Abstract
To date two classes of shikimate dehydrogenases have been identified and characterized, YdiB and AroE. YdiB is a bifunctional enzyme that catalyzes the reversible reductions of dehydroquinate to quinate and dehydroshikimate to shikimate in the presence of either NADH or NADPH. In contrast, AroE catalyzes the reversible reduction of dehydroshikimate to shikimate in the presence of NADPH. Here we report the crystal structure and biochemical characterization of HI0607, a novel class of shikimate dehydrogenase annotated as shikimate dehydrogenase-like. The kinetic properties of HI0607 are remarkably different from those of AroE and YdiB. In comparison with YdiB, HI0607 catalyzes the oxidation of shikimate but not quinate. The turnover rate for the oxidation of shikimate is ~1000-fold lower compared with that of AroE. Phylogenetic analysis reveals three independent clusters representing three classes of shikimate dehydrogenases, namely AroE, YdiB, and this newly characterized shikimate dehydrogenase-like protein. In addition, mutagenesis studies of two invariant residues, Asp-103 and Lys-67, indicate that they are important catalytic groups that may function as a catalytic pair in the shikimate dehydrogenase reaction. This is the first study that describes the crystal structure as well as mutagenesis and mechanistic analysis of this new class of shikimate dehydrogenase.
The shikimate pathway occupies a central position for aromatic biosynthesis in microbes and plants but is not present in humans and other higher animals. The absence of the shikimate pathway in animals makes it an ideal target for herbicide and anti-microbial drug design. Recently the shikimate pathway was identified in apicomplexan parasites, including Toxoplasma gondii and Plasmodium falciparum, which has renewed interest in better understanding the enzymes in the pathway (1, 2). The importance of the shikimate pathway is exemplified by the common herbicide glyphosate, which inhibits the enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Recent studies have also shown that glyphosate blocks the shikimate pathway in apicomplexan parasites and is effective in controlling their growth (3). The shikimate pathway consists of seven enzymatic steps initiated by the condensation of phosphoenolpyruvate and erythrose-4-phosphate by 3-deoxy-d-arabino-heptolosonate 7-phosphate synthase. The last enzymatic step produces the branch point intermediate, chorismate, which serves in turn as the precursor for a number of pathways including those involved in aromatic amino acid, phytoalexin, flavanoid, and lignin biosynthesis.
The fourth enzyme in the pathway, shikimate dehydrogenase (shikimate:NADP+ oxidoreductase; EC 1.1.1.25), is involved in the NADPH-dependent reduction of dehydroshikimate to shikimate. This enzymatic reaction proceeds in both the forward and reverse direction with similar rates and similar Michaelis constants for substrates in either direction. Kinetic studies with substrate analogues and isotope exchange demonstrated that the shikimate dehydrogenase-catalyzed reaction is consistent with an ordered sequential mechanism (4).
Two types of shikimate dehydrogenases from bacteria have been characterized to date, AroE and YdiB (quinate/shikimate dehydrogenase, EC 1.1.1.282). Although AroE has been established as the enzyme responsible for flux through the main trunk of the shikimate pathway, YdiB has been implicated in a branch point involving the metabolism of quinate (5, 6). YdiB was first characterized in fungi and was shown to play a significant role in the quinate utilization pathway rather than in the shikimate pathway itself (7, 8). This pathway consists of three enzymes (quinate/shikimate dehydrogenase, 3-dehydroquinase, and dehydroshikimate dehydratase) that catabolize quinate into protocatechuic acid. In addition, quinate/shikimate dehydrogenase can also catalyze the reduction of 3-dehydroshikimate to shikimate. The structures of both the AroE and YdiB proteins have been determined (9).
In this study we identified a novel class of shikimate dehydrogenases termed shikimate dehydrogenase-like (SDH-L).1 Phylogenetic and kinetic analyses show that SDH-L is distinct from both AroE and YdiB, yet, all three classes have similar three-dimensional structures. This class of shikimate dehydrogenase was annotated by sequencing projects as shikimate dehydrogenase-like because of the lack of supporting biological and biochemical data. Because the shikimate pathway enzymes are ideal targets for the development of antimicrobial and herbicidal compounds, identification and characterization of novel enzyme(s) will enhance our understanding of the path-way and may reveal other avenues for drug and herbicide design. In this study we present the crystal structure and kinetic analysis of the shikimate dehydrogenase-like enzyme (HI0607) from Haemophilus influenzae.
MATERIALS AND METHODS
Chemicals
Shikimic acid was a generous gift from Professor John Frost at Michigan State University. All other reagents are of molecular biology grade and were purchased from Sigma, Bioshop, or BDH.
Cloning, Expression, and Purification
The gene (gene identifier 38233601) encoding the hypothetical shikimate 3-dehydrogenase-like protein HI0607 was amplified by polymerase chain reaction from H. influenzae genomic DNA and cloned into the MCSG7 vector as described elsewhere (10). Expression and purification of HI0607 were conducted according to the published protocol by Christendat et al. (11). In brief, HI0607 recombinants were expressed in the E. coli strain BL21 Gold in 1 liter of Luria-Bertani medium supplemented with 50 µg/ml kanamycin and 50 µg/ml ampicillin and incubated at 37 °C with shaking until the culture reached an A of 0.7 at 600 nm. The culture was then induced with 0.4 mm isopropyl-β-d-thiogalactopyranoside for 3 h at 37 °C and allowed to grow overnight with shaking at 24 °C. Cells were harvested by centrifugation and then disrupted by sonication, and the insoluble cellular material was removed by centrifugation. HI0607 was purified from other contaminating proteins using nickel-nitrilotriacetic acid affinity chromatography. Purified HI0607 was concentrated and quantified at 280 nm using the extinction coefficient of 16640 m−1 cm−1 from a contribution of its aromatic amino acids.
Selenomethionine-labeled protein was prepared by supplementing a methionine auxotroph BL21 Escherichia coli strain (B834 DE3 from Novagen) with selenomethionine in its growth media. The protein was purified and quantified as described above for native HI0607 with the addition of 5 mm β-mercaptoethanol in all buffers during the purification process.
