Abstract
The crystal structure of the product of the Bacillus subtilis ykuD gene was solved by the multiwavelength anomalous dispersion (MAD) method and refined using data to 2.0 Å resolution. The ykuD protein is a representative of a distinctly prokaryotic and ubiquitous family found among both pathogenic and nonpathogenic Gram-positive and Gram-negative bacteria. The deduced amino acid sequence reveals the presence of an N-terminal LysM domain, which occurs among enzymes involved in cell wall metabolism, and a novel, putative catalytic domain with a highly conserved His/Cys-containing motif of hitherto unknown structure. As the wild-type protein did not crystallize, a double mutant was designed (Lys117Ala/Gln118Ala) to reduce excess surface conformational entropy. As expected, the structure of the LysM domain is similar to the NMR structure reported for an analogous domain from Escherichia coli murein transglycosylase MltD. The molecular model also shows that the 112-residue-long C-terminal domain has a novel tertiary fold consisting of a β-sandwich with two mixed sheets, one containing five strands and the other, six strands. The two β-sheets form a cradle capped by an α-helix. This domain contains a putative catalytic site with a tetrad of invariant His123, Gly124, Cys139, and Arg141. The stereochemistry of this active site shows similarities to peptidotransferases and sortases, and suggests that the enzymes of the ykuD family may play an important role in cell wall biology.
Keywords: hydrolase, crystal structure, catalytic triad, structural genomics, protein crystallization
INTRODUCTION
In spite of rapid advances in microbial genomics and an expanding set of complete prokaryotic genomes, a significant portion of bacterial genes code for nonannotated proteins of unknown structure and function. As the search for novel antibiotic targets intensifies in the face of an advancing threat of revival in infectious diseases, structural characterization of such orphan proteins or entire families of bacterial proteins is of particular importance.
One such structurally uncharacterized, ubiquitous family of exclusively prokaryotic proteins comprises putative enzymes containing the so-called ErfK/YbiS/YhnG domain (Pfam03734.8) as identified by the SMART database.1 These proteins are found in a vast variety of bacteria, ranging from pathogenic Bacillus anthracis, Vibrio cholera, Clostridium tetani, and others, to nonpathogenic Escherichia coli, Thermotoga maritima, Bacillus subtilis, and so forth. All ErfK/YbiS/YhnG domains contain a highly conserved stretch of amino acids (ΦGΦHGTX10(S/T)XGCΦR(M/L), where Φ is a hydrophobic residue and X denotes any amino acid). The presence of absolutely conserved His and Cys residues is suggestive of a catalytic function, probably as hydrolases or transferases (Fig. 1). Many of these proteins also contain the so-called LysM domain, which appears to have a peptidoglycan-binding function and is found in enzymes involved in cell wall degradation, and other proteins that could be associated with bacterial cell walls.2 Interestingly, homologous domains have also been discovered in a family of plant transmembrane receptor kinases, which are involved in the recognition of bacteria secreting lipochitin oligosaccharide signal molecules.3 Only one structural study of a LysM domain has been reported to date.2 The association of the ErfK/YbiS/YhnG domain with the LysM module suggests that the former may also play a role in cell wall degradation or, more specifically, in peptidoglycan metabolism. This is interesting because such unique prokaryotic proteins constitute a particularly suitable target for antibiotics.
Fig. 1.
Sequence alignment of the fingerprint motif in pathogenic and nonpathogenic bacteria (Φ is a hydrophobic residue and X denote any amino acid in consensus sequence).
The B. subtilis ykuD protein was selected as one of the targets for high-throughput structure determination by the Midwest Center for Structural Genomics (MCSG). It expressed well in E. coli but failed to crystallize despite extensive screening. We have therefore applied the recently proposed approach of surface conformational entropy reduction to generate on the protein’s surface patches that would be able to mediate crystal contacts.4 Accordingly, Lys117 and Gln118 were mutated to alanines, and the purified double mutant yielded high-quality crystals.
In this article we describe the details of the ykuD structure. Aside from the expected LysM module, the structure reveals a novel tertiary fold for the C-terminal ErfK/YbiS/YhnG domain. The stereochemical features of the amino acids in the fingerprint, conserved stretch (i.e., of His123, Gly124, Cys139, and Arg131), confirm that the ErfK domain is very likely to have catalytic properties. Based on the crystal structure, we propose a possible catalytic mechanism for the enzyme and speculate on the nature of the substrate. Our work sets the stage for functional and biochemical characterization of a novel and probably important family of ubiquitous bacterial enzymes.
