between the malignant and normal adipose tissue spectra is that malignant tissue has relatively higher fluorescence at 460 nm and lower fluorescence at 520 nm, as compared to the normal adipose tissue. Also the measured spectra and the PLS fits are very similar, and the residual are within 5% of the normalized measured spectra across the spectrum.
Table II shows the correlation of the primary PC at each of the excitation wavelengths within 300–400 nm, with %adipose, %fibrous, and %ducts/lobules in the nonmalignant tissues. The variance accounted by each PC is also listed at the end of the table. The 300 nm PC1 showed a positive correlation with %adipose tissue content but a negative correlation with %fibrous content. The 320 nm PC4, 340 nm PC1, and 360 nm PC1 are negatively correlated with %adipose content and positively correlated with the %fibrous and %ducts/lobules tissue content. The 380 nm PC1 is negatively correlated with %adipose content but positively correlated with the %fibrous content.
TABLE II.
Correlation of PCs (ones that Account for Largest Amount of Variance in Spectra) with %Adipose, %Fibrous, and %Ducts/Lobules Tissue Content
| Excitation λ | %Adipose | %Fibrous | %Duct/lobules | Variance accounted |
|---|---|---|---|---|
| 300 nm PC1 |
0.26 (p < 0.05) |
−0.32 (p < 0.005) |
Ø | 50.0% |
| 320 nm PC4 |
−0.38 (p < 0. 0005) |
0.30 (p < 0.005) |
0.40 (p<le-4) |
38.3% |
| 340 nm PC1 |
−0.53 (p<le-6) |
0.47 (p<le-5) |
0.36 (p < 5e-4) |
59.0% |
| 360 nm PC1 |
−0.51 (p<le-6) |
0.47 (P< le-5) |
0.28 (p<0.01) |
66.4% |
| 380 nm PC1 |
−0.32 (p < 0.005) |
0.31 (p < 0.005) |
Ø | 57.8% |
| 400 nm PC1 |
Ø | Ø | Ø | 95.5% |
Table III shows the PCs identified from Wilcoxon rank-sum test that displayed statistically significant differences (p < 0.01) between the various tissue types: 1) malignant versus fibrous/benign; 2) malignant versus adipose; and 3) malignant versus nonmalignant. The PCs marked with * have a p-value of p < 0.001, and the PCs marked with ** have p < 0.0001. Eight PCs obtained from fluorescence spectra at excitation wavelengths of 300–380 nm, displayed statistically significant differences between malignant and fibrous/benign breast tissues, while far more PCs (at all excitation wavelengths) displayed statistically significant differences between malignant and adipose breast tissues. The PCs that showed statistical differences between malignant and nonmalignant (including fibrous, benign and adipose, and mix of fibrous and adipose) tissues is a subset of the combined PCs that displayed differences between malignant and adipose tissues or between malignant and fibrous/benign tissues.
TABLE III.
PCs Obtained from PLS Analysis of Fluorescence Spectra at Each Excitation Wavelength, Which Show Differences (p < 0.01) between (a) Malignant and Fibrous/Benign, (b) Malignant and Adipose, and (c) Malignant and Nonmalignant Tissue Samples
| Excitation λ | (a) Mal. (n=27) vs. Fibr./Benign (n=47) | (b) Mal. (n=27) vs. Adip. (n=37) | (c) Mal. (n=27) vs. Non-mal. (n=94) |
|---|---|---|---|
| 300 nm | PC1 | PC3 | |
| 320 nm | PC1, PC5 | PC1*, PC4* | PC1*, PC4 |
| 340 nm | PC2, PC4** | PC1**, PC4 | PC1*, PC2, PC4* |
| 360 nm | PC4* | PC1**, PC2, PC4 | PC1*, PC4 |
| 380 nm | PC1, PC3* | PC1*, PC2, PC3* | PC1*, PC3*, PC4 |
| 400 nm | PC1** | PC1* |
Note: PCs marked with
p< 0.001, PCs marked with
: p < 0.0001.
Table IV lists the PCs that are least correlated and used for the classification between: 1) malignant and fibrous/benign; 2) malignant and adipose; and 3) malignant and nonmalignant breast tissues. The PCs marked with * have a p-value of <0.001, and the PCs marked with ** have p < 0.0001. Two sets of PC scores were considered least correlated if: 1) p > 0.01 for the Pearson correlation or 2) their corresponding PCs have distinct (i.e., nonoverlapping) spectral line shapes that should characterize different fluorophores even though the scores may have a correlation of p < 0.01. This yielded four to six inputs to SVM classifiers to discriminate malignant from other nonmalignant breast tissues.
TABLE IV.
