Abstract
Lysyl hydroxylase (EC 1.14.11.4), a homodimer, catalyzes the formation of hydroxylysine in collagens. Recently, an isoenzyme termed lysyl hydroxylase 2 has been cloned from human sources [M. Valtavaara, H. Papponen, A.-M. Pirttilä, K. Hiltunen, H. Helander and R. Myllylä (1997) J. Biol. Chem. 272, 6831–6834]. We report here on the cloning of a third human lysyl hydroxylase isoenzyme, termed lysyl hydroxylase 3. The cDNA clones encode a 738 amino acid polypeptide, including a signal peptide of 24 residues. The overall amino acid sequence identity between the processed human lysyl hydroxylase 3 and 1 polypeptides is 59%, and that between the processed lysyl hydroxylase 3 and 2 polypeptides is 57%, whereas the identity to the processed Caenorhabditis elegans polypeptide is only 45%. All four recently identified critical residues at the catalytic site, two histidines, one aspartate, and one arginine, are conserved in all these polypeptides. The mRNA for lysyl hydroxylase 3 was found to be expressed in a variety of tissues, but distinct differences appear to exist in the expression patterns of the three isoenzyme mRNAs. Recombinant lysyl hydroxylase 3 expressed in insect cells by means of a baculovirus vector was found to be more soluble than lysyl hydroxylase 1 expressed in the same cell type. No differences in catalytic properties were found between the recombinant lysyl hydroxylase 3 and 1 isoenzymes. Deficiency in lysyl hydroxylase 1 activity is known to cause the type VI variant of the Ehlers–Danlos syndrome, and it is therefore possible that deficiency in lysyl hydroxylase 3 activity may lead to some other variant of this syndrome or to some other heritable connective tissue disorder.
Lysyl hydroxylase (EC 1.14.11.4), a homodimer with a subunit molecular weight of about 82,000, catalyzes the formation of hydroxylysine in collagens and more than 10 additional proteins with collagen-like sequences by the hydroxylation of lysine residues in -X-Lys-Gly- sequences. This cotranslational and posttranslational modification plays an important role in the synthesis of collagens, since the hydroxylysine residues serve as attachment sites for carbohydrate units and are essential for the stability of the intermolecular collagen crosslinks (for a recent review, see ref. 1). Lysyl hydroxylase has now been cloned from human (2, 3), rat (4), and chicken (5) sources, and the catalytic properties of a recombinant enzyme expressed in insect cells have been shown to be virtually identical to those of an enzyme isolated from vertebrate tissues (4, 6, 7).
Deficiency in lysyl hydroxylase activity leads to a connective tissue disorder known as the type VI variant of the Ehlers–Danlos syndrome (1, 8, 9). Several mutations in the lysyl hydroxylase gene have recently been characterized in families with this disease (10–15). One interesting aspect of Ehlers–Danlos syndrome type VI is that the deficiency in hydroxylysine shows a wide variation between tissues (1, 8). This has suggested the existence of lysyl hydroxylase isoenzymes. An isoenzyme termed lysyl hydroxylase 2, found to be highly expressed in pancreas and muscle tissues, has recently been cloned from human sources (16), and the previously known isoenzyme is now correspondingly known as lysyl hydroxylase 1. We report here on the cloning and characterization of an additional human isoenzyme, termed lysyl hydroxylase 3, which appears to be expressed in a variety of tissues. Lysyl hydroxylase 3 shows 59% amino acid sequence identity with isoenzyme 1 and 57% identity with isoenzyme 2.
MATERIALS AND METHODS
Isolation of cDNA Clones.