MATERIALS AND METHODS
Expression, Purification, and Crystallization Screens of ykuD
The ykuD gene was cloned by polymerase chain reaction (PCR) into the pMCSG7 vector.5 The vector encodes a leader sequence consisting of a hexahistidine affinity tag followed by an eight–amino acid spacer and the tobacco etch virus (TEV) protease recognition site. The protein was expressed in the E. coli strain BL21 (DE3) and purified by Ni(2+)-nitrilotriacetic acid (Ni-NTA) metal-affinity chromatography. The affinity tag was removed during incubation with recombinant tobacco etch virus (rTEV) protease for 24 h. Protein was dialyzed against 20 mM 2-morpholinoethanesulfonic acid (MES) (pH 6.5), 300 mM NaCl, and 1.4 mM β-mercaptoethanol; and concentrated to 15 mg/mL for crystallization. Extensive screens yielded no results. Consequently, suitable sites for surface mutagenesis to enhance crystallizability were identified based on secondary structure prediction (http://www.bioinfo.pl). The dipeptide Lys-Gln (residues 117 and 118) was identified as one such potentially solvent exposed site and both residues were mutated to Ala with use of the QuikChange™ mutagenesis kit (Stratagene). Crystals of this mutant were obtained at 21°C from 0.1 M acetate (pH 4.5), 0.2 M Li2SO4, 30% polyethylene glycol (PEG) 8000, and 10 mM CdCl2. The selenomethionine (Se-Met)-labeled protein was expressed in E. coli strain B843 and purified in a manner identical to that described for the native protein. Crystals of the labeled protein were obtained from 0.1 M acetate (pH 5.1), 0.2 M Li2SO4, 30% PEG 3350, and 10 mM CdCl2.
Data Collection and Structure Solution
Data collection was performed at the National Synchrotron Light Source (NSLS), beamline X9B, at the Brookhaven National Laboratory. All data were processed and scaled using HKL2000 software package.6 The ykuD structure was solved at 2.05 Å resolution by multiwavelength anomalous dispersion (MAD) using data collected at the absorption peak, inflection point, and high-energy remote wavelengths. Data statistics are shown in Table I. The crystal belongs to space group P212121; the unit cell parameters are as follows: a = 56.2 Å, b = 63.9 Å, c = 93.6 Å. The Matthews coefficient is 2.4 Å3/Da assuming two molecules per asymmetric unit. SHELXD7 was used to solve the anomalous substructure. Four selenium sites were used for phase calculation in SHARP.8 The phases were used as an input to RESOLVE,9 which automatically built 297 out of 328 residues present in the asymmetric unit. The remaining portions of the model were build manually with O10 and COOT.11 The refinement of the ykuD model was carried out with REFMAC512 using the high-energy remote data to 2.0 Å resolution and individual isotropic displacement parameters (B factors).
TABLE I.
Data Collection and Refinement Statistics
| Data collectiona | |||||||
|---|---|---|---|---|---|---|---|
| Data type | Wavelength (Å) |
Resolution (Å) |
No. of unique reflections |
Redundancy | Completeness (%) |
Rmergeb (%) |
I/σ(I) |
| λ1remote | 0.97178 | 2.04 | 45,851 | 2.3 | 99.1 (99.0) | 5.7 (34.2) | 15.5 (2.8) |
| λ2peak | 0.97919 | 2.18 | 34,329 | 2.2 | 98.9 (93.7) | 4.3 (26.6) | 19.0 (2.8) |
| λ3inflection | 0.97946 | 2.07 | 43,799 | 2.1 | 98.4 (91.1) | 4.9 (34.4) | 16.0 (2.1) |
| Phasing statistics | |||||||
| Parameter (unit) | Value | ||||||
| F.O.M. SHARP | 0.42 | ||||||
| F.O.M. Resolve | 0.66 | ||||||
| Phasing power, isomorphous/anomalousc | |||||||
| λ1 | −/0.85 | ||||||
| λ2 | 0.72/1.55 | ||||||
| λ3 | 1.0/0.55 | ||||||
| Rcullis, isomorphous/anomalousd | |||||||
| λ1 | −/0.89 | ||||||
| λ2 | 0.75/0.71 | ||||||
| λ3 | 0.68/0.94 | ||||||
| Refinement statistics | |||||||
| Parameter (unit) | Value | ||||||
| Resolution range (Å) | 30.0–2.05 | ||||||
| Reflections (working) | 20,691 | ||||||
| Reflections (test) | 1114 | ||||||
| Rwork (%)eRfree (%)e | 21.1/26.9 | ||||||
| No. of nonhydrogen atoms | |||||||
| Protein/waters | 2469/150 | ||||||
| Mean B value (Å2) | 23.6 | ||||||
| Root-mean-square (r.m.s.) deviations | |||||||
| Bond length (Å)/bond angle (°) | 0.012/1.441 | ||||||
Values in parentheses correspond to the last shell.