PCs that Are Least Correlated and Used for Tissue Classification. Two Sets of PC Scores were Considered Least Correlated if 1) p > 0.01 for Pearson Correlation, or 2) Their Corresponding PCs have Disctinct Spectral Line Shapes even though Scores May Have a Correlation of p < 0.01
| Excitation λ | (a) Mal. (n=27) vs. Fibr./Benign (n=47) | (b) Mal. (n=27) vs. Adip. (n=37) | (c) Mal. (n=27) vs. Non-mal. (n=94) |
|---|---|---|---|
| 300 nm | PC1 (50.0%) | ||
| 320 nm | PC1 (8.5%) | PC1* (8.5%) | PC1* (8.5%) PC4 (38.3%) |
| 340 nm | PC4** (2.0%) | PC1** (59.0%) PC4 (2.0%) |
PC4* (2.0%) |
| 360 nm | PC2 (19.8%) | ||
| 380 nm | PC1 (57.8%) | PC1* (57.8%) PC4 (2.4%) |
|
| 400 nm | PC1** (95.5%) | PCI* (95.5%) |
Note: PCs marked with
p < 0.001, PCs marked with
p < 0.0001
Table V shows the results from leave-one-out cross validation of: 1) a linear SVM and 2) a RBF kernel SVM, for discriminating malignant from nonmalignant tissue types, using the least correlated and diagnostically significant PCs shown in Table IV. Using a linear SVM classifier, the malignant breast samples can be discriminated from fibrous/benign samples with a cross-validated sensitivity and specificity of 70.4% and 86.8%, respectively, and discriminated from adipose tissue samples with a cross-validated sensitivity and specificity of 81.5% and 81.0%, respectively. In the nonlinear RBF-SVM classification, the cross-validated sensitivity and specificity for discriminating between malignant and fibrous/benign tissue samples are 81.5% and 86.8%, respectively, while that for discriminating between malignant and adipose tissue samples are 74.1% and 81.0%, respectively. In these SVM classifications, the negative predictive value (NPV) was over 80%, i.e., over 80% of samples with negative test results were correctly diagnosed. The positive predictive value (PPV) was over 70% when differentiating malignant from fibrous/benign or adipose tissues, and 58%–67% for discriminating between malignant and nonmalignant tissues.
TABLE V.
Leave-one-out Cross Validation of (a) Linear SVM and (b) RBF Kernel SVM, for Discriminating Malignant from other Nonmalignant Tissue Types, Using Least Correlated and Diagnostically Significant PCs Shown in Table IV
| (a) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Training |
Testing |
|||||||
| C rate | SE | SP | C rate | SE | SP | PPV | NPV | |
| Mal. (n=27) vs. Non-mal. (n=94) | 83.1±0.8 | 28.1±4.7 | 98.9±0.7 | 79.3 | 14.8 | 97.9 | 67 | 80 |
| Mal. (n=27) vs. Fibr./Benign (n=47) | 81.5±0.8 | 74.1±1.6 | 86.8±0.6 | 80.0 | 70.4 | 86.8 | 75 | 84 |
| Mal. (n=27) vs. Adip. (n=37) | 82.4±0.9 | 84.5±1.7 | 81.0±0.9 | 81.2 | 81.5 | 81.0 | 76 | 86 |
| (b) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Training |
Testing |
|||||||
| C rate | SE | SP | C rate | SE | SP | PPV | NPV | |
| Mal. (n=27) vs. Non-mal. (n=94) | 98.4±0.2 | 96.5±0.9 | 98.9±0.1 | 81.0 | 55.6 | 88.3 | 58 | 87 |
| Mal. (n=27) vs. Fibr./Benign. (n=47) | 89.5±1.0 | 89.1±1.2 | 89.8±1.2 | 84.6 | 81.5 | 86.8 | 78 | 89 |
| Mal. (n=27) vs. Adip. (n=37) | 89.8±0.6 | 88.8±0.9 | 90.4±0.7 | 78.3 | 74.1 | 81.0 | 74 | 81 |
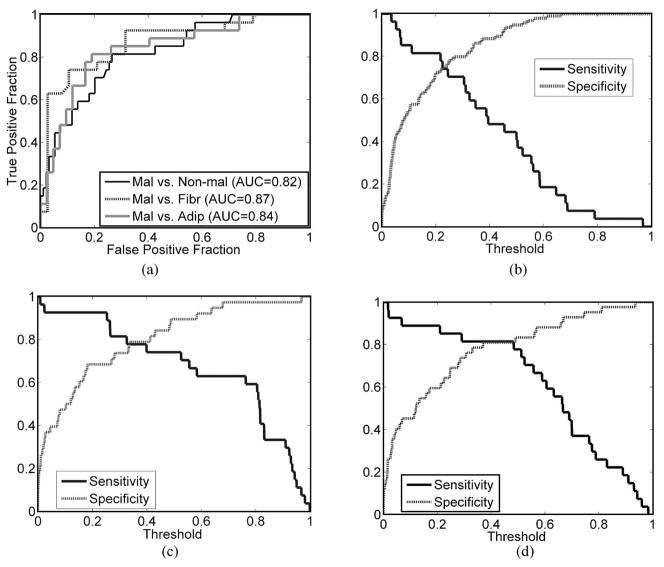
Fig. 4(a) shows the ROC graphs for the classification between malignant versus nonmalignant, malignant versus fibrous/benign, and malignant versus adipose samples. Fig. 4(b)–(d) shows the sensitivity and specificity produced in each classification: (b) malignant versus nonmalignant; (c) malignant versus fibrous/benign; and (d) malignant versus adipose. Each LR model was selected as the one in which all the predictor variables were significant for an adequate prediction of the tissue categories of interest (p < 0.05), and which yielded the maximum AUC. In