The database of human expressed sequence tags was searched with the lysyl hydroxylase 1 cDNA sequence (2), and a 343-bp sequence (AA 340 606) homologous to the 3′ end of the coding region of the lysyl hydroxylase 1 cDNA was found in a fetal human kidney expressed sequence tag database. PCR primers F1 (5′-ACT TTG TGG TTC GCT ACC G-3′) and R3 (5′-GCG TGT GCC CCA GGT CGT-3′) were designed based on this sequence and used to obtain a 236-bp product from an adult human kidney λgt-10 cDNA library (CLONTECH). The purified product was used to screen fetal (λgt-11) and adult (λgt-10) human kidney and human placenta (λgt-11) cDNA libraries (CLONTECH). Altogether, 5 positive clones were obtained from the 2 kidney libraries and 16 from the placenta library. Seven of these clones were characterized in detail. To obtain the 5′ end of the cDNA, two new probes, NP and SL, were generated by PCR from the placenta library by primers LH3seqN (5′-GCT GGT CGT CGT CAT CAT C-3′), LH3seqP (5′-GCA GAG AGC TTC TGC TGG-3′), LH3seqS (5′-CAC TGT GCG GAC CCT GGG CC-3′), and LH3seqL (5′-GAT GAT CAT ATC CTC CCG GT-3′) and used for rescreening this library. Eight positive clones were obtained, four of which were characterized in detail. In addition, 5′-RACE (rapid amplification of cDNA ends) analysis was performed using pooled cDNAs from human placenta (Marathon–Ready cDNA, CLONTECH) as a template and primers AP1 (5′-CCA TCC TAA TAC GAC TCA CTA TAG GGC-3′, a linker specific primer from the Marathon kit), and LH3seqL.
DNA Sequencing and Northern Blot Analysis.
DNA sequencing was performed by using an automated sequencer (Abi Prism 377, Applied Biosystems). dnasis and prosis version 6.00 software (Pharmacia) were used to analyze the sequence data.
Human multitissue Northern blots I and III and a fetal blot II (CLONTECH) containing 2 μg of poly(A)+ RNA per sample and samples of 15 μg of total RNA [prepared by using RNeasy Midi Kit, Qiagen (Chatsworth, CA)] from cultured adult human skin fibroblasts and human Saos-2 osteosarcoma cells were hybridized by using ExpressHyb solution (CLONTECH) under the stringent conditions specified by the manufacturer. A 352-bp PCR fragment from the 3′ end of the cDNA was used as a probe.
Generation of a Recombinant Baculovirus.
To construct a full-length cDNA, two pairs of sequence-specific primers, LH3–5′B (5′-GTC AGG ATC CGT GCT GTC TGG GCC CGC TCC-3′) and LH3–5′N (5′-GCT CAG GGC GCC CCA GAA GT-3′), and correspondingly LH3–3′X (5′-CCA TTC TAG AGG CAC AAT GGC AGG GCA GG-3′) and LH3–3′N (5′-ACT TCT GGG GCG CCC TGA GC-3′) were designed. LH3–5′B contained an artificial BamHI site, LH3–5′N and LH3–3′N a natural NarI site, and LH3–3′X an artificial XbaI site. The 5′ end of the cDNA (nucleotides 37–1360) was amplified with the first pair of primers and the 3′ end (nucleotides 1361–2317) with the second pair. The 5′ product was digested with BamHI and NarI and the 3′ product with NarI and XbaI. The BamHI–NarI and NarI–XbaI fragments were ligated to the BamHI–XbaI site of the baculovirus transfer vector pVL1393 (17) and the whole construct was sequenced.
Spodoptera frugiperda Sf9 cells (Invitrogen) were cultured as monolayers in TNM-FH medium (Sigma) supplemented with 10% fetal bovine serum (FBS) (GIBCO) at 27°C. The recombinant baculovirus transfer vector was cotransfected into Sf9 cells with a modified Autographa californica nuclear polyhedrosis virus DNA (BaculoGold, PharMingen) by calcium phosphate precipitation (18), and the resultant viral pool was collected 4 days later and amplified twice (17). The other baculovirus used coded for lysyl hydroxylase 1, as described previously (6).
Expression and Analysis of Recombinant Proteins.