Rmerge = Σ|Ii−〈I〉|/ΣI, where Ii is the intensity of the ith observation, and 〈I〉 is the mean intensity of the reflections. The values are for unmerged Friedel pairs.
Phasing power = r.m.s.(|Fh|/E), where |Fh| is the heavy atom structure factor amplitude, and E is residual lack of closure error.
Rcullis = Σ ||Fh.obs| − ||Fh.calc||/Σ|Fh| for acentric reflections, where |Fh.obs| is the observed heavy atom structure factor amplitude, and |Fh.calc| is the calculated heavy atom structure factor amplitude.
Rwork = Σ||Fobs| − |Fcalc||/Σ|Fobs|, crystallographic R factor, and Rfree = Σ||Fobs| − |Fcalc|| /Σ|Fobs|, where all reflections belong to a test set of randomly selected data.
Accession Code of the Structure
The atomic coordinates and structure factors of the ykuD protein have been deposited in the Protein Data Bank (PDB) under the accession code 1Y7M.
RESULTS AND DISCUSSION
Quality of the Model
The refined crystallographic model consists of two ykuD molecules in the asymmetric unit, each made up of 164 residues in a single polypeptide chain, 2 Cd2+ ions, 4 sulfate ions, and 150 water molecules. The stereochemical quality of the model was analyzed with PROCHECK13: 88.6% of nonglycine residues are in the favored regions, and 11.4% are in the allowed regions of the Ramachandran plot. The side-chains of 18 amino acids (11 from molecule A, and 7 from molecule B) are not visible in the σA weighted 2mFobs-DFcalc electron density map contoured at 1 σ, and were consequently excluded from refinement. The average B factors for the two LysM domains are consistently about 15% higher than those for the catalytic domains, consistent with a degree of dynamic disorder in these modules. The final values of R and Rfree factors were 21.1 and 26.9 %, respectively. The refinement statistics are shown in Table I.
Overall Structure
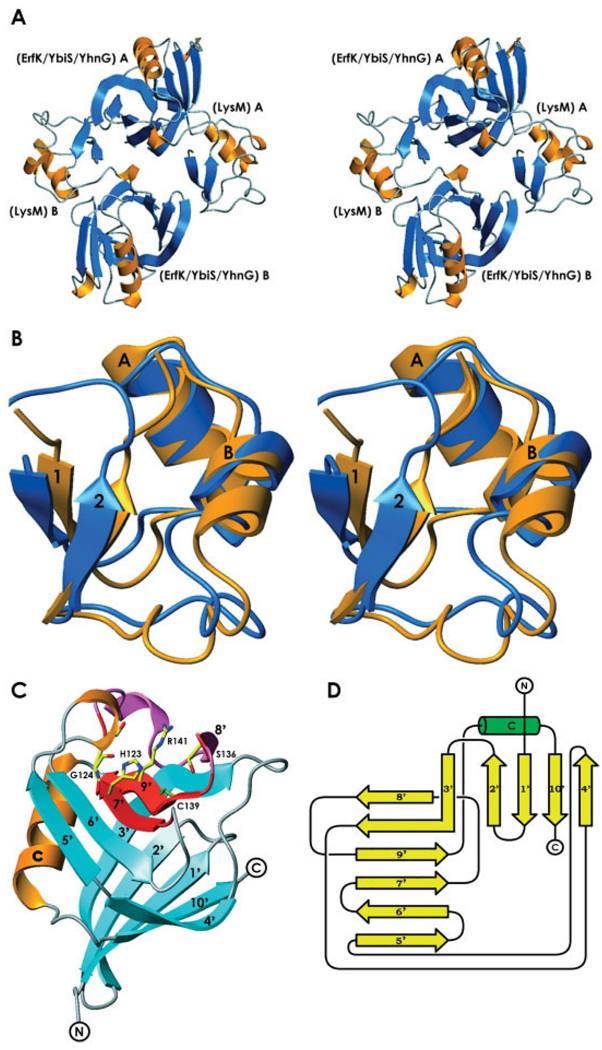
The ykuD protein is made up of two distinct domains: the 52-amino acid-long N-terminal LysM domain and the 112-amino acid C-terminal putative catalytic domain. The two ykuD molecules form a noncrystallographic, head-to-tail dimer with the local two fold axis nearly parallel to the crystallographic c axis [Fig. 2(A)]. Gel filtration indicated a monomer as a dominant species in solution; consequently, the noncrystallographic dimer has no functional implications.
Fig. 2.
The molecular architecture of ykuD. (A) A stereodiagram showing the head-to-tail noncrystallographic dimer of ykuD molecules; the two molecules are labeled A and B. (B) A comparison of the NMR structure of the LysM domain from murein transglycosylase D (PDB entry: 1E0G), orange, and the LysM domain of ykuD, blue; for details see text. (C) The tertiary structure of the ErfK/YbiS/YhnG domain of ykuD with the fingerprint conserved stretch shown in red; the side-chains of His123, Gly124, Ser136, Cys139, and Arg141 are shown in full. (D) A schematic representation of the teriary fold of the ErfK/YbiS/YhnG domain.