the LR model for malignant versus nonmalignant samples, the optimal PCs included were 320 nm PC1, 320 nm PC4, and 340 nm PC4, and the area under the ROC curve was 0.82. PCs included in the LR model for malignant versus fibrous/benign samples were 300 nm PC1, 320 nm PC1, 340 nm PC4, and 380 nm PC1, and the area under the ROC curve was 0.87. The selected LR model for malignant versus adipose samples included 340 nm PC1, 340 nm PC4, and 360 nm PC2, and the AUC was 0.84. These AUCs were obtained from the training set. No cross-validation was performed using the LR model.
Fig. 4.
(a) ROC graphs for the classification between malignant versus. nonmalignant, malignant versus fibrous/benign, and malignant versus adipose samples. Figure (b)–(d) shows the sensitivity and specificity produced in each classification: (b) malignant versus nonmalignant, (c) malignant versus fibrous/benign, and (d) malignant versus adipose.
In Fig. 4(b), at a sensitivity of 55.6%, which is the cross-validated sensitivity for discriminating malignant from nonmalignant breast tissue using RBF-SVM, the corresponding specificity of the LR model classification is 86%–88%, and the corresponding PPV and NPV are 54%–58% and 87%, respectively. In Fig. 4(c), at a sensitivity of 81%, which is the cross-validated sensitivity for discriminating malignant from fibrous/benign samples using RBF-SVM, the corresponding specificity from the LR model classification ranges between 71% and 74%, and the corresponding PPV and NPV are 68% and 85%, respectively. In Fig. 4(d), at a sensitivity of 74%, which is the cross-validated sensitivity for discriminating malignant from adipose samples using RBF-SVM, the corresponding specificity from the LR model classification is 82%, and the corresponding PPV and NPV are 75% and 85%, respectively. Most of these figures are comparable with those obtained in RBF-SVM classification, except for the specificity for discriminating between malignant and fibrous/benign samples, where RBF-SVM yielded higher specificity than the LR model.
IV. Discussion
In this study, we explored the use of a side-firing fiber-optic probe to perform fluorescence spectroscopy in vivo on patients undergoing percutaneous image-guided breast biopsy. The goal of this study is to investigate the feasibility of in vivo fluorescence spectroscopy as an adjunct diagnostic tool to assist the sampling of malignant breast lesions. Fluorescence spectra of breast tissues were measured in vivo from patients through the bore of a nine-gauge vacuum-assisted core biopsy needle. The spectral features observed in the in vivo fluorescence EEMs (as shown in Fig. 2) were similar to that observed in the fluorescence EEMs measured ex vivo from freshly excised breast tissues [22], [23].
Four fluorescing components or fluorophores were characterized in the in vivo fluorescence EEM. The fluorescence peak at excitation–emission pair of (300, 340) nm is likely attributed to tryptophan, the one at (340, 385) nm is likely attributed to collagen, and the one at (340/360, 460) nm coincide with that of NADH fluorescence. The shoulder shown in the adipose tissue EEM at the excitation wavelengths of 340–360 nm, and emission wavelength of 460–510 nm may be attributed to retinol whose precursor β-carotene is primarily stored in fatty tissues. The spectral features of malignant tissues and normal fibrous/benign tissues were similar, while that of normal adipose tissues were distinct. The average fluorescence EEM of malignant and fibrous/benign tissue displayed higher collagen and NADH fluorescence, as compared to that of normal adipose tissues, while the retinol fluorescence was found to be significant in the adipose tissue EEM.