Site-directed Mutagenesis
Site-directed mutagenesis was carried out using the QuikChange™ protocol (Stratagene) in which the following complimentary oligonucleotides containing the required mutation (indicated by boldfaced codons) for the HI0607 active site were used: K67H, 5′-GCTGTTTCAATGCCATTCCACGAAACTTGTATGCC-3′; K67N, 5′-GCTGTTTCAATGCCATTCAACGAAACTTGTATGCC-3′; K67A, 5′-GCTGTTTCAATGCCATTCGCAGAAACTTGTATGCC-3′; D130N, 5′-GCGTGCATATAACACTAACTACATTGCCATCG-3′; and D103A, 5′-GCGTGCATATAACACTGCATACATTGCCATCG-3′. DNA encoding wild-type HI0607 was used as a template for the polymerase chain mutagenesis reaction. Briefly, 25 ng of template DNA were incubated with the appropriate mutagenic primers, dNTPs, and Pfu DNA polymerase using the cycling parameters recommended in the supplier’s technical manual. Following completion of the amplification reaction, 10 units of DpnI were added to each reaction mixture and incubated at 37 °C for 6 h, and 1 µl of the resulting mix was then transformed into XL2-Blue cells as recommended by the supplier. Plasmid DNA was purified from the resulting colonies using the Mini Prep Kit (Qiagen), and all mutations were verified by DNA sequencing. Mutant proteins were expressed and purified using the identical protocol as that used for the wild-type protein described above.
Enzyme Kinetics
The enzymatic activity of HI0607 was assayed by monitoring the reduction of NADP+ at 340 nm and 30 °C in the presence of either shikimate or quinate. The Km and Vmax values were determined by varying the concentrations of either shikimate or NADP+ while keeping the other substrate at saturation; 4 mm shikimate and 2 mm NADP+ were considered saturating. These substrate concentrations were at least 10 times higher than their respective Km values. The ability of HI0607 to utilize NAD+ was determined by assaying the enzyme at NAD+ concentrations up to 10 mm with shikimate at saturation or with varying the concentration of quinate up to 100 mm. The effect of divalent metals on the enzyme-catalyzed reaction rate was determined with either 5 mm MgCl2, ZnCl2, Zn(OAc)2, MnCl2, or CaCl2 in the reaction cuvette and saturating amounts of NADP+ and shikimate.
The pH rate profile was prepared by conducting Michaelis-Menten saturation kinetics at each pH value, and both kcat and Km values were determined. To determine the effect of extreme pH on enzyme stability, the enzyme was preincubated at pH 7.5 and 9.5 and assayed for loss of activity after a specific time at pH 8.5. In addition, enzyme activity was also monitored at the two pH extremes over a 30-min period to determine whether a time-dependent inactivation was occurring.
Data Analysis
The kinetic data were fitted to the following rate equations using the computer programs of Cleland or GraFit®. Initial velocity data were obtained by varying the concentration of either shikimate or NADP+ (A) and fitted to Equation 1, shown here,
| (1) |
to yield maximal velocity values (V) and the Michaelis constant (K). The variation of the values for V and V/K as a function of pH were fitted to the log form of Equation 2 or Equation 3, shown here,
| (2) |
| (3) |
where Y represents the value of V or V/K at a particular pH value, C represents the pH-independent value of the parameters, KA and KB are acid dissociation constants, and H is the hydrogen ion concentration.
Crystallization, Data Collection, Processing, and Refinement
Crystals of HI0607 were grown using the vapor diffusion hanging drop approach (12). The crystallization condition contained 1 µl of protein (20 mg/ml) and 1 µl of crystallization solution containing 0.1 m sodium acetate, pH 4.6, 2.0 m NaCl, and 10% (v/v) polyethylene glycol 400. The crystals belong to space group P21 with unit cell parameters of a = 90.6 Å, b = 66.6 Å, c = 96.9 Å, and β = 109.6°, and four protein molecules per asymmetric unit. The resulting protein crystal diffracted to 1.75-Å resolution, and the diffraction data were collected from a single crystal of native protein at a temperature of 100 K in a nitrogen stream. Crystals of selenomethionine protein diffracted to 2.6-Å resolution.
Structure Determination
A multiwavelength anomalous dispersion experiment was conducted, and two wavelength data sets were collected at the inflection point and the peak of the selenium absorption edge (Table I). The diffraction data were processed and scaled with the HKL2000 software package (13). Selenium positions and phase information were obtained with the software package SOLVE, followed by density modification with the program RESOLVE. The initial protein model was built with RESOLVE and refined with REFMAC as implemented in the software package RESOLVE version 2.03 (14, 15). The resulting protein model from the selenium-labeled protein was combined with the high resolution data from the native protein crystal and used to build 98% of the total protein structure with the software package ARP/warp (16).
TABLE I.
Summary of phasing and HI0607 model refinement statistics
| X-ray data | Crystal 1 (Se-Met) | Crystal 2 (native) | |
|---|---|---|---|
| Peak | Inflection | ||
| Wavelength (Å) | 0.97921 | 0.97935 | 0.97892 |
| Resolution (Å) | 50–2.6 | 50–2.6 | 50–1.75 |
| Rmerge (%)a | 9.9 (44.8) | 10.7 (59.6) | 5.9 (36) |
| Completeness (%)a | 100 (99.9) | 99.8 (97.6) | 98.5 (88) |
| I/Σ〈I〉 at 1.75 Å | 2.7 | 2.3 | 1.8 |
| Refinement | |
|---|---|
| Resolution (Å) | 30–1.75 |
| No. protein non-H atoms | 8,328 |
| No. acetate molecules | 3 |
| No. water molecules | 1,186 |
| No. reflections | 102,713 |
| No. reflection test set (5%) | 5,397 |
| R-value (%)a | 17.9 (22.0) |
| Free R-value (%)a | 21.9 (26.7) |
| R.m.s.d.b | |
| Bonds (Å) | 0.008 |
| Angles (°) | 1.7 |
| Overall B-factor (Å2) | 16.5 |
Values for highest resolution shell (1.75–1.8 Å for native and 2.6–2.69 Å for Se-Met-containing crystals) are shown in parentheses.