The LysM domain was identified in ykuD based on sequence analysis, and indeed its structure is similar to the solution (NMR) structure of the isolated LysM domain from the E. coli membrane-bound lytic murein transglycosylase D.2 Ours is the first reported crystal structure of any protein containing LysM. The domain is known to have a general peptidoglycan binding function and is characterized by a βααβ tertiary structure with two α-helices located on the same side of a two stranded, antiparallel β-sheet. Figure 2(B) represents the LysM domain of ykuD superimposed on its structural homolog. Although both structures are very similar, there are some discrepancies, including part of helix B and the downstream loop (residues 22 to 36). The average B factors for that part of the structure are higher than for the rest of the domain (~25 Å2 vs ~22 Å2), consistent with higher intrinsic mobility of the loop. Moreover, the LysM domain of ykuD does not form any significant crystal contacts, which may be a reason for its reduced stability.
The 112–amino acid ErfK/YbiS/YhnG domain represents a novel fold. No structure similar to this domain was detected using DALI.14 It is a β-sandwich, with two mixed β-sheets, one containing five, and the other, six β-strands [Fig. 2(C and D)]. Strand 3 is involved in both sheets, while strands 4 and 5 are almost contiguous, except for Ile 98, which has atypical secondary conformation (ϕ = −100.6, ψ = −50.9 ) and creates a kink between them. A single α-helix is found between strands 9 and 10. The most conserved residues of the fingerprint sequence [i.e., 120ΦGΦHGT125 and 136(S/T)XGCΦR(M/L)142] are located on the solvent-exposed concave face within strands 7 and 9 [Fig. 2(C)].
The Putative Catalytic Site
Because the fingerprint of the ErfK/YbiS/YhnG domain family (Fig. 1) contains strictly invariant His and Cys residues, often found in hydrolytic enzymes and transferases, the SMART database1 suggests that the domain has a catalytic function; consequently, we will henceforth refer to it as such. As pointed out above, the most conserved residues cluster within the seventh and the ninth strands of the β-sheet. We note that the disposition of three residues—His123 and Gly124, located on β-strand 7, and Cys 139, located on β-strand 9—has distinct similarities to so-called catalytic triads present in a wide range of enzymes, including serine proteases, cysteines proteases, esterases, lipases, and a number of more esoteric, nonhydrolytic enzymes.15 The role of a nucleophile is typically reserved for a Ser or a Cys, which interacts with the most invariant residue in a triad, a histidine, which in turn is hydrogen-bonded to a neutral