In the spectral analysis, the in vivo tissue spectra were dimensionally reduced using PLS analysis. A set of PCs that not only describe as much of the data variance as possible, but also are most relevant to the known histological diagnosis (malignant or nonmalignant) of each sample were extracted to represent the tissue spectra with dramatically reduced dimension. The primary PCs, which account for the largest portion of data variances, characterized the differences in the spectra observed between malignant and nonmalignant samples. The 300 nm PC1 is primarily characteristic of tryptophan fluorescence. The 320 nm PC4 and 340 nm PC1 both captured the features of collagen and retinol fluorescence at these excitation wavelengths in malignant and nonmalignant tissues. The 360 nm PC1 and 380 nm PC1 characterized primarily the difference in NADH and retinol fluorescence in malignant and nonmalignant breast tissues.
In nonmalignant tissue samples, the scores of most PCs above were correlated with the histological composition of the tissue sample. The scores of 320 nm PC4 and 340 nm PC1 showed negative correlation with %adipose tissue content and a positive correlation with %fibrous tissue content. Both PCs have positive values over the emission wavelength range of collagen and negative values over the emission wavelength range of retinol, suggesting that these PCs reflect increasing collagen and decreasing retinol fluorescence. This agrees with the fact that fibrous stroma is rich in collagen while adipose tissue is rich in fat [30]. The score of 300 nm PC1, which likely characterizes tryptophan fluorescence, was positively correlated with %adipose tissue content, while negatively correlated with %fibrous content. Alimova et al. [31] performed fluorescence and phosphorescence measurements on malignant and normal adipose and glandular breast tissues, and showed that the ratio of intensity at 345 nm (tryptophan fluorescence) to the intensity at 500 nm (tryptophan phosphorescence) is five times greater in adipose tissues compared to that in glandular tissues, indicating greater tryptophan fluorescence from fat compared to glandular tissues. Our results are consistent with these findings. The 360 nm PC1 and 380 nm PC1 are both characteristic of NADH fluorescence. Their negative correlation with %adipose and positive correlation with %fibrous suggested that adipose has lower NADH fluorescence while fibrous tissue has higher NADH fluorescence. The 360 nm PC1 also positively correlated with %duct/lobules tissue content, but the 380 nm PC1 did not show statistically significant correlation with %duct/lobules. In normal breast, NADH may increase with the increased metabolism associated with the duct proliferation and secretory activities of the breast lobules, while adipose cells have an indolent metabolism [30]; thus, lower NADH fluorescence would be expected from adipose tissues.
We have applied both linear and nonlinear SVM classifiers to discriminate malignant from other nonmalignant tissues. The linear SVM performed better than the nonlinear RBF-SVM for discriminating between malignant and adipose tissues, while the nonlinear RBF-SVM performed better for the discrimination between malignant and fibrous/benign tissues. This might be due to the fact that the PC scores of malignant and adipose tissues are more linearly separable than that of malignant and fibrous/benign tissues (as evidenced in Table III), and a linear SVM works better for linearly separable cases while a nonlinear SVM works better for linearly less separable cases in the original data space. This may also explain the better classification accuracy achieved using a nonlinear SVM for discrimination between malignant and nonmalignant samples, as compared to that achieved using a linear SVM. For a poorly or nonlinearly separable case in the original data space, the nonlinear SVM uses a kernel function to map the data from the original data space to a higher dimensional feature space, where the mapped feature data are linearly more separable and a better linear decision hyperplane may be obtained [27].
The SVM classification between malignant and nonmalignant tissue samples has lower sensitivity compared to specificity. This may be attributed to the unbalanced sample size of each tissue category that was investigated in this study. The sample set investigated in this study consisted of 27 malignant and 94 nonmalignant breast tissue samples. The number of nonmalignant samples is more than three times the number of malignant samples. This unbalanced sample size is inherently related to the fact that approximately 80% of the breast biopsies turned out to be nonmalignant on histopathology. An SVM classifier built on such an unbalanced sample set may be skewed toward the class with larger sample size, in our case the class of non-malignant tissues. The principal idea of an SVM is to determine an optimal hyperplane that maximizes the margin between two classes, such that the distance of the misclassified samples in each class from the separation boundary are equalized. When the numbers of elements in the two classes are unbalanced, especially for the case when the two classes are not completely separable, a large number of samples from each class become support vectors (i.e., marginal samples), and it is likely that most of samples from the class with smaller sample size (in this case, the malignant tissue class) will be misclassified. For the discrimination between malignant and fibrous/benign, and that between malignant and adipose tissue samples, the sample size of each tissue type is more balanced (i.e., 27 malignant versus 47 fibrous/benign, and 27 malignant versus 37 adipose), and the sensitivity and specificity do not seem to bias toward any tissue type.