Root mean square deviation from ideal geometry.
The final model was manually rebuilt and visualized with the program O (17) and refined with the program REFMAC (18). Statistics for the data collection and structure refinement are shown in Table I. A Ramachandran plot of all the amino acids in the final model revealed that 92% of these residues have excellent stereochemistry, and no residues were found with generally allowed or disallowed geometry as determined with the program PROCHECK (19). All x-ray diffraction data were collected at beamline 19ID of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory.
RESULTS AND DISCUSSION
Sequence Analysis
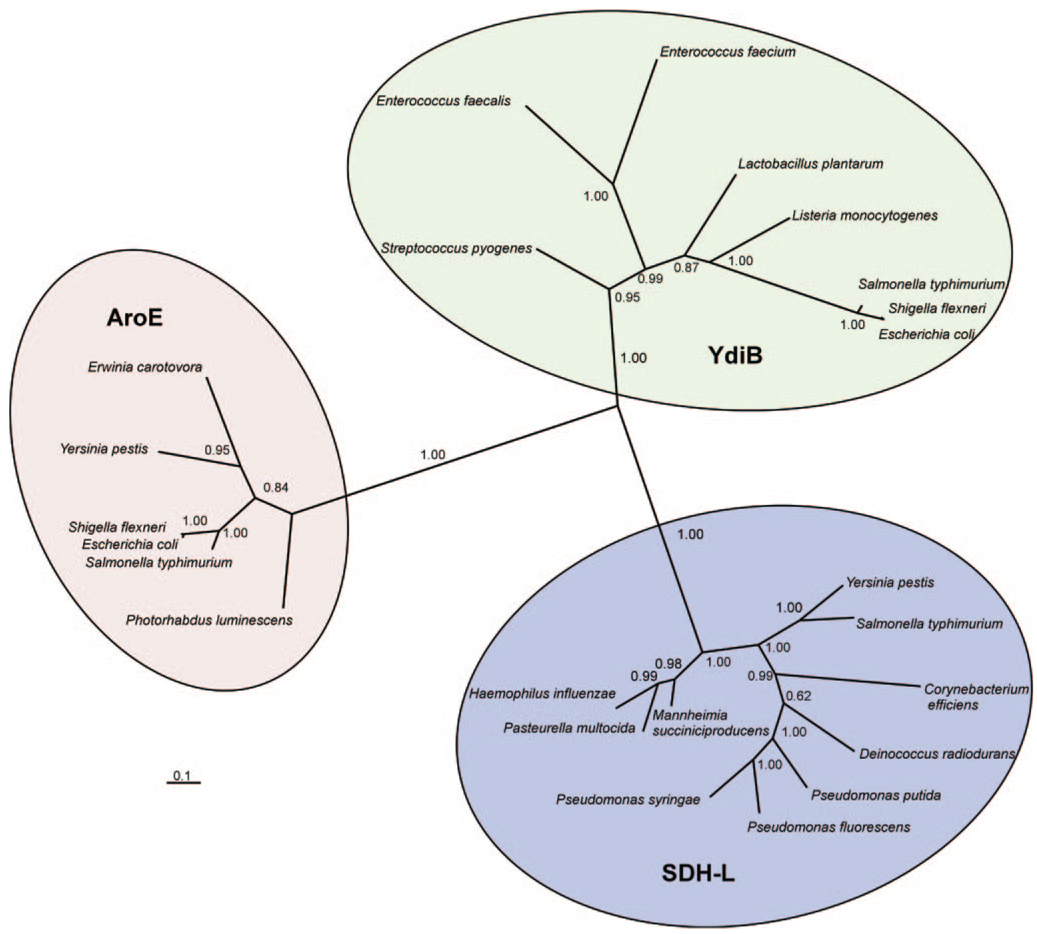
Amino acid sequence analyses of HI0607-identified proteins belonging to the three functional classes of shikimate dehydrogenases; the first was functionally annotated as a SDH-L enzyme, the second as shikimate dehydrogenase (encoded by the aroE gene), and the third as quinate/shikimate dehydrogenase (encoded by the ydiB gene). Subsequently, phylogenetic analysis revealed three distinct groups, YdiB, AroE, and SDH-L, thus indicating a distinct evolutionary relationship (Fig. 1). The SDH-L protein was identified only in a small group of organisms, including species of Pseudomonas, Corynebacterium, Salmonella, Yersinia, and Haemophilus. In contrast, AroE was identified in most microbial species, and YdiB was represented in a lower percentage of organisms. Sequence analysis identified a number of conserved residues in SDH-L as well as residues that are common to both AroE and YdiB; two of these latter residues, Lys-67 and Asp-103, were studied by site-directed mutagenesis.
FIG. 1. Bayesian-inferred, unrooted phylogeny of the shikimate dehydrogenase family of enzymes.
The radial tree, produced by MRBAYES version 3.04b, clearly indicates three distinct subgroups, which are AroE, YdiB, and SDH-L. Genetic distances (calculated with MEGA2) between the classes are calculated to be 1.89 (AroE–YdiB), 1.79 (SDH-L–YdiB), and 2.22 (SDH-L–AroE); genetic distances within each group are 0.468 (AroE), 0.601 (SDH-L), and 0.842 (YdiB). NCBI protein data base accession numbers for the proteins of the SDH-L subgroup are as follows: H. influenzae, ZP_00154645; Mannheimia succiniciproducens, YP_089507; Pasteurella multocida, AAK03513; Yersinia pestis, NP_405191; Salmonella typhimurium, NP_462758; Deinococcus radiodurans, NP_293803; Pseudomonas fluorescens, ZP_00263633; Pseudomonas putida, AAN69362; Corynebacterium efficiens, NP_737804; and Pseudomonas syringae, ZP_00123871. NCBI protein data base accession numbers for the proteins of the YdiB subgroup are as follows: E. coli, NP_310426; Shigella flexneri, NP_837377; S. typhimurium, NP_460325; Lactobacillus plantarum, NP_786702; Streptococcus pyognes, YP_060639; Enterococcus faecalis, NP_815278; Enterococcus faecium, ZP_00285321; and Listeria monocytogenes, ZP_00232331. NCBI protein data base accession numbers for the proteins of the AroE subgroup are as follows: E. coli, NP_417740; S. flexneri, NP_709069; S. typhimurium, NP_462305; Y. pestis, NP_403898; Ewinia carotovora, YP_052082; and Photorhabdus luminescens, CAE17063. The branch confidence values are given as posterior probabilities. The distance scale is shown at the bottom left.