or negatively charged H-bond acceptor. The details vary from family to family.15 The interaction of Ser with His is in most cases mediated in the ground state by a weak H-bond between the hydroxyl and imidazol’s unprotonated Nϵ2. During the acylation step, the hydrogen released by Ser is captured by His, which adopts the protonated, imidazolium form. In papainlike cysteine proteinases, the ground state enzyme contains an ion pair in which both Cys and His are found in the ionized forms as a thiolate and imidazolium, enhancing the nucleophilic potential of cysteines.16 This pattern is not universal though, because viral 3C proteases operate by a general base mechanism, similar to serine proteases. 17 The conformation of the catalytic histidine is stabilized in all cases by additional interactions. In serine hydrolases, the Nδ1 group of the imidazole is suitably positioned to donate an H-bond to Asp or Glu,18,19 and in two notable cases the accepting group is a backbone carbonyl20,21; in the papain family of cysteine proteases, the Nϵ2 of the catalytic histidine donates an H-bond to Asn, although examples of enzymes with aspartates in that position are also known.22,23
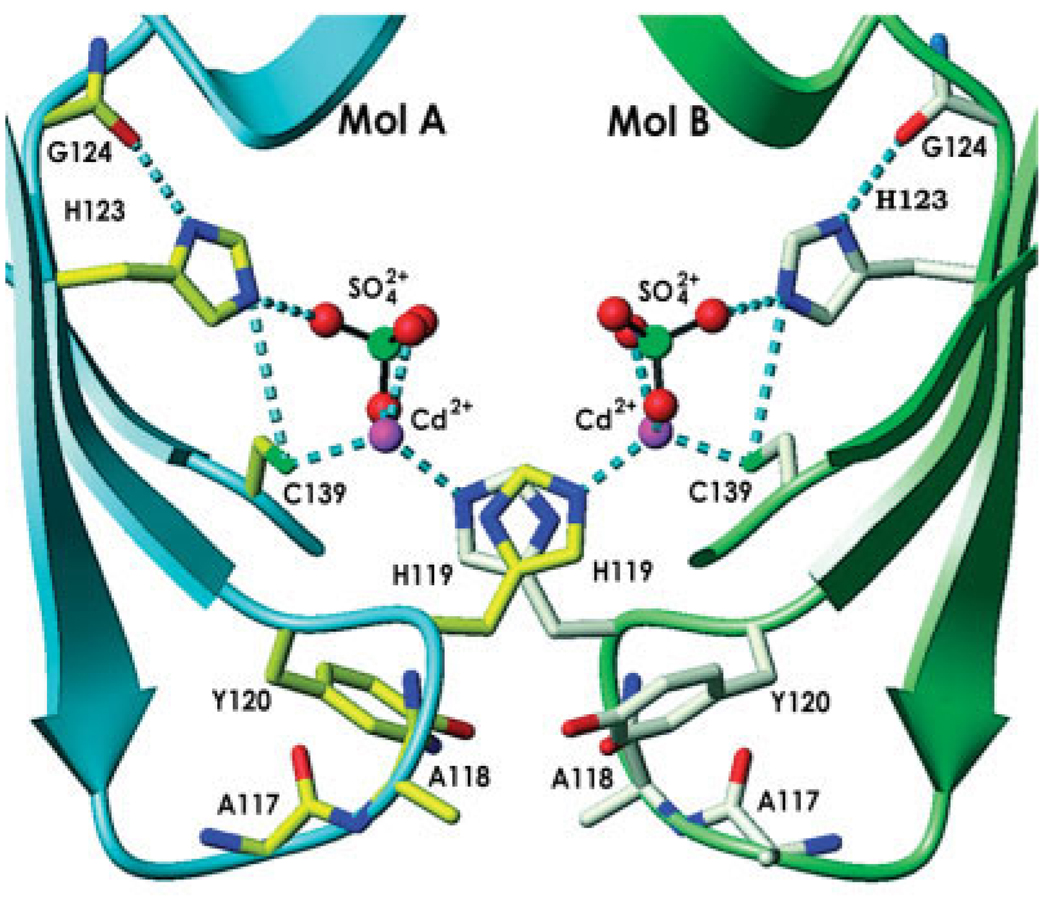
In ykuD, the three amino acids of the putative triad are capable of forming similar interactions, albeit in the crystal structure, the side-chains of Cys139 and His123 in both molecules are perturbed by an intimate symmetric crystal contact that traps two Cd2+ ions and two sulfates at the intermolecular interface (Fig. 3). Cys139 directly coordinates Cd2+ (sulfur–metal distance of 2.33 Å), which is also coordinated by Nϵ2 of His119 from an adjacent protein molecule and two oxygen atoms of a sulfate ion, to complete a tetrahedral complex. The third oxygen of the sulfate accepts an H-bond from Nϵ2 of the catalytic His123. Thus, the thiolate of Cys139 actually points away from the imidazole of His123, not as expected for a catalytic triad. However, we believe that this is a crystallization artifact as a rotation around the χ1 torsion angle of Cys139 is sufficient to generate a canonical interaction with Nϵ2 of His123. The Nδ1 of His123 donates an H-bond to the main chain carbonyl of Gly124, in a manner similar to other triads. Overall, the mutual disposition of Cys139, His123 and Gly124 strongly suggest that this group constitutes a variant of the ubiquitous catalytic triad [Fig. 4(A)].
Fig. 3.
The ykuD putative active site involved in a crystal contact. Side-chains of select residues, as well as Gly124, are shown in full; sulfate ions are represented by spheres: red (oxygen) and green (sulfur).
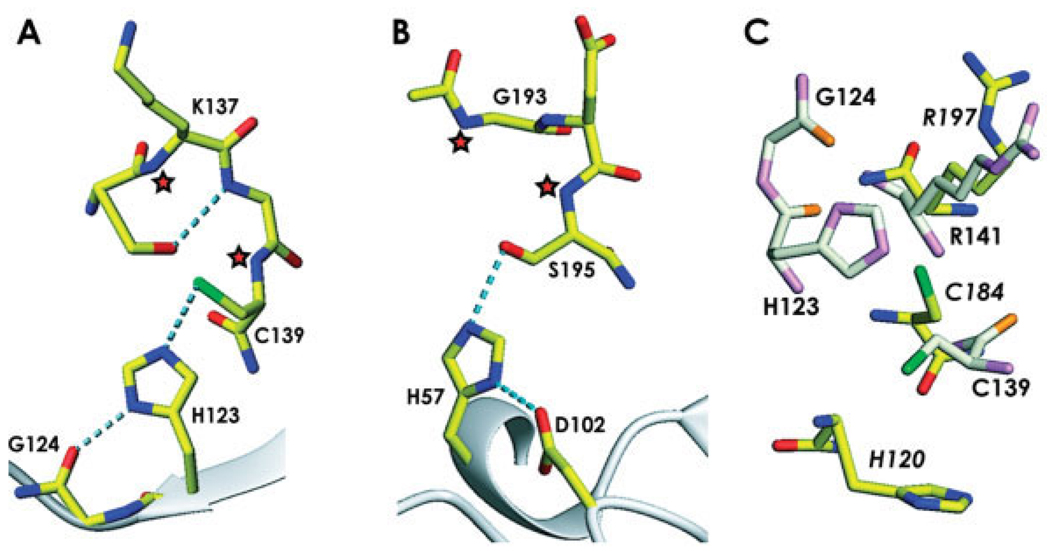
Fig. 4.