An SVM classifier provides a “hard” classification rule (i.e., fixed separation boundary) for assigning an individual to one of two classes. Using this classification rule, a tissue sample is assigned as either class 1 (malignant) or class 0 (other nonmalignant tissue type) based on the tissue measurements. However, in the clinical settings, it may be more practical to have a probabilistic diagnosis, i.e., what is the likelihood or probability that the tissue being probed is malignant. Thus, a probability predictor may be more amenable in clinical applications such as providing guidance to the optimal tissue sites for breast biopsy. Also this will enable a “soft” classification rule (varying thresholds) for assigning the membership of an individual, and thus allows one to choose the threshold to achieve certain sensitivity or specificity that is desired for the problem. In this study, we have implemented a probability-based predictive model based on LR. At the same sensitivity for discriminating malignant from adipose, fibrous/benign, or all nonmalignant tissue samples, the specificity, and therefore, the PPV and NPV were all comparable to that obtained from RBF-SVM classification, which suggests that the SVM classifier may achieve comparable diagnostic accuracy as that yielded from LR.
The study demonstrated the feasibility of performing in vivo fluorescence spectroscopy of breast tissues during a clinical core biopsy procedure; however, there are many challenges that call for further investigation and improvement. First, there are experimental obstacles, which significantly affect the quality of fluorescence measurements and the histological diagnosis of the biopsy cores. Bleeding during the biopsy procedure is a primary concern, which is very common in the study subjects due to the insertion of the biopsy needle and the removal of breast tissues. The use of a local anesthetic lidocaine-epinephrine in the lesion was effective in reducing bleeding; however, patient bleeding still persists. The blood absorbs light and fills the gap between the probe and tissue surface, which may cause significant attenuation of both excitation and emission light. Although a vacuum was applied to suck out the blood in the cavity and in the needle during biopsy, residual blood and persisting bleeding is still a concern. In addition, the cavity resulting from biopsy may also affect the probe–tissue contact, which can potentially impact the quality of fluorescence measurements.
Another experimental obstacle is that sometimes the biopsy cores break into pieces during the biopsy or suction. A total of 58 samples broke into pieces in this study, among which 15 have %adipose of less than 40% (average 18.7%), 12 have %adipose between 40% and 60% (average 46.3%), and 31 have %adipose of 60% or above (average 70.5%). A large portion of broken samples have %adipose content greater than 60%, suggesting adipose tissues are more likely to break into pieces. The fragmentation of tissue samples makes it difficult to determine with certainty the region of interest (i.e., the center of the core) for the histological diagnosis. This consequently imposes difficulties in correlating the tissue measurements with the corresponding histological diagnosis. When a biopsy core was broken into pieces, the histopathological analysis of the entire core was used to describe the histological composition of the optical tissue sample, and the tissue histology was designated as being the predominant tissue type presented in the sample. The histological diagnosis of the entire sample may thus not accurately represent the histology of the tissue site on which the optical measurement was actually made. However, among the total of 23 samples misclassified in cross-validation using RBF-SVM, nine were fragmented samples (accounting for 15.3% of the total fragmented samples) and 14 were intact samples (accounting for 22.2% of the total intact samples). This suggests that fragmentation did not significantly contribute to the misclassification and the fragmented samples did not seem to be more likely misclassified than nonfragmented samples.
In addition to the experimental obstacles, there are also hardware limitations for the fluorescence measurements. The side-firing fiber-optic probe employs obliquely polished fiber tips to reflect the light and a quartz cap to encase all the fibers. There may be cross-talk between the illumination and collection fibers due to the internal reflection from the inner and outer surface of the quartz cap. This cross-talk can bring interfering excitation light to the measured fluorescence and such interference cannot be completely removed through background calibration. One way to reduce the internal reflection could be adding a thin-film antireflection (AR) coating on the inner surface of the quartz cap to reduce the unwanted reflection. A selected multilayer AR coating may eliminate the reflection at multiple excitation wavelengths. Offsetting the illumination and collection beams may also help reduce the cross-talk between the two fibers. In the current probe design, the illumination and collection fibers were aligned in one plane, and part of the collection fiber may block the light beam from the illumination fiber; thus, a small fraction of excitation light may be collected by the collection fiber. Pulling the collection fiber backward and off the illumination plane may help remove the collection fiber out of the illumination light beam, thus reducing the interference from excitation light.
In conclusion, we have demonstrated the feasibility of in vivo fluorescence spectroscopy of breast tissues during a clinical core needle breast biopsy and the potential of this technique for diagnosing breast malignancy. The outcome of this preliminary study sets the precedent for the next-generation probe design and a larger scale clinical study. It is expected that with the accrual of clinical data, we will be able to further test the value and capability of the fluorescence spectroscopy technique as an adjunct real-time diagnostic tool to assist the biopsy of malignant breast lesion.
Acknowledgments
This work was supported in part by the National Institutes of Health under Grant R01CA100559-01 and by the Department of Defense under Pre-Doctoral Traineeship W81XWH-05-1-0380.