Structure Description
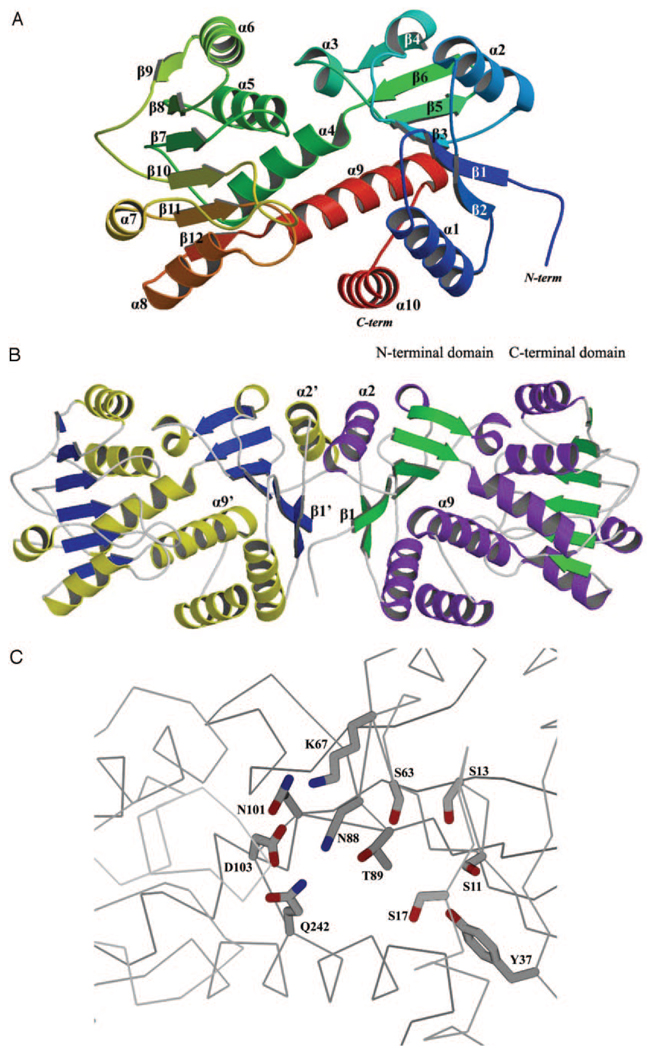
HI0607 consists of two domains, both of which belong to the structural class of α/β proteins. These two domains are linked by α-helices α4 and α9 and are important in keeping the N- and C-terminal domains together (Fig. 2A). The first 100 residues constitute the N-terminal domain of HI0607 with a secondary structure arrangement consisting of a central, mainly parallel six-stranded β-sheet with the strand order β2, β1, β3, β5, β6, and β4 flanked by four α-helices, α1, α2, α3, and α10. The C-terminal domain consists of a central six-stranded parallel β-sheet with the strand order β9, β8, β7, β10, β11, and β12 surrounded by five α-helices (helices α4, α5, α6, α7, and α8) (Fig. 2A). A deep central cavity is found in the middle of the N-terminal domain, which is formed partly by the β-sheet (strands β3, β5, and β6) and an α-helix (α2) forming the back wall of the cavity (Fig. 2A). The root mean square deviation between the N-terminal domains of the four molecules ranges from 0.17 to 0.19 Å, and between the C-terminal α/β domains it ranges from 0.19 to 0.39 Å. The overall root mean square deviation between the N- and C-terminal domains ranges from 0.42 to 0.59 Å, which indicates that flexibility exists between N-and C-terminal domains at the two hinge regions around residues Thr-102 and Gly-235. Further evidence of this behavior came from the Cα atom of Ala-173, which moves ~3.5 Å upon superposition of the N-terminal domains of the A and C monomers.
FIG. 2.
A, ribbon diagram of HI0607 monomer. The protein is essentially composed of two distinct domains, a nucleotide domain and a substrate-binding domain, which are linked by two long helices, helices 4 and 9. B, the biological dimer of HI0607. Dimerization is mediated via hydrophobic interactions between the N-terminal α2 and β1 structures of each monomer. C, HI0607 active site. In addition to the two ionizable groups, Lys-67 and Asp-103, there are a number of polar groups in the active site (Gln-242, Asn-88, and Asn-101). Toward the back of the pocket one encounters a cluster of serines (Ser-11, Ser-13, Ser-17, and Ser-63), Thr-89, and other polar groups including tyrosines (Tyr-37, for example).
Oligomeric Structure
Gel filtration studies performed on HI0607 indicate that the protein is a dimer in solution, which may also correspond to its biological state. This hypothesis is further supported by the crystal structure; intersubunit contacts are observed predominantly in a pairwise manner, which is consistent with the protein being a dimer (Fig. 2B). Subunit contacts between molecules A and D involve 2153 Å2, and between molecules B and C these contacts involve 2145 Å2 of the solvent-accessible surface area. The N-terminal domains of these molecules form a network with extensive hydrophobic interactions within the dimer.
