Abstract
Structural proteomics projects are generating three-dimensional structures of novel, uncharacterized proteins at an increasing rate. However, structure alone is often insufficient to deduce the specific biochemical function of a protein. Here we determined the function for a protein using a strategy that integrates structural and bioinformatics data with parallel experimental screening for enzymatic activity. BioH is involved in biotin biosynthesis in Escherichia coli and had no previously known biochemical function. The crystal structure of BioH was determined at 1.7 Å resolution. An automated procedure was used to compare the structure of BioH with structural templates from a variety of different enzyme active sites. This screen identified a catalytic triad (Ser82, His235, and Asp207) with a configuration similar to that of the catalytic triad of hydrolases. Analysis of BioH with a panel of hydrolase assays revealed a carboxylesterase activity with a preference for short acyl chain substrates. The combined use of structural bioinformatics with experimental screens for detecting enzyme activity could greatly enhance the rate at which function is determined from structure.
The protein complement of both prokaryotes and eukaryotes remains largely uncharacterized. At least 30% of all proteins have no known biochemical function, and a larger percentage have sequence similarity to proteins of known biochemical activity (e.g. most predicted protein kinases) but for which the physiological role is unknown. The challenge in the post-genomic era is to define both the biochemical and physiological functions of all proteins as rapidly as possible.
Structural proteomics, the large scale determination of protein structure, is expected to provide insight into the fundamental mechanisms by which a protein sequence adopts a defined three-dimensional structure. Most of the organized efforts in structural proteomics (Ref. 1; rcsb.org/pdb/strucgen.html) specifically target protein sequences for which there is no known structural homologue in the public data bases at a level of 30% sequence identity. One aim of this effort is to more fully define the universe of protein folds. Importantly, because protein structure is often conserved in the absence of detectable sequence homology, the comparison of new protein structures with those of known proteins will likely provide clues to biochemical function.
The discovery of biochemical function from a new protein structure begins with automated searches for structural homologues of known function. The results of these comparisons are provided as lists with significance scores. The methods of comparison are now used routinely in the structural community and have proved invaluable for detecting structural conservation and for providing the basis for hypotheses (2). However, the interpretation of the results from structural comparisons often consumes a significant amount of time and is influenced by the extent to which the investigator is able to scour the literature.
In an effort to improve the process by which function is derived from structure, we have combined two methods to facilitate functional studies. First, we have employed a data base of structural templates derived from the active sites of 189 different classes of enzymes.1 This exploits the fact that the chemistry of the reaction restricts the types and the topological arrangement of the catalytic amino acids and hence results in strong conservation of their spatial arrangement, even where the protein folds are very different (3). By focusing on the catalytic moieties, functional similarities can be detected in cases where there is no similarity in sequence, fold, or secondary structure. Second, we have created and used a panel of generic biochemical assays to test the functional hypotheses raised by the structural comparisons. These assays are based on simple, often nonphysiological, substrates; the experiment is designed to reveal the chemistry of the active site and not the cellular substrate.
Here we present the results of the combined structural, bioinformatic, and enzymatic analysis of Escherichia coli BioH, a target within the Midwest Center for Structural Genomics (www.mcsg.anl.gov). By comparing the crystal structure of BioH with other known enzymes, we found that BioH is a member of the protein hydrolase superfamily and contains a classical Ser-His-Asp catalytic triad. A screen with different hydrolase substrates revealed that BioH has significant carboxylesterase activity, with a preference for short acyl chain substrates, and weak thioesterase activity. The strategy used for BioH might facilitate analysis of novel, uncharacterized proteins and structures arising form structural proteomics projects.
EXPERIMENTAL PROCEDURES
BioH Expression and Purification
The open reading frame of bioH was amplified by PCR from E. coli DH5α genomic DNA. The gene was cloned as previously described (4) into the NdeI and BamHI sites of a modified form of pET15b (Novagen) in which a TEV protease cleavage site replaced the thrombin cleavage site and a double stop codon was introduced downstream from the BamHI site. The fusion protein was overexpressed and purified using nickel affinity chromatography as previously described (4).
For the preparation of the selenomethionine enriched protein, BioH was expressed in the E. coli methionine auxotroph strain B834 (DE3) (Novagen) in supplemented M9 medium. The sample was prepared under the same conditions as the native protein except for the addition of 5 mm 2-mercaptoethanol to the purification buffers.
Crystallization
BioH was crystallized by vapor diffusion in hanging drops (ratio of 2 µl of protein to 2 µl of precipitant) equilibrated against reservoir containing 1.2 m sodium citrate trihydrate and 0.1 m Tris-HCl (pH 8.0). X-ray quality crystals grow at 21 °C in 2–5 days. For diffraction studies, the crystals were stabilized with the crystallization buffer supplemented with 15% ethylene glycol as a cryoprotectant and flash frozen in liquid nitrogen.