Biographies

Changfang Zhu (S′05–M′08) received the B.S. degree in biomedical engineering from Zhejiang University, Hangzhou, China, in 1998, the M.S. degree in biomedical engineering from Tsinghua University, Beijing, China, in 2001, and the M.S. and Ph.D. degrees in electrical engineering from the University of Wisconsin, Madison, in 2005 and 2007, respectively.
She is currently with Boston Scientific, Valencia, CA.
Dr. Zhu was the recipient of a 2004 International Society for Optical Engineering (SPIE) Educational Scholarship, a 2005 American Society for Laser Medicine and Surgery (ASLMS) Student Research Grant, and a 2005–2008 Department of Defense Breast Cancer Research Program Pre-Doctoral Fellowship.
Elizabeth S. Burnside received the B.S. degree from Dartmouth College, Hanover, NH, the M.D. degree combined with M.S. in public health from Tufts University School of Medicine, Boston, MA, and the M.S. degree in medical informatics from Stanford University, Stanford, CA.
He completed a residency at the University of California, San Francisco. He is currently with the Department of Radiology, University of Wisconsin, Madison.
Dr. Burnside was elected a Fellow of the Society of Breast Imaging and is a member of the American College of Radiology Commission on Breast Imaging Education.
Gale A. Sisney received the M.D. degree from the University of Illinois, Chicago.
She completed the Radiology residency at Northwestern University. She was the Director of Breast Imaging at the University of Colorado, Boulder, and Georgetown University Hospital. She is currently the Director of Breast Imaging at the University of Wisconsin Hospital and Clinics, Madison.
Lonie R. Salkowski received the B.S. degree in biology and chemistry from Mount Mary College, Milwaukee, Wisconsin and the M.D. degree from the Medical College of Wisconsin, Milwaukee.
He completed the Transitional and Nuclear Medicine residency at St. Luke’s Medical Center, Milwaukee, WI, and the Radiology residency and Breast Imaging Fellowship at the Medical College of Wisconsin. He is currently with the Department of Radiology, University of Wisconsin, Madison.
Josephine M. Harter received the B.S. degree in cell and molecular biology and the M.D. degree from the University of Michigan, Ann Arbor, in 1989 and 1994, respectively.
She completed the residency in Family and Community Medicine at Duke University, Durham, NC. She also completed the residency in Pathology at the University of Wisconsin, Madison, where she is currently with the Department of Pathology.
Bing Yu received the B.S. and M.S. degrees in optoelectronics technology from the University of Electronic Science and Technology of China, Chengdu, China, in 1989 and 1994, respectively, and the Ph.D. degree in electrical engineering from Virginia Polytechnic Institute and State University, Blacksburg, in 2005.
He is currently a Senior Research Scientist in biomedical engineering at Duke University, Durham, NC.
Dr. Yu is a member of the Optical Society of America (OSA) and the International Society for Optical Engineering (SPIE).
Nirmala Ramanujam received the B.S. and M.S. degrees in mechanical engineering, and the Ph.D. degree in biomedical engineering from the University of Texas, Austin, in 1989, 1992, and 1995, respectively.
She is currently an Associate Professor of biomedical engineering at Duke University, Durham, NC. She has more than 15 years of experience in optical spectroscopy, light–tissue interaction, and technology development. Her current research interests include developing light-based technologies for breast cancer. She has authored or coauthored more than 50 peer-reviewed publications and holds ten patents.
Dr. Ramanujam was named by Massachusetts Institute of Technology (MIT)’s prestigious Technology Review as one of the top 100 young innovators in technology in 2003, named a Department of Defense (DOD) Era of Hope Scholar for Breast Cancer research in 2004, and won the Global Indus Technovators award from MIT in 2005.
Contributor Information
Changfang Zhu, Email: czhu@uwalumni.com, Department of Electrical and Computer Engineering, University of Wisconsin, Madison, WI 53706 USA.
Elizabeth S. Burnside, Department of Radiology, University of Wisconsin, Madison, WI 53706 USA
Gale A. Sisney, Department of Radiology, University of Wisconsin, Madison, WI 53706 USA
Lonie R. Salkowski, Department of Radiology, University of Wisconsin, Madison, WI 53706 USA
Josephine M. Harter, Department of Pathology, University of Wisconsin, Madison, WI 53706 USA
Bing Yu, Department of Biomedical Engineering, Duke University, Durham, NC 27708 USA.
Nirmala Ramanujam, Email: nimmi@duke.edu, Department of Biomedical Engineering, Duke University, Durham, NC 27708 USA.