High Five insect cells (Invitrogen) were cultured at 27°C as monolayers in TNM-FH medium supplemented with 10% FBS (Life Technologies, Gaithersburg, MD). The cells were infected at a multiplicity of 5, harvested 24–72 hr after infection, washed with a solution of 0.15 M NaCl and 0.02 M phosphate (pH 7.4), and homogenized in a solution of 1% Nonidet P-40 (NP-40), 0.1 M glycine, and 0.02 M Tris⋅HCl (pH 7.8) (termed NP-40 buffer), and centrifuged at 10,000 × g for 10 min. The insoluble pellets were homogenized further in a 50% glycerol, 0.6 M NaCl, 1% NP-40, 0.1 M glycine, 100 μM DTT, and 0.06 M Tris⋅HCl buffer (pH 7.8) (glycerol buffer), incubated on ice for 30–60 min, and centrifuged at 10,000 × g for 20 min (6). Aliquots of the supernatants were analyzed by SDS/8% PAGE under reducing conditions and assayed for lysyl hydroxylase activity. The remaining pellets were solubilized in 1% SDS and also analyzed by SDS/8% PAGE.
Enzyme Activity Assays.
Lysyl hydroxylase activity was assayed by a procedure based on the hydroxylation-coupled decarboxylation of 2-oxo[1-14C]glutarate with 0.75 mg/ml of (Ile-Lys-Gly)3 as the peptide substrate (19). Km values were measured as described (6, 19), the pooled soluble fractions of the cell homogenate being used as the source of enzyme. Protein concentrations were determined with a Bio-Rad assay kit.
RESULTS AND DISCUSSION
Isolation of cDNA Clones for Human Lysyl Hydroxylase 3.
To study the possible existence of additional isoenzymes for human lysyl hydroxylase, the database of human expressed sequence tags was searched with the lysyl hydroxylase 1 sequence (2). A 343-bp sequence was found in a fetal kidney database that showed 81% identity to the 3′ end of the coding region of the lysyl hydroxylase 1 and 77% identity to the 3′ end of the coding region of the lysyl hydroxylase 2 cDNA at the nucleotide level. Two PCR primers based on this sequence were used to obtain a 236-bp product from an adult human kidney cDNA library, and this product was used to screen fetal and adult human kidney and human placenta cDNA libraries. Seven out of the total of 21 positive clones were characterized in detail. The 5′ end of the cDNA was obtained by two independent methods: (i) rescreening of the placenta library with two new PCR products, which gave eight positive clones, and (ii) 5′-RACE analysis with pooled human placenta cDNAs.
The cDNA clones cover 61 nucleotides of the 5′ untranslated sequence, the whole coding region, and 295 nucleotides of the 3′ untranslated sequence of the corresponding lysyl hydroxylase 3 mRNA. The 3′ untranslated sequence contains the canonical polyadenylation signal AATAAA, which is accompanied 11 bp downstream by a poly(A)+ tail of 46 nucleotides (these cDNA sequences are not shown but have been deposited in the GenBank/EMBL data base).
Amino Acid Sequence of Human Lysyl Hydroxylase 3 and Its Comparison with Those of Human Lysyl Hydroxylase 1 and 2 and the Caenorhabditis elegans Enzyme.
The cDNA clones encode a 738-amino acid polypeptide. A putative signal peptide is present in its N terminus, the most likely first amino acid of the mature polypeptide, based on the computational parameters of von Hejne (20) being serine. Thus, the size of the signal peptide is likely to be 24 amino acids and that of the processed polypeptide 714 amino acids (Fig. 1); the calculated molecular weight of the processed polypeptide being 82,380 kDa, i.e., about 0.81 kDa less than that of lysyl hydroxylase 1 (2).
Figure 1.