Mass Spectrometry
All of the mass spectrometry data were acquired and analyzed using Masslynx 3.5 (Micromass, Manchester, UK). Electrospray ionization mass spectrometry (ESI-MS)2 was performed on a Micromass Q-Tof2 mass spectrometer. Positive ion mode ESI-MS of the whole protein was achieved in 50:50 acetonitrile:water with 0.1% formic acid. Exact mass MS was performed in negative ion mode regular ESI-MS using 10% aqueous methanol containing 1% ammonia as a carrier solvent. Tryptic digestions were performed overnight in 100 mm ammonium bicarbonate (pH 7.8) or in 100 mm ammonium bicarbonate buffer (pH 6.4) for 1.5 h followed by MALDI-MS analysis. MALDI-MS was performed on a Micro-mass MALDI-R mass spectrometer (Micromass) using an m/z range of 500–4000. ESI-MS and MS/MS analysis of the low pH tryptic digest were performed on a Micromass Q-Tof2 mass spectrometer using nano-LC with a C18 column (0.3 × 5 mm; LC Packings). Data-dependent acquisition parameters were set to select the doubly and triply charged unmodified and modified precursor ions corresponding to residues 78–100 of the protein. MS-MS spectra were processed by base-line subtraction and deconvoluted using the Max-Ent3 module of MassLynx 3.5. The peptide sequences were determined semi-automatically from the resulting singly charged, deisotoped spectra using PepSeq, version 3.3 supplied with MassLynx 3.5.
Enzyme Assays
Rapid screening for enzyme activities were performed using the following procedures: (a) fatty acid esterase activity was measured spectrophotometrically at 37 °C using p-nitrophenyl (pNP) acetate or pNP esters of other fatty acids (C3–C18) as substrates (5), (b) thioesterase activity was measured spectrophotometrically using CoA thioesters of fatty acids (acetyl-CoA, malonyl-CoA, and palmitoyl-CoA) as described earlier (6), (c) lipase activity (with sonicated olive oil as substrate) was measured spectrophotometrically by the copper soap assay after extraction of released free fatty acids with chloroform: heptane:methanol mixture (7), (d) protease activity was measured using l-leucine p-nitroanilide (aminopeptidase activity) or Nα-benzoyl-l-arginine p-nitroanilide (trypsin-like endopeptidase activity) as described (8, 9), (e) phosphatase activity was determined spectrophotometrically using 5 mm p-nitrophenyl phosphate in 50 mm HEPES-K (pH 7.5) buffer at 37 °C (10), and (f) bromoperoxidase activity was measured spectrophotometrically with phenol red or monochlorodimedon as described previously (11).
Crystallographic Data Collection
A two-wavelength multiple-wave-length anomalous dispersion experiment was carried out on the 19ID line of the Structural Biology Center at Advanced Photon Source (Argonne, IL). All of the crystallographic data were collected at 110 K on one crystal containing selenomethionine-substituted protein. The crystal belongs to the tetragonal space group P43 with unit cell dimensions a = b = 75.2 Å, c = 49.3 Å, α = β = γ = 90°. The multiple-wavelength anomalous dispersion data set was colleted using inverse beam strategy at the selenium absorption peak energy (0.97947 Å) and at a remote wavelength (0.95373 Å). The absorption edge was determined from the x-ray fluorescence spectrum and the f′ and f″ plots versus energy obtained with the program CHOOCH (12). High resolution data were collected from the unexposed part of the same crystal, which had been stored in liquid nitrogen. All of the data were measured with the CCD detector (13) 210 × 210-mm2 sensitive area and fast duty cycle. Control of the experiment, data collection and visualization was done with d*TREK (14), and all of the data were integrated and scaled with the program package HKL2000 (15). Some of the basic statistics of data collection and processing are given in Table I.
TABLE I.
Basic statistics of data collection and processing
| Number of residues/A.U. | 256 | ||
| Number of selenomethionine/A.U. | 6 | ||
| Number of molecules/A.U. | 1 | ||
| Crystal lattice | P43 a = b = 75.21Å, c = 49.26Å, α = β = γ = 90° | ||
| Crystal 1 (MAD) | Crystal 1 | ||
| Peak | Remote | High | |
| Wavelength (Å) | 0.97947 | 0.95373 | 1.03321 |
| Resolution (Å) | 50.0–1.87 | 50.0–1.82 | 50.0–1.63 |
| Number of observationsa | 334548 | 364998 | 114995 |
| Number of unique reflectionsa | 44558 | 49079 | 33538 (3061) |
| Completeness (%)b | 99.9 (99.8) | 99.4 (94.8) | 96.8 (82.5) |
| I/σ(I)b | 22.2 (3.0) | 22.3 (2.0) | 16.5 (1.1) |
| Rsymb | 0.11 (0.50) | 0.108 (0.66) | 0.075 (0.595) |
Bijvoet pairs for scaling the MAD data sets were kept separately.
In the last resolution shell.
Structure Determination
Multiple-wavelength anomalous dispersion phasing of BioH data was carried out with the program CNS (16). Experimental phases were extended from 2.5 to 2.0 Å resolution with density modification, using data collected at the f″ peak wavelength. With these improved phases, the initial model was built with the program ARP/wARP (17). The high quality of the phases allowed 94% of the main chain to be built automatically and most of the side chains to be placed with a confidence level of 79%. The remainder of the model was built, and all of the side chains were corrected manually using the program O (18). This model was then refined against the 1.7 Å resolution data with several macro cycles of CNS, including simulated annealing, B-factor, and positional refinements. After each macro cycle, the model was inspected, and corrections and/or additions were made manually, with the programs O and QUANTA (Accelrys, Inc.). All subsequent refinement was carried out with REFMAC (19) within the CCP4 (20) suite of programs. The phasing and refinement parameters are shown in Table II.