References
- 1.Parker SH, Lovin JD, Jobe WE, Burke BJ, Hopper KD, Yakes WF. Nonpalpable breast lesions: Stereotactic automated large-core biopsies. Radiology. 1991;180:403–407. doi: 10.1148/radiology.180.2.1648757. [DOI] [PubMed] [Google Scholar]
- 2.Parker SH, Burbank F, Jackman RJ, Aucreman CJ, Cardenosa G, Cink TM, Coscia JL, Jr, Eklund GW, Evans WP, III, Garver PR, Gramn HF, Haas DK, Jacob KM, Kelly KM, Killebrew LK, Lechner MC, Perlman SJ, Smid AP, Tabar L, Taber FE, Wynn RT. Percutaneous large-core breast biopsy: A multi-institutional study. Radiology. 1994;193:359–364. doi: 10.1148/radiology.193.2.7972743. [DOI] [PubMed] [Google Scholar]
- 3.Pfarl G, Helbich TH, Riedl CC, Wagner T, Gnant M, Rudas M, Liberman L. Stereotactic 11-gauge vacuum-assisted breast biopsy: A validation study. AJR Amer J Roentgenol. 2002;179:1503–1507. doi: 10.2214/ajr.179.6.1791503. [DOI] [PubMed] [Google Scholar]
- 4.Helbich TH, Mayr W, Schick S, Youssefzadeh S, Rudas M, Taucher S, Wagner T, Kelkar P, Wolf G, Thurnher M, Mostbeck GH. Coaxial technique: Approach to breast core biopsies. Radiology. 1997;203:684–690. doi: 10.1148/radiology.203.3.9169689. [DOI] [PubMed] [Google Scholar]
- 5.Elvecrog EL, Lechner MC, Nelson MT. Nonpalpable breast lesions: Correlation of stereotaxic large-core needle biopsy and surgical biopsy results. Radiology. 1993;188:453–455. doi: 10.1148/radiology.188.2.8327696. [DOI] [PubMed] [Google Scholar]
- 6.Dershaw DD, Morris EA, Liberman L, Abramson AF. Nondiagnostic stereotaxic core breast biopsy: Results of rebiopsy. Radiology. 1996;198:323–325. doi: 10.1148/radiology.198.2.8596825. [DOI] [PubMed] [Google Scholar]
- 7.Jackman RJ, Nowels KW, Rodriguez-Soto J, Marzoni FA, Jr, Finkelstein SI, Shepard MJ. Stereotactic, automated, large-core needle biopsy of nonpalpable breast lesions: False-negative and histologic underestimation rates after long-term follow-up. Radiology. 1999;210:799–805. doi: 10.1148/radiology.210.3.r99mr19799. [DOI] [PubMed] [Google Scholar]
- 8.Kettritz U, Rotter K, Schreer I, Murauer M, Schulz-Wendtland R, Peter D, Heywang-Kobrunner SH. Stereotactic vacuum-assisted breast biopsy in 2874 patients: A multicenter study. Cancer. 2004;100:245–251. doi: 10.1002/cncr.11887. [DOI] [PubMed] [Google Scholar]
- 9.Meyer JE, Smith DN, Lester SC, DiPiro PJ, Denison CM, Harvey SC, Christian RL, Richardson A, Ko WD. Large-needle core biopsy: Nonmalignant breast abnormalities evaluated with surgical excision or repeat core biopsy. Radiology. 1998;206:717–720. doi: 10.1148/radiology.206.3.9494490. [DOI] [PubMed] [Google Scholar]
- 10.Jackman RJ, Burbank F, Parker SH, Evans WP, 3rd, Lechner MC, Richardson TR, Smid AA, Borofsky HB, Lee CH, Goldstein HM, Schilling KJ, Wray AB, Brem RF, Helbich TH, Lehrer DE, Adler SJ. Stereotactic breast biopsy of nonpalpable lesions: Determinants of ductal carcinoma in situ underestimation rates. Radiology. 2001;218:497–502. doi: 10.1148/radiology.218.2.r01fe35497. [DOI] [PubMed] [Google Scholar]
- 11.Parikh J, Tickman R. Image-guided tissue sampling: Where radiology meets pathology. Breast J. 2005;11:403–409. doi: 10.1111/j.1075-122X.2005.00130.x. [DOI] [PubMed] [Google Scholar]
- 12.American Cancer Society. Cancer Reference Information-Breast cancer. 2007 [Google Scholar]
- 13.Liberman L, Benton CL, Dershaw DD, Abramson AF, LaTrenta LR, Morris EA. Learning curve for stereotactic breast biopsy: How many cases are enough? AJR Amer J Roentgenol. 2001;176:721–727. doi: 10.2214/ajr.176.3.1760721. [DOI] [PubMed] [Google Scholar]
- 14.Richards-Kortum R, Sevick-Muraca E. Quantitative optical spectroscopy for tissue diagnosis. Annu Rev Phys Chem. 1996;47:555–606. doi: 10.1146/annurev.physchem.47.1.555. [DOI] [PubMed] [Google Scholar]