Alignment of the amino acid residues of the processed human lysyl hydroxylase 3 (Lh3), 1 (Lh1), and 2 (Lh2) polypeptides, and the C. elegans lysyl hydroxylase (C.e.Lh) polypeptide. Gaps (small dots) are introduced for the maximal alignment. Positions of cysteine residues (large dots) and potential N-glycosylation sites (solid lines) in any of the polypeptides are indicated above the sequence. Residues required for the binding of the Fe2+ atom and the C-5 carboxyl group of 2-oxoglutarate are indicated by asterisks. White letters on a black background show identity and black letters on a grey background show similarity. Similar amino acids: G, A, S; A, V; V, I, L, M; I, L, M, F, Y, W; L, K, H; D, E, Q, N; S, T, E, N.
The processed lysyl hydroxylase 3 polypeptide is very similar in size to the corresponding human polypeptides 1 (2) and 2 (16) and the C. elegans polypeptide (21), which have 709, 712, and 714 amino acids, respectively (Fig. 1). The overall amino acid sequence identity between the human polypeptides 3 and 1 is 59%, and that between the human polypeptides 3 and 2 57%, whereas the identity between the human polypeptide 3 and the C. elegans polypeptide is only 45%. Identity is highest within the catalytically important C-terminal region (1, 6), the 87 extreme C-terminal residues being 72% identical between human polypeptides 3 and 1, 69% between polypeptides 3 and 2, and 64% between human polypeptide 3 and the C. elegans polypeptide (Fig. 1). All four recently identified critical residues at the catalytic site, two histidines and one aspartate that bind the Fe2+ atom (6) and an arginine that binds the C-5 carboxyl group of the 2-oxoglutarate (K.P., J. Myllyharju, Asta Pirskanen, and K.I.K., unpublished observations) are conserved in all these polypeptides; these residues being His-643, Asp-645, His-695, and Arg-705 in the lysyl hydroxylase 3 polypeptide (Fig. 1). The closely related enzymes, prolyl 4-hydroxylase isoforms 1 and 2, also contain the corresponding histidine and aspartate residues (22–24), but interestingly, the residue that binds the C-5 carboxyl group of 2-oxoglutarate is lysine (24) in both prolyl 4-hydroxylase isoforms (1, 25, 26) and not arginine.
All three human lysyl hydroxylase polypeptides contain nine cysteine residues in conserved positions. Polypeptides 1 and 2 contain an additional cysteine conserved between the two but not found in polypeptide 3, and polypeptides 2 and 3 each contain still another unique cysteine (Fig. 1). The C. elegans polypeptide contains seven cysteine residues, six of which are conserved in all three human polypeptides, whereas its extreme N-terminal cysteine is conserved only in the human polypeptide 3 (Fig. 1). The six cysteines that are conserved in all four polypeptides may be especially important structurally.
All lysyl hydroxylase isoforms have potential attachment sites for asparagine-linked oligosaccharides, human polypeptide 3 having two such sites, polypeptides 1 and 2 four and seven sites, respectively, whereas the C. elegans polypeptide has only one site (Fig. 1). The position of the first site in polypeptide 3 is identical to that in polypeptide 2, and the position of the second site is identical to that of the third site in polypeptide 1. Site-directed mutagenesis studies have suggested that glycosylation of the second site present in the human polypeptide 1 may be required for full catalytic activity of lysyl hydroxylase 1 (6). This position also has a potential N-glycosylation site in polypeptide 2, but not in polypeptide 3 or the C. elegans polypeptide, suggesting that the various lysyl hydroxylase isoforms may have different glycosylation requirements for their catalytic activity.
Expression of Lysyl Hydroxylase 3 mRNA in Various Human Tissues.