A comparison of the catalytic site of ykuD with other enzymes. (A) The ykuD catalytic triad with the side-chain of Cys139 rotated to achieve the best stereochemistry; H-bonds are shown by dotted lines, putative oxyanion hole forming backbone amides are denoted by asterisks. (B) Trypsin (PDB entry: 1YYY), with oxyanion hole indicated by asterisks. (C) Comparison of the ykuD and sortase A (PDB entry: 1T2O ) putative Cys-Arg dyads; ykuD residues are shown in light colors and the labels indicating the sortase A amino acids are in italics.
A triad is not sufficient to account for all the catalytic potential of serine- and cysteines-dependent hydrolases and transferases. The stabilization of the incipient oxyanion following the acylation step is accomplished within a so-called oxyanion hole, a positively charged surface crevice with two or three strong H-bond donors.24,25 Our atomic model reveals that a similar structure is also found within ykuD. The aforementioned sulfate molecule coordinated by the Cd2+ ion fortuitously serves as an approximate model for the tetrahedral intermediate. One of its oxygens forms H-bonds with backbone amides of Lys137, the absolutely conserved Gly138, and Cys139. The distances are 3.19 Å, 3.24 Å, and 3.03 Å, respectively. Two or three of these amides could act as the oxyanion hole, forming hydrogen bonds with the negatively charged oxygen of the tetrahedral intermediate of proteolytic reaction [Fig. 4(A)]. This observation further supports the notion that the putative active site’s stereochemistry in ykuD is consistent with canonical general base mechanism for serine/cysteine hydrolases.
Similarity to Sortases
Although the stereochemistry of the putative active site in ykuD is consistent with it having a hydrolytic or transferase function, akin to that of a protease or an esterase, the tertiary fold of the catalytic domain is not similar to any of the triad-containing families, such as chymotrypsin, subtilisin, papain or α/β hydrolase proteins. However, we note some limited topological similarities to the recently characterized structures of sortases (i.e., transpeptidases located in the cell envelope of Gram-positive bacteria) that catalyze the anchoring of specific cell-surface proteins via their unique LPXTG motif to a pentaglycine cross-bridge of peptidoglycans.26–29 In either sortase, the putative active site is located in a shallow depression on a concave side of a β-sheet, with the Cys residue found at the end of a β-strand. Interestingly, even though an involvement of a His residue has been postulated in sortases, the crystal structures suggest that it is a dyad of Cys and an invariant Arg that constitutes the active ensemble of residues.30 This has been corroborated by biochemical studies of the Staphylococcus aureus sortase A, which showed that replacement of the relevant Arg197 with other amino acids severely impedes catalysis.31 It has been postulated that Arg197 may facilitate the formation of a thiolate during the transpeptidation reaction catalyzed by the sortases.31
Close inspection of the ykuD active site also reveals the presence of the side-chain of an arginine, Arg141, in the active site, in an immediate proximity of Cys139. The mutual disposition of the two amino acids is very similar to that seen in sortases [Fig. 4(B)]. Arg141 is conserved in all sequences of the ErfK/YbiS/YhnG domains (Fig. 1). Thus, ykuD and its homologs appear to have within a short stretch of amino acids all the active site components found in cysteines-dependent proteinases and transpeptidases: a triad of Cys, His, and an H-bond acceptor stabilizing the imidazol, a well- formed oxyanion hole stabilizing the tetrahedral intermediate, as well as a positively charged residue enhancing the nucleophilicity of the Cys. It is tempting to speculate that the structural similarities between ykuD constitute a reflection of functional parallels.
Substrate Specificity and Biological Function of ykuD
The biological function of ykuD, or indeed of any of its homologs containing the ErfK/YbiS/YhnG domain, remains unknown. However, the presence of the LysM domain in ykuD indicates that the enzyme may be active when bound to peptidoglycan, although we note that in a number of proteins containing the ErfK/YbiS/YhnG domains the LysM domain is absent. It is known that ykuD is expressed in B. subtilis among spore proteins, and that its transcription is regulated by the σK factor, which is active primarily during bacterial sporulation.32 However, the function of the protein cannot be exclusive to sporulation, because it is found in a variety of Gram-negative bacteria as well.