Dali (Distance Matrix Alignment) Analysis
Comparative structure analysis of HI0607 was conducted by searching the Protein Data Bank with the Dali algorithm to identify structural neighbors of HI0607. In addition to other shikimate dehydrogenases, we have identified a number of proteins with high structural similarity, having a Z score of 6 or higher, to that of HI0607. Other proteins with highly conserved structural features to that of HI0607 include tetrahydrofolate dehydrogenase/cyclohydrolase (1A4I), NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase (1EE9), alcohol dehydrogenase (2OHX), and NADP(H)-dependent ketose reductase (1E3J). In each case, structural conservation is localized mainly in the cofactor-binding domain as expected for nucleotide-binding proteins. The substrate-binding domain, however, adopts a fold specific to the shikimate dehydrogenase family (20).
Identification of the Nucleotide and Substrate-binding Domains
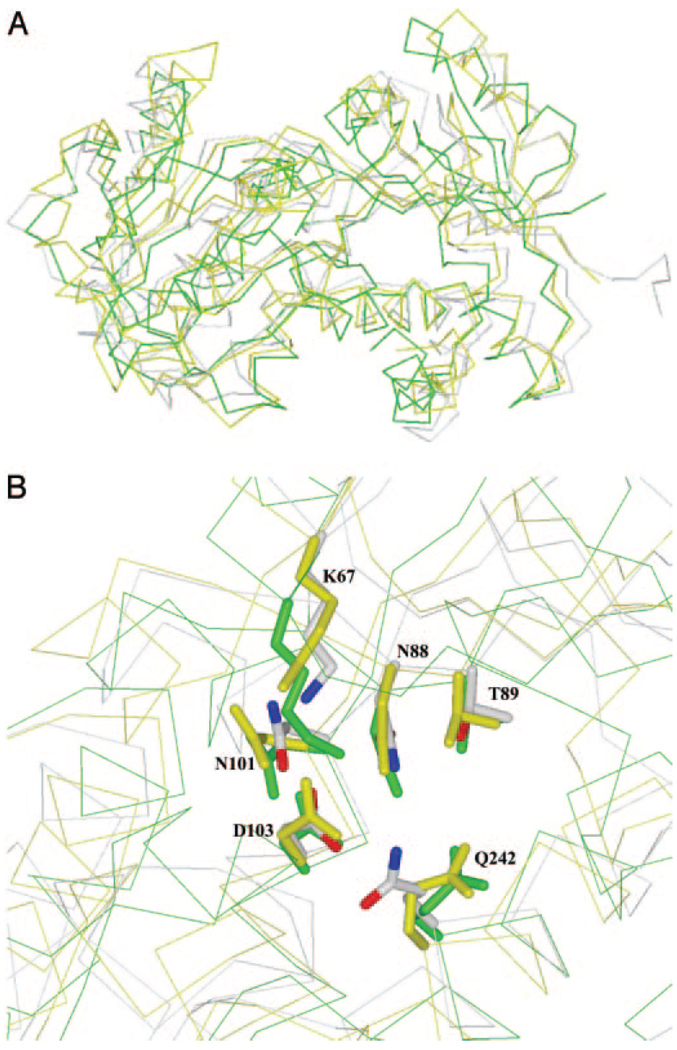
The crystal structures of five shikimate dehydrogenases were recently determined and are now available in the Protein Data Bank. These include the structures of the AroE enzymes from E. coli (1NYT), H. influenzae (1P74 and 1P77), and Methanococcus jannaschii (1NVT), the YdiB enzyme from E. coli (1O9B and 1NPD), and HI0607, the shikimate-dehydrogenase-like enzyme from H. influenzae (1NPY) that is discussed here. Superposition of the dehydrogenase structures with that of HI0607 indicated that their substrate and nucleotide-binding sites are highly conserved (Fig. 3, A and B). The nucleotide-binding sites of E. coli and H. influenzae AroE have been identified by co-crystallization with either NADP+ (E. coli AroE) or a nucleotide analog, 2′-monophosphoadenosine-5′-diphosphate (H. influenzae AroE). Arg-151 of HI0607 is conserved among AroE and some SDH-L proteins from different organisms. The crystal structure of the E. coli AroE-NADP+ complex revealed that this conserved Arg is interacting with the 2′-phosphate of NADP+. Such an interaction is important for discriminating between NAD+ and NADP+ (21, 22). In addition, its alkyl side chain stacks with the adenine ring, and such interactions have been shown to improve the binding affinity for the nucleotide (23). This observation, i.e. the interaction of Arg-151 with the 2′phosphate of NADP+, is in line with the enzymatic reaction, which indicated that HI SDH-L requires NADP+ for activity, but not NAD+. Although several crystal structures of shikimate dehydrogenases are now available, analysis of the substrate-binding site is still lacking, and important mechanistic information that can be obtained from these structures has not been attempted.
FIG. 3.
A, Cα superimposition of the E. coli shikimate dehydrogenases AroE (green) and YdiB (yellow) with HI0607 (gray), indicating a conserved three-dimensional structure for proteins in the shikimate dehydrogenase family. B, superimposition of active site residues from E. coli AroE (green) and YdiB (yellow) with HI0607 (gray). Residue numbering is according to the HI0607 sequence with single letter amino acid abbreviations. Lys-67, Asp-103, Asn-88, Asn-101, Thr-89, and Gln-242 are conserved in all dehydrogenases, indicating a role in either catalysis or substrate binding.
Architecture of the Substrate-binding Site
The substrate-binding site of HI0607 is ~14 × 10-Å wide and 12-Å deep. The bottom of this pocket is lined by a number of polar groups including Tyr-37, Tyr-250, His-24, and Gln-246. Down into this cleft at the right are a number of hydrophobic groups including Ala-243, Phe-19, Phe-23, and Phe-247. On the immediate left of this cleft are mainly polar groups, including a cluster of serines (Ser-11, Ser-13, Ser-17, and Ser63). Closer to the domain interface, on the wall of this substrate-binding pocket, are a number of polar groups including Thr-89, Thr-102, Asn-88, Asn-101, and Gln-242 (Fig. 2C). Most of these residues are conserved among the different shikimate dehydrogenases, suggesting that they are functionally important (Fig. 3B). Asp-103 and Lys-67 are invariant across the three classes of shikimate dehydrogenases and are found in the substrate-binding pocket (Fig. 2C and Fig 3B). Asp-103 and Lys-67 are most likely involved in the catalytic mechanism of shikimate dehydrogenase, because they are the only two ionizable groups in the substrate-binding site close to the C4 position of nicotinamide. We superposed the structures of shikimate dehydrogenases with that of HI0607 and searched a 6.5-Å radius around the C4 position of the superposed nicotinamide. We would expect the reaction center of shikimate to be positioned ~2.8 Å away from this catalytic pair (Asp-103 and Lys-67), which is involved in acid base chemistry, and another 2.8 Å away from the C4 of nicotinamide, via which hydride transfer would occur.