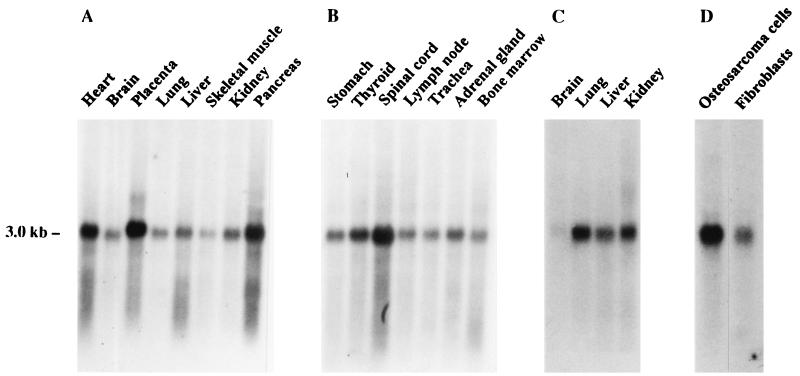
The mRNAs for lysyl hydroxylase 1 and 2 have been found to be expressed in a variety of human tissues (16, 27). The highest expression levels of the isoenzyme 1 mRNA have been found in the liver and skeletal muscle (27), whereas those for isoenzyme 2 mRNA have been found in the pancreas, placenta, heart, and skeletal muscle (16). Northern hybridization with a cDNA probe for lysyl hydroxylase 3 indicated the presence of only a single mRNA of about 3.0 kb, which was expressed in a variety of tissues (Fig. 2). This mRNA is slightly smaller than the 3.2 kb mRNA for isoenzyme 1 (2) and considerably smaller than the 4.2 kb mRNA for isoenzyme 2 (16). The highest expression levels among the tissues studied were found in the placenta, pancreas, spinal cord, and heart (Fig. 2). Fetal lung, kidney, and liver had higher expression levels than the corresponding adult tissues (Fig. 2 A and C). The expression pattern differs from that of the isoenzyme 1 mRNA, especially in that the relative expression level of isoenzyme 3 mRNA was very much lower in the adult liver, and from that of isoenzyme 2 mRNA in that the relative expression level was much lower in the skeletal muscle (Fig. 2 and refs. 16 and 27). Comparison of mRNA levels between human Saos-2 osteosarcoma cells and fibroblasts indicated a considerably higher value in the former (Fig. 2). It should be noted that comparison of mRNA levels in whole tissues does not demonstrate the possible existence of major differences between certain cell types, as has already been demonstrated in the case of the mRNAs for the two human prolyl 4-hydroxylase α-subunit isoforms (28). Further immunofluorescence and in situ hybridization studies will thus be required to elucidate the possible presence of cell type-specific differences.
Figure 2.
Northern blot analysis of lysyl hydroxylase 3 mRNA in human tissues and cells. Each lane in A–C contains 2 μg of poly(A)+ RNA from the adult (A and B) or fetal (C) tissue indicated. Each lane in D contains 15 μg of total RNA from adult human Saos-2 osteosarcoma cells or skin fibroblasts. Blots were hybridized with a 354-bp PCR fragment from the 3′ end of the cDNA. Autoradiography time was 4 (A–C) or 7 (D) days.
Expression of Recombinant Lysyl Hydroxylase 3 in Insect Cells.
A baculovirus coding for human lysyl hydroxylase 3 was generated and used to infect High Five cells. The cells were harvested 24–72 hr after infection, homogenized in a buffer containing 1% NP-40 (NP-40 buffer), and centrifuged. The cell pellet was homogenized further in a buffer containing 50% glycerol, 0.6 M NaCl, and 1% NP-40 (glycerol buffer), as described for recombinant human lysyl hydroxylase 1 (6), incubated on ice, and centrifuged. The remaining pellet was solubilized in 1% SDS, and samples were studied by SDS/PAGE under reducing conditions followed by Coomassie staining.