To our knowledge, there are no structures known for Cys-dependent, triad-containing esterases, thioesterases, or lipases. It appears that cysteines only function in proteases and transpeptidases. The stereochemistry of the putative active site in ykuD, and in particular the presence of a well-developed oxyanion hole, supports the notion that the enzyme utilizes a general base mechanism and undergoes acylation. This is consistent with either a peptidase (protease) activity or a transpeptidase activity. Although we were unable to confidently model an acceptor peptide (such as pentaglycine) into the active site, there appears room for such an acceptor, so we cannot rule out that the enzyme in fact is a transpeptidase similar to sortases, anchoring proteins in the spore’s coat, and performing similar function in the cell wall of Gram-negative bacteria.
Further studies are necessary to answer all the pending questions regarding the function and activity of the ErfK/YbiS/YhnG domains. The determination of the ykuD crystal structure is an important step in this process and paves the way for the future studies of this family of proteins. Once a substrate is identified and a functional assay is available, it will be possible to dissect the structure–function relationships in the putative active site using site-directed mutagenesis.
Crystal Contacts
The determination of the crystal structure of ykuD has been possible due to the application of the conformational surface entropy reduction concept, because the wild-type protein did not crystallize. It is therefore instructive to analyze the crystal contacts and assess the impact of surface mutagenesis on the lattice-forming interactions.
There are extensive contacts crystallographically related to dimers in the crystal structure, but there are specifically two crystal contacts that appear to stem directly from the surface mutations and crystallization conditions. The first contact is formed between two polypeptide chains related by noncrystallographic symmetry that belong to two adjacent noncrystallographic dimers (Fig. 3). Not surprisingly, it involves the two mutated residues—Ala117 and Ala118. These amino acids are located, as predicted, on a solvent-exposed loop. Although the two alanines are not involved in any direct intermolecular interactions, the wild-type side-chains of Lys and Gln within this loop would have almost certainly caused a steric obstacle, preventing two ykuD molecules from interacting. The third residue involved in this contact is His119, which coordinates a cadmium ion (Nϵ2–Cd2+ distance of 2.27 Å) trapped between two protein molecules at the active site. The second crystal contact is mediated by sulfate ion sequestered between two arginines (Arg159) in the center of the noncrystallographic dimer. The distances between sulfate oxygen and Nη1 of arginines from molecule A and molecule B are 2.78 Å and 2.65 Å, respectively. Moreover, Nη2 of Arg159 form molecule A creates one more hydrogen bond (3.40 Å) with sulfate oxygen. This interaction is additionally stabilized by His57 from molecule A and by Tyr96 from molecule B.
Successful crystallization of ykuD depends on the mutations (wild-type protein does not crystallize), as well as the presence of both sulfate and Cd2+ ions in the precipitant. The structure shows that the sulfate ions help stabilize a noncrystallographic dimer, while the Cd2+ ions and mutations are critical in forming cross bridges between the dimers generating layers of molecules perpendicular to the c axis. We note that this crystal architecture has similarities to other successful cases of surface mutagenesis, where noncrystallographic dimers are found in the asymmetric unit.33
CONCLUSION
This is the first crystallographic structural study of a representative of a ubiquitous family of exclusively prokaryotic proteins that contain both the LysM module, which mediates interactions with cell wall peptidoglycan, and the ErfK/YbiS/YhnG domain. The structure of the B. subtilis ykuD reveals a well-formed putative catalytic site, identifying potential functional roles for specific conserved residues such as His123, Cys139, and Arg141. The stereochemistry of the active site strongly suggests that the enzyme is a peptidase or transpeptidase, although we cannot be certain of the substrate. The presence of ykuD homologs in a variety of pathogenic bacteria, and the likelihood that these proteins play a role in the maintenance of cell wall, will hopefully lead to a thorough functional characterization of these interesting enzymes.
Acknowledgments
Grant sponsor: National Institute of General Medical Sciences; Grant numbers: GM62615 (to Z. S. Derewenda) and P-50-GM62414 (to A. Joachimiak).