HI0607 Catalyzes the Oxidation of Shikimate
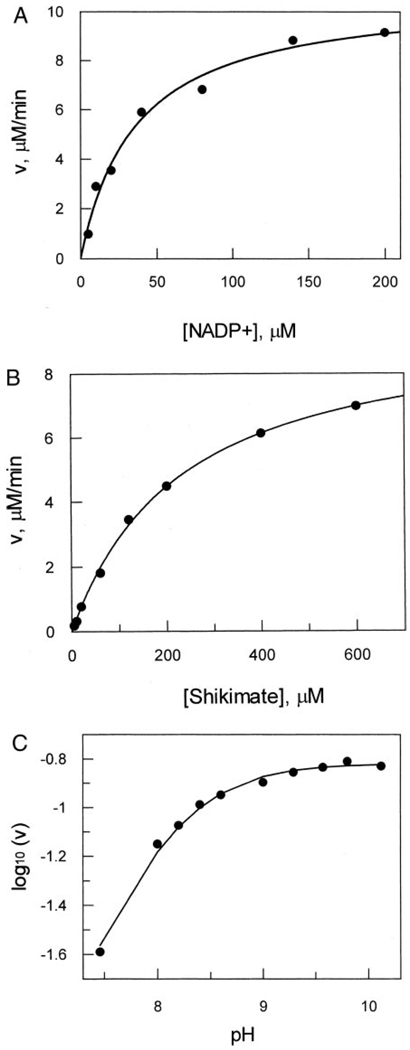
Because sequence analysis revealed that HI0607 may belong to a completely new class of shikimate dehydrogenases (Fig. 1), we set out to determine the kinetic properties of HI0607 for comparative analysis. Aside from HI0607, the AroE (HI0655) enzyme was also identified in H. influenzae; however a YdiB homologue has not been identified. We compared the substrate requirement and kinetic properties of HI0607 with those of the E. coli AroE and YdiB enzymes. We observed that HI0607 catalyzes only the oxidation of shikimate in the presence of NADP+ with a kcat of 0.2 s−1 and Michaelis constants of 234 µm ± 14 µm for shikimate and 37 µm ± 6 µm for NADP+ (Fig. 4). Interestingly, the catalytic rate for shikimate was 1000-fold lower than that of the published rate for E. coli AroE (236.7 s−1), but it was comparable with the published rate for E. coli YdiB (0.117 s−1) (9). This low turnover rate can be attributed to substrate specificity, indicating that shikimate may not be the biological substrate for the SDH-L enzyme. We also determined that the cofactor specificities of HI0607 were comparable with those of AroE, because HI0607 was very specific for NADP+ and did not utilize NAD+ as a cofactor. In addition, HI0607 did not accept quinate as a substrate in the presence of either cofactor NADP+ or NAD+, unlike YdiB. These observations are consistent with our sequence analysis showing that this shikimate dehydrogenase-like enzyme clusters separately from the AroE and YdiB shikimate dehydrogenases. Furthermore, the ability of HI0607 to catalyze the oxidation of shikimate to dehydroshikimate in the presence of NADP+, the reverse reaction, is further evidence that this enzyme belongs to the shikimate dehydrogenase class of enzymes. Comparison of the structures of shikimate dehydrogenase reveals small amino acid variations among the active sites of the three classes of shikimate dehydrogenases. As such, it is likely that a number of subtle variations of active site residues may contribute to the inherent kinetic properties of these enzymes. These observations indicate that the SDH-L protein shares kinetic and structural properties with both AroE and YdiB shikimate dehydrogenases but may have an unrelated biological function.
FIG. 4. Saturation kinetic profiles for the determination of wild-type HI0607 kinetic parameters.
A, increasing concentration of NADP+ with shikimate at saturation, Km(NADP+) = 34 µm. B, increasing concentration of shikimate with NADP+ at saturation, Km(shikimate) = 234 µm. The enzymatic rate, v, was calculated by monitoring the rate of reduction of NADP+ at 340 nm (ϵ = 6220 m−1 cm−1). In each case Vmax was calculated to be 10 µm/min. C, pH rate profile of HI0607 under saturating conditions for both shikimate and NADP+. The shape of the profile (with a pH optimum of 8.8 and a calculated pK of 8.1) indicates the dependence on deprotonation of a group for enzyme catalysis.
Divalent Metal Ion Requirement
The 1000-fold lower kcat for HI0607 as compared with that of the E. coli AroE protein can also indicate a metal cofactor requirement for activity. To address this question, we looked at the dependence of a number of divalent cations (Zn2+, Ca2+, Mg2+, and Mn2+) to determine whether they would affect the kinetic parameters of HI0607; however, at concentrations up to 5 mm no significant rate enhancement was observed. We also monitored the effect of removing any endogenous metal(s) from the protein by the addition of up to 10 mm EDTA in the reaction mixture. EDTA did not affect the reaction rate, indicating that there is no divalent metal dependence on the reaction mechanism.