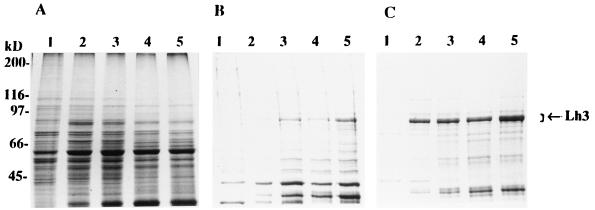
The infected cells were found to express a new 80–85 kDa polypeptide that was not to be seen 24 hr after infection (Fig. 3). This polypeptide was seen in all three fractions, i.e., the NP-40 and glycerol buffer extracts and the samples solubilized with 1% SDS, the highest levels being found in the SDS soluble fraction (Fig. 3). The polypeptide migrated in multiple bands, probably because of heterogeneity in glycosylation, as has previously been demonstrated for the recombinant isoenzyme 1 (6).
Figure 3.
Analysis of the expression of the human lysyl hydroxylase 3 polypeptide in insect cells by SDS/PAGE under reducing conditions. (A) NP-40 soluble proteins. (B) Glycerol buffer soluble proteins. (C) Proteins solubilized from the remaining pellets with 1% SDS. Lanes 1–5, samples from High Five cells infected with the virus coding for lysyl hydroxylase 3 and harvested 24, 40, 48, 64, and 72 hr after infection, respectively. Arrow indicates the location of the lysyl hydroxylase 3 polypeptides. Gels were stained with Coomassie brilliant blue.
The NP-40 and glycerol buffer extracts were analyzed for lysyl hydroxylase activity with an assay based on hydroxylation-coupled decarboxylation of 2-oxo[1-14C]glutarate with the synthetic peptide (Ile-Lys-Gly)3 as a substrate (19). The NP-40 buffer extract, but not the glycerol extract, was found to give a high 2-oxo[1-14C]glutarate decarboxylation rate even in noninfected cells (Table 1). A slight increase in enzyme activity level was already seen in both extracts 24 hr after infection. The activity in the NP-40 extract reached its maximum value at 48 hr, whereas that in the glycerol extract continued to increase throughout the experiment (Table 1). These data differ from those obtained in similar experiments with a virus coding for lysyl hydroxylase 1 in that little, if any, of polypeptide 1 can be extracted with the NP-40 buffer and only about 10% of the recombinant enzyme 1 is soluble in the glycerol buffer 72 hr after infection, 90% being found in the 1% SDS soluble portion at that point in time (6). Lysyl hydroxylase has previously been reported to be a luminally oriented peripheral membrane protein in the endoplasmic reticulum (29), part of its activity being tightly bound and part loosely bound (1). The present data suggest that the heterogeneity in lysyl hydroxylase solubilization from various cells and tissues may be in part because of the existence of isoenzymes, lysyl hydroxylase 3 being solubilized more easily than lysyl hydroxylase 1.
Table 1.
Time course of the appearance of lysyl hydroxylase 3 activity in High Five cells infected with a recombinant baculovirus coding for the human enzyme
| Time after infection, hr | Lysyl hydroxylase activity, dpm/mg*
|
|
|---|---|---|
| NP-40 extract | Glycerol buffer extract | |
| 0† | 5,800 | <100 |
| 24 | 8,900 | 700 |
| 40 | 15,900 | 2,600 |
| 48 | 18,900 | 2,900 |
| 64 | 18,400 | 4,500 |
| 72 | 17,700 | 6,100 |
Values are given as dpm/mg for two duplicate experiments.
Noninfected High Five cells harvested with the 48-hr samples.
Catalytic Properties of Recombinant Lysyl Hydroxylase 3.
Km values for Fe2+, 2-oxoglutarate, ascorbate, and two peptide substrates were determined with the soluble cell extracts as sources of the enzyme. All these values were found to be essentially identical to those determined for recombinant human lysyl hydroxylase 1 (Table 2).
Table 2.