- 15.Bigio IJ, Bown SG, Briggs G, Kelley C, Lakhani S, Pickard D, Ripley PM, Rose IG, Saunders C. Diagnosis of breast cancer using elastic-scattering spectroscopy: Preliminary clinical results. J Biomed Opt. 2000;5:221–228. doi: 10.1117/1.429990. [DOI] [PubMed] [Google Scholar]
- 16.van Veen RL, Amelink A, Menke-Pluymers M, Van Der Pol C, Sterenborg HJ. Optical biopsy of breast tissue using differential path-length spectroscopy. Phys Med Biol. 2005;50:2573–2581. doi: 10.1088/0031-9155/50/11/009. [DOI] [PubMed] [Google Scholar]
- 17.Yu B, Burnside ES, Sisney GA, Harter JM, Zhu C, Dhalla A, Ramanujam N. Feasibility of near-infrared diffuse optical spectroscopy on patients undergoing image-guided core-needle biopsy. Opt Exp. 2007;15:7335–7350. doi: 10.1364/oe.15.007335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bigio IJ, Mourant JR. Ultraviolet and visible spectroscopies for tissue diagnostics: Fluorescence spectroscopy and elastic-scattering spectroscopy. Phys Med Biol. 1997;42:803–814. doi: 10.1088/0031-9155/42/5/005. [DOI] [PubMed] [Google Scholar]
- 19.Gupta PK, Majumder SK, Uppal A. Breast cancer diagnosis using N2 laser excited autofluorescence spectroscopy. Lasers Surg Med. 1997;21:417–422. doi: 10.1002/(sici)1096-9101(1997)21:5<417::aid-lsm2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 20.Majumder SK, Gupta PK, Jain B, Uppal A. UV excited autofluorescence spectroscopy of human breast tissues for discriminating cancerous tissue from benign tumor and normal tissue. Lasers Life Sci. 1999;8:249–264. [Google Scholar]
- 21.Palmer GM, Ramanujam N. Diagnosis of breast cancer using optical spectroscopy. Med Laser Appl. 2003;18:233–248. [Google Scholar]
- 22.Palmer GM, Zhu C, Breslin TM, Xu F, Gilchrist KW, Ramanujam N. Comparison of multiexcitation fluorescence and diffuse reflectance spectroscopy for the diagnosis of breast cancer (March 2003) IEEE Trans Biomed Eng. 2003 Nov;50(11):1233–1242. doi: 10.1109/TBME.2003.818488. [DOI] [PubMed] [Google Scholar]
- 23.Zhu C, Palmer GM, Breslin TM, Xu F, Ramanujam N. The use of a multi-separation fiber optic probe for the optical diagnosis of breast cancer. J Biomed Opt. 2005;10:024032-1–024032-13. doi: 10.1117/1.1897398. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q, Zhu C, Ramanujam N. Experimental validation of Monte Carlo modeling of fluorescence in tissues in the UV-visible spectrum. J Biomed Opt. 2003;8:223–236. doi: 10.1117/1.1559057. [DOI] [PubMed] [Google Scholar]
- 25.Welch AJ, van Gemert MJC. Optical-Thermal Response of Laser-Irradiated Tissue. New York: Plenum; 1995. [Google Scholar]
- 26.Zhu C, Palmer GM, Breslin TM, Harter J, Ramanujam N. Diagnosis of breast cancer using diffuse reflectance spectroscopy: Comparison of a Monte Carlo versus partial least squares analysis based feature extraction technique. Lasers Surg Med. 2006;38:714–724. doi: 10.1002/lsm.20356. [DOI] [PubMed] [Google Scholar]
- 27.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines: And Other Kernel-Based Learning Methods. Cambridge, New York: Cambridge Univ. Press; 2000. [Google Scholar]
- 28.Hand D, Mannila H, Smyth P. Principles of Data Mining. Cambridge, MA: MIT Press; 2001. [Google Scholar]
- 29.Collett D. Modelling Binary Data. 2. London, U.K./Boca Raton, FL: Chapman & Hall/CRC; 2003. [Google Scholar]
- 30.Thomsen S, Tatman D. Physiological and pathological factors of human breast disease that can influence optical diagnosis. Ann N Y Acad Sci. 1998;838:171–193. doi: 10.1111/j.1749-6632.1998.tb08197.x. [DOI] [PubMed] [Google Scholar]
- 31.Alimova A, Katz A, Sriramoju V, Budansky Y, Bykov AA, Zeylikovich R, Alfano RR. Hybrid phosphorescence and fluorescence native spectroscopy for breast cancer detection. J Biomed Opt. 2007;12:014004–014009. doi: 10.1117/1.2437139. [DOI] [PubMed] [Google Scholar]