pH Dependence Studies
The pH rate profile of HI0607 was determined by measuring the maximal enzyme rate within the pH range of 7.5–9.8. The rate data from this analysis was fitted with the pH rate-dependence equation (3) described under “Materials and Methods.” The enzyme displayed a half-bell pH rate profile, with the lower activity limb residing at the acidic pH range and maximal activity at pH 8.8 (Fig. 4C). The rate profile indicated that a group, with a pK of ~8.1, is involved in the catalytic reaction and needs to be deprotonated for activity. Detailed kinetic and pH rate analysis for AroE shikimate dehydrogenase from the pea also displayed a similar pH rate profile (24). Active site lysine and histidine residues are known to titrate in this region (range pH 6.5 to 8.5); however, other active site groups, such as aspartate, can also titrate in this pH range depending on their environment (25). This observation is in agreement with the crystal structure of HI0607, which showed that Lys-67 and Asp-103 are located at the entrance of the substrate-binding pocket, the expected position for a catalytic group in this protein (Fig. 2C). Moreover, the positioning of the Asp-103 and Lys-67 with respect to each other suggests that they may function as a catalytic dyad in the reaction mechanism. Consequently, the role of these two active site groups was investigated by site-directed mutagenesis. Although Asp-103 and Lys-67 are the only two ionic groups in the active site, other polar groups in the active site can easily be ionized at this pH range, depending on their microenvironment. Therefore, detailed mechanistic analysis in terms of pH dependence and mutagenesis studies is being undertaken on the Arabidopsis thaliana AroE protein.
Site-directed Mutagenesis
If Asp-103 and Lys-67 function as a catalytic pair, then the mutation of either group will produce an inactive enzyme. However, if these two groups function independently, we should observe different kinetic properties, perhaps varying levels of inactivation of HI0607 with each mutation, and a varying effect on the pH rate profile. We mutated these residues to alanine and asparagine independently; we also mutated Lys-67 to a histidine. This mutation, however, is not conservative in terms of the van der Waals volume of its side chain; therefore, an asparagine mutation was also introduced as a conservative replacement. This mutation should help to compensate for the size of the side chain and its ability to hydrogen bond.
Characterization and Kinetic Properties of Mutant Proteins
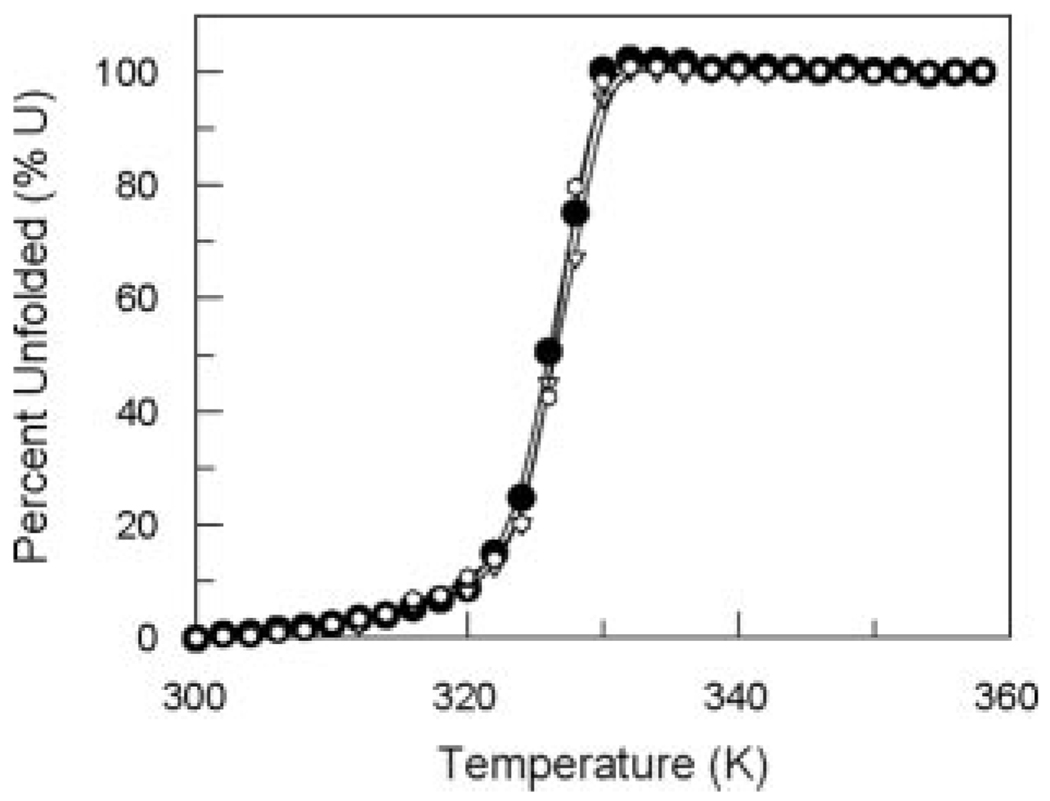
The expression and purification properties of the mutant enzymes were similar to those of the wild-type, which indirectly indicated that there were no global conformational changes due to these mutations. In addition, the circular dichroism profile (from 204 to 360 nm) of each mutant protein was similar to that of the wild-type protein. Further biophysical analysis showed that these proteins denature with similar melting temperatures (Tm) compared with the wild-type protein (Fig. 5). How-ever, these studies are only true indicators for major structural effects.
FIG. 5. Thermal denaturation of HI0607 and mutants monitored with circular dichroism at 222 nm.
HI0607 active site mutants D103N (∇) and K67N (□) exhibit similar temperature-dependent denaturation profiles and melting temperatures (Tm = 327 K) as those observed for the wild-type enzyme (●). U, units.