Km values for cosubstrates and two peptide substrates of the recombinant human lysyl hydroxylases 3 and 1
| Cosubstrate or substrate |
Km, μM
|
|
|---|---|---|
| Lysyl hydroxylase 3 | Lysyl hydroxylase 1 | |
| Fe2+ | 2 | 2 |
| 2-Oxoglutare | 100 | 100 |
| Ascorbate | 300 | 350 |
| Peptide substrate L-1* | 600 | 500 |
| Peptide substrate (IKG)3 | 800 | 700 |
Ala-Arg-Gly-Ile-Lys-Gly-Ile-Arg-Gly-Phe-Ser-Gly (19).
CONCLUSIONS
Lysyl hydroxylase was for a long time thought to have no isoenzymes, and it was difficult to understand why the deficiency in its activity observed in Ehlers–Danlos syndrome type VI showed a wide variation between tissues (1, 8). The recent cloning of lysyl hydroxylase 2 (16) and the present data indicating the existence of a lysyl hydroxylase 3 explain these findings. Studies at the tissue level showed distinct differences in the expression patterns of the three lysyl hydroxylase mRNAs, suggesting that there may be major tissue and cell type-specific differences in the expression of the three isoenzymes. There also appear to be definite differences in solubility between the isoenzymes, in that recombinant lysyl hydroxylase 1 cannot be extracted from insect cells in any significant amounts with the NP-40 buffer (6), whereas large amounts of enzyme 2 (16) and enzyme 3 activity were found in the NP-40 extracts. No differences in catalytic properties were found here between lysyl hydroxylase 3 and 1, but the data do not exclude the possibility that differences might exist with respect to the hydroxylation of various collagen types. Further studies are thus needed to elucidate possible differences between the three lysyl hydroxylase isoenzymes with respect to their expression in various cell types and to the hydroxylation of various collagens. Studies are also needed to find out whether deficiencies in the activities of enzymes 2 and 3 lead to a subtype of the Ehlers–Danlos syndrome or to some other type of connective tissue manifestations.
Acknowledgments
We thank Anu Myllymäki and Jaana Jurvansuu for their expert technical assistance and Ari-Pekka Kvist for helping with the computational work. This work was supported by grants from the Research Council for Health within the Academy of Finland.
ABBREVIATION
- NP-40
Nonidet P-40
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF068229).
References
- 1.Kivirikko K I, Pihlajaniemi T. Adv Enzymol Relat Areas Mol Biol. 1998;72:325–398. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 2.Hautala T, Byers M G, Eddy R L, Shows T B, Kivirikko K I, Myllylä R. Genomics. 1992;13:62–69. doi: 10.1016/0888-7543(92)90202-4. [DOI] [PubMed] [Google Scholar]
- 3.Yeowell H N, Ha V, Walker L C, Murad S, Pinnell S R. J Invest Dermatol. 1992;99:864–869. doi: 10.1111/1523-1747.ep12614840. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong L C, Last J A. Biochim Biophys Acta. 1995;1264:93–102. doi: 10.1016/0167-4781(95)00130-9. [DOI] [PubMed] [Google Scholar]
- 5.Myllylä R, Pihlajaniemi T, Pajunen L, Turpeenniemi-Hujanen T, Kivirikko K I. J Biol Chem. 1991;266:2805–2810. [PubMed] [Google Scholar]
- 6.Pirskanen A, Kaimio A-M, Myllylä R, Kivirikko K I. J Biol Chem. 1996;271:9398–9402. doi: 10.1074/jbc.271.16.9398. [DOI] [PubMed] [Google Scholar]