REFERENCES
- 1.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- 3.Spaink HP. Specific recognition of bacteria by plant LysM domain receptor kinases. Trends Microbiol. 2004;12:201–204. doi: 10.1016/j.tim.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 6.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 7.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 8.de la Fortelle E, Bricogne G. Maximum-likelihood heavy atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 9.Terwilliger TC. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr D Biol Crystallogr. 2003;59:45–49. doi: 10.1107/S0907444902018048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 11.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 12.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 13.Laskowski RA, McArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:282–291. [Google Scholar]
- 14.Holm L, Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 16.Lewis SD, Johnson FA, Shafer JA. Effect of cysteine-25 on the ionization of histidine-159 in papain as determined by proton nuclear magnetic resonance spectroscopy: evidence for a his-159–Cys-25 ion pair and its possible role in catalysis. Biochemistry. 1981;20:48–51. doi: 10.1021/bi00504a009. [DOI] [PubMed] [Google Scholar]
- 17.Huang C, Wei P, Fan K, Liu Y, Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 18.Sprang S, Standing T, Fletterick RJ, Stroud RM, Finer-Moore J, Xuong NH, Hamlin R, Rutter WJ, Craik CS. The three-dimensional structure of Asn102 mutant of trypsin: role of Asp102 in serine protease catalysis. Science. 1987;237:905–909. doi: 10.1126/science.3112942. [DOI] [PubMed] [Google Scholar]
- 19.Schrag JD, Li Y, Wu S, Cygler M. Ser-His-Glu triad forms the catalytic site of the lipase from Geotrichum candidum. Nature. 1991;351:761–764. doi: 10.1038/351761a0. [DOI] [PubMed] [Google Scholar]
- 20.Wei Y, Contreras JA, Sheffield P, Osterlund T, Derewenda U, Kneusel RE, Matern U, Holm C, Derewenda ZS. Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase. Nat Struct Biol. 1999;6:340–345. doi: 10.1038/7576. [DOI] [PubMed] [Google Scholar]
- 21.Serre L, Verbree EC, Dauter Z, Stuitje AR, Derewenda ZS. The Escherichia coli malonyl-CoA:ACP transacylase at 1.5Å resolution. J Biol Chem. 1995;270:12961–12964. doi: 10.1074/jbc.270.22.12961. [DOI] [PubMed] [Google Scholar]
- 22.Wenig K, Chatwell L, von Pawel-Rammingen U, Bjorck L, Huber R, Sondermann P. Structure of the streptococcal endopeptidase IdeS, a cysteine proteinase with strict specificity for IgG. Proc Natl Acad Sci USA. 2004;101:17371–17376. doi: 10.1073/pnas.0407965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 Å resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews DA, Alden RA, Birktoft JJ, Freer ST, Kraut J. X-ray crystallographic study of boronic acid adducts with subtilisin BPN′ (Novo): a model for the catalytic transition state. J Biol Chem. 1975;250:7120–7126. [PubMed] [Google Scholar]
- 25.Martinez C, Nicolas A, van Tilbeurgh H, Egloff MP, Cudrey C, Verger R, Cambillau C. Cutinase, a lipolytic enzyme with a preformed oxyanion hole. Biochemistry. 1994;33:83–89. doi: 10.1021/bi00167a011. [DOI] [PubMed] [Google Scholar]
- 26.Novick RP. Sortase: the surface protein anchoring transpeptidase and the LPXTG motif. Trends Microbiol. 2000;8:148–151. doi: 10.1016/s0966-842x(00)01741-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R, Wu R, Joachimiak G, Mazmanian SK, Missiakas DM, Gornicki P, Schneewind O, Joachimiak A. Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site. Structure. 2004;12:1147–1156. doi: 10.1016/j.str.2004.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem. 2004;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 29.Zong Y, Mazmanian SK, Schneewind O, Narayana SV. The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall. Structure. 2004;12:105–112. doi: 10.1016/j.str.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Dessen A. A new catalytic dyad regulates anchoring of molecules to the Gram-positive cell wall by sortases. Structure. 2004;12:6–7. doi: 10.1016/j.str.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Marraffini LA, Ton-That H, Zong Y, Narayana SV, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus: a conserved arginine residue is required for efficient catalysis of sortase A. J Biol Chem. 2004;279:37763–37770. doi: 10.1074/jbc.M405282200. [DOI] [PubMed] [Google Scholar]
- 32.Kodama T, Takamatsu H, Asai K, Ogasawara N, Sadaie Y, Watabe K. Synthesis and characterization of the spore proteins of Bacillus subtilis YdhD, YkuD, and YkvP, which carry a motif conserved among cell wall binding proteins. J Biochem. 2000;128:655–663. doi: 10.1093/oxfordjournals.jbchem.a022798. [DOI] [PubMed] [Google Scholar]
- 33.Devedjiev Y, Surendranath Y, Derewenda U, Gabrys A, Cooper DR, Zhang RG, Lezondra L, Joachimiak A, Derewenda ZS. The structure and ligand binding properties of the B.subtilis YkoF gene product, a member of a novel family of thiamin/HMP-binding proteins. J Mol Biol. 2004;343:395–406. doi: 10.1016/j.jmb.2004.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]