We observed that all of the mutations independently inactivated HI0607. We worked at very high protein concentrations in our reaction vessel, from 10 to 200 µg of mutant protein in a 1-ml reaction mixture, but did not observe a reaction rate with any of the mutants. It is possible that these mutations may have a localized effect on the active site architecture and, therefore, may affect substrate binding. To compensate for this possibility we increased the concentration of substrates; shikimic acid was increased to 10 mm and NADP+ to 10 mm, which is at least 40 times their respective Km values, and we monitored the reaction rate but were still unable to restore enzyme activity. If these mutations affected substrate binding, we should have been able to partially restore enzyme activity at such high substrate concentrations. We analyzed the corresponding mutants of the Arabidopsis thaliana AroE homologue (NP_187286); the K385N and D423N mutants independently resulted in a 1000-fold reduction in shikimate dehydrogenase activity.2 As such, we should expect a similar magnitude reduction of HI0607 enzymatic activity for the corresponding mutants. Assuming a reduced activity of 1000-fold for a given HI0607 mutant, the expected observable rate would be only 5.8 × 10−4 absorbance units per minute (with 200 µg mutant protein), which is below the detectable limit of a UV-visible spectrometer using the extinction coefficient value of 6220 m−1 cm−1 for NADPH. In general, the reduced enzyme activity observed for these active site mutants, for both Arabidopsis AroE and HI0607, is consistent with our expectation for catalytic residues. Taking together the location of these two ionizable groups in the enzyme active site, the pH profile indicating an ionizable group of pK 8.0, and the mutagenesis studies that inactivated HI0607 and the Arabidopsis thaliana AroE homologue, we conclude that Lys-67 and Asp-103 are key catalytic groups and may function as a catalytic pair for HI0607 activity. We are currently investigating this notion further, in addition to the roles of a number of other conserved active site residues.
Biological Role of HI0607
The experimental evidence presented here supports the current sequence annotation of HI0607 as being a shikimate dehydrogenase-like protein. Its crystal structure is similar to those of AroE and YdiB, it catalyzes the oxidation of shikimate with NADP+, and functionally important active site residues are conserved among this protein and the two other classes of shikimate dehydrogenases. However, AroE and YdiB have distinct biological roles. AroE functions primarily in shuttling metabolites thorough the main trunk of the shikimate pathway in both plants and microbes. On the other hand, YdiB is a branch point enzyme for quinate biosynthesis from the shikimate pathway. Because SDH-L catalyzes the oxidation of shikimate and is only present in distinct classes of pathogenic organisms, it is likely that this enzyme is involved in a novel branch from the shikimate pathway.
Acknowledgments
We thank all members of the Structural Biology Center at Argonne National Laboratory for help in conducting experiments, Dr. Joanne Turnbull for critically reading the manuscript, Dr. Vivian Saridakis for assistance in the preparation of the manuscript, and John Stavrinides for assistance with the phylogenetic analysis.
Footnotes
This work was supported by National Institutes of Health Grant GM62414-01, a research grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), and the United States Department of Energy, Office of Biological and Environmental Research, under contract W-31-109-Eng-38.
The atomic coordinates and structure factors (code 1NPY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: SDH-L, shikimate dehydrogenase-like; HI, Haemophilus influenzae.
S. Singh and D. Christendat, unpublished data.
REFERENCES
- 1.McConkey GA. Exp. Parasitol. 2000;94:23–32. doi: 10.1006/expr.1999.4467. [DOI] [PubMed] [Google Scholar]
- 2.Campbell SA, Richards TA, Mui EJ, Samuel BU, Coggins JR, McLeod R, Roberts CW. Int. J. Parasitol. 2004;34:5–13. doi: 10.1016/j.ijpara.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Roberts CW, Roberts F, Lyons RE, Kirisits MJ, Mui EJ, Finnerty J, Johnson JJ, Ferguson DJ, Coggins JR, Krell T, Coombs GH, Milhous WK, Kyle DE, Tzipori S, Barnwell J, Dame JB, Carlton J, McLeod R. J. Infect. Dis. 2002;185 Suppl. 1:S25–S36. doi: 10.1086/338004. [DOI] [PubMed] [Google Scholar]
- 4.Balinsky D, Dennis AW, Cleland WW. Biochemistry. 1971;10:1947–1952. doi: 10.1021/bi00786a032. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann KM. Plant Cell. 1995;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrmann KM, Weaver LM. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- 7.Giles NH, Case ME, Baum J, Geever R, Huiet L, Patel V, Tyler B. Microbiol. Rev. 1985;49:338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler KA, Lamb HK, Hawkins AR. Biochem. J. 1996;315:195–205. doi: 10.1042/bj3150195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel G, Roszak AW, Sauve V, Maclean J, Matte A, Coggins JR, Cygler M, Lapthorn AJ. J. Biol. Chem. 2003;278:19463–19472. doi: 10.1074/jbc.M300794200. [DOI] [PubMed] [Google Scholar]
- 10.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. Protein Expression Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 11.Christendat D, Saridakis V, Dharamsi A, Bochkarev A, Pai EF, Arrowsmith CH, Edwards AM. J. Biol. Chem. 2000;275:24608–24612. doi: 10.1074/jbc.C000238200. [DOI] [PubMed] [Google Scholar]
- 12.Jankarik J, Kim SH. J. Appl. Crystallogr. 1991;24:409–411. [Google Scholar]
- 13.Minor W, Tomchick D, Otwinowski Z. Structure Fold. Des. 2000;8:R105–R110. doi: 10.1016/s0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 14.Terwilliger TC. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger TC, Berendzen J. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrakis A, Morris R, Lamzin VS. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 17.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project Number 4. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. [Google Scholar]
- 19.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 20.Vogan E. Structure (Camb.) 2003;11:902–903. doi: 10.1016/s0969-2126(03)00165-5. [DOI] [PubMed] [Google Scholar]
- 21.Lesk AM. Curr. Opin. Struct. Biol. 1995;5:775–783. doi: 10.1016/0959-440x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S, Kodaki T, Makino K. J. Biol. Chem. 2005;280:10340–10349. doi: 10.1074/jbc.M409443200. [DOI] [PubMed] [Google Scholar]
- 23.Ahvazi B, Coulombe R, Delarge M, Vedadi M, Zhang L, Meighen E, Vrielink A. Biochem. J. 2000;349:853–861. [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis AW, Balinsky D. Int. J. Biochem. 1972;3:93–102. [Google Scholar]
- 25.Christendat D, Saridakis VC, Turnbull JL. Biochemistry. 1998;37:15703–15712. doi: 10.1021/bi981412b. [DOI] [PubMed] [Google Scholar]