- 7.Krol B J, Murad S, Walker L C, Marshall M K, Clark W L, Pinnell S R, Yeowell H N. J Invest Dermatol. 1996;106:11–16. doi: 10.1111/1523-1747.ep12326956. [DOI] [PubMed] [Google Scholar]
- 8.Steinmann B, Royce P M, Superti-Furga A. In: Connective Tissue and Its Heritable Disorders. Royce P M, Steinmann B, editors. New York: Wiley–Liss; 1993. pp. 351–407. [Google Scholar]
- 9.Byers P H. J Invest Dermatol. 1994;103:47S–52S. doi: 10.1111/1523-1747.ep12399038. [DOI] [PubMed] [Google Scholar]
- 10.Hyland J, Ala-Kokko L, Royce P, Steinmann B, Kivirikko K I, Myllylä R. Nat Genet. 1992;2:228–231. doi: 10.1038/ng1192-228. [DOI] [PubMed] [Google Scholar]
- 11.Hautala T, Heikkinen J, Kivirikko K I, Myllylä R. Genomics. 1993;15:399–404. doi: 10.1006/geno.1993.1074. [DOI] [PubMed] [Google Scholar]
- 12.Ha V T, Marshall M K, Elsas L J, Pinnell S R, Yeowell H N. J Clin Invest. 1994;93:1716–1721. doi: 10.1172/JCI117155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pousi B, Hautala T, Heikkinen J, Pajunen L, Kivirikko K I, Myllylä R. Am J Hum Genet. 1994;55:899–906. [PMC free article] [PubMed] [Google Scholar]
- 14.Heikkinen J, Toppinen T, Yeowell H N, Krieg T, Steinmann B, Kivirikko K I, Myllylä R. Am J Hum Genet. 1997;60:48–56. [PMC free article] [PubMed] [Google Scholar]
- 15.Pajunen L, Suokas M, Hautala T, Kellokumpu S, Tebbe B, Kivirikko K I, Myllylä R. DNA Cell Biol. 1998;17:117–123. doi: 10.1089/dna.1998.17.117. [DOI] [PubMed] [Google Scholar]
- 16.Valtavaara M, Papponen H, Pirttilä A-M, Hiltunen K, Helander H, Myllylä R. J Biol Chem. 1997;272:6831–6834. doi: 10.1074/jbc.272.11.6831. [DOI] [PubMed] [Google Scholar]
- 17.Luckow V A, Summers M D. Virology. 1989;170:31–39. doi: 10.1016/0042-6822(89)90348-6. [DOI] [PubMed] [Google Scholar]
- 18.Gruenwald S, Heitz J. Baculovirus Expression Vector System: Procedures and Methods Manual. San Diego: PharMingen; 1994. [Google Scholar]
- 19.Kivirikko K I, Myllylä R. Methods Enzymol. 1982;82:245–304. doi: 10.1016/0076-6879(82)82067-3. [DOI] [PubMed] [Google Scholar]
- 20.von Hejne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 22.Myllylä R, Gunzler V, Kivirikko K I, Kaska D D. Biochem J. 1992;286:923–927. doi: 10.1042/bj2860923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamberg A, Pihlajaniemi T, Kivirikko K I. J Biol Chem. 1995;270:9926–9931. doi: 10.1074/jbc.270.17.9926. [DOI] [PubMed] [Google Scholar]
- 24.Myllyharju J, Kivirikko K I. EMBO J. 1997;16:1173–1180. doi: 10.1093/emboj/16.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helaakoski T, Annunen P, Vuori K, MacNeil I A, Pihlajaniemi T, Kivirikko K I. Proc Natl Acad Sci USA. 1995;92:4427–4431. doi: 10.1073/pnas.92.10.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko K I. J Biol Chem. 1997;272:17342–17348. doi: 10.1074/jbc.272.28.17342. [DOI] [PubMed] [Google Scholar]
- 27.Heikkinen J, Hautala T, Kivirikko K I, Myllylä R. Genomics. 1994;24:464–471. doi: 10.1006/geno.1994.1654. [DOI] [PubMed] [Google Scholar]
- 28.Annunen P, Autio-Harmainen H, Kivirikko K I. J Biol Chem. 1998;273:5989–5992. doi: 10.1074/jbc.273.11.5989. [DOI] [PubMed] [Google Scholar]
- 29.Kellokumpu S, Sormunen R, Heikkinen J, Myllylä R. J Biol Chem. 1994;269:30524–30529. [PubMed] [Google Scholar]