TABLE II. Phasing and refinement statistics for BioH structure.
FOM after phase extension with density modification in 50–2.0 Å shell was 0.95 (0.92).
| Phasing wavelength | Resolutiona | Number of reflectionsa |
Phasing powera |
FOMa | |
|---|---|---|---|---|---|
| Å | |||||
| Peak | 42–2.5 (2.6–2.5) | 18206 (2027) | 2.92 (2.28) | 0.50 (0.44) | |
| Remote | 42–2.5 (2.6–2.5) | 18210 (2024) | 2.18 (1.67) | 0.41 (0.35) | |
| Overall | 42–2.5 (2.6–2.5) | 18414 (2047) | 0.72 (0.63) | ||
| Refinement | |||||
| Resolution (Å) | 75–1.70 (1.79–1.70) | ||||
| Number of reflections | 27141 (3631) | ||||
| R factor (%) | 14.7 (21.2) | ||||
| R free (%) | 18.9 (24.6) | ||||
| Correlation | 97.1 | ||||
| Correlation free | 95.2 | ||||
| Number of all atoms | 2419 | ||||
| Number of solvent atoms | 242 | ||||
| Mean B factor | 15.13 | ||||
| Deviations from ideal | Refined | Target | |||
| Covalent bonds | 0.022 | 0.021 | |||
| Bond angles | 1.905 | 1.950 | |||
| Planarity | 0.011 | 0.020 | |||
| Chiral centers | 0.131 | 0.20 | |||
| Torsion angle 1 | 6.4 | 5.0 | |||
| Torsion angle 3 | 18.74 | 15.0 | |||
| VDW contacts | 0.261 | 0.20 | |||
Last resolution shell.
Coordinates
The coordinates have been deposited in the Protein Data Bank under accession code 1M33.
RESULTS AND DISCUSSION
The BioH Crystal Structure
The final model of the BioH crystal structure consists of 256 residues, two molecules of ethylene glycol, and 240 water molecules. The last two residues of the model, Gly257 and Ser258, were appended to the protein as a result of the cloning strategy. The first two residues of the native sequence, Met1 and Asn2, were not included because of the absence of the corresponding electron density. Two molecules of ethylene glycol from the cryoprotectant were bound to the protein molecule mostly because of hydrophobic interactions. Residue 100 was unambiguously identified as Arg, instead of Gln, in electron density maps and is likely a PCR-induced mutation. The side chains of residues Glu116, Lys121, Asp123, Phe136, Glu152, and Lys213 have incomplete electron density.
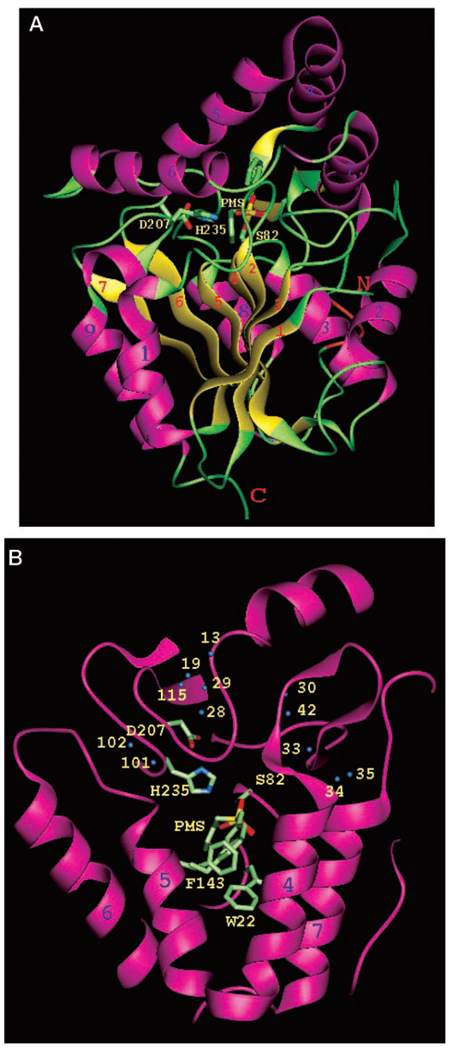
BioH is a two-domain protein (Fig. 1A). The α/β/γ three layer sandwich of the large domain (residues 5–109 and 188–256; see below) consists of a twisted β-sheet formed by seven mostly parallel strands β1↓ (residues 5–9), β3↑ (residues 14–19), β2↑ (residues 41–46), β4↑ (residues 76–81), β5↑ (residues 101–105), β6↑ (residues 198–203) and β7↑ (residues 225–230) and flanked on both sides by five α-helices α1 (residues 31–39), α2 (residues 60–70), α3 (residues 83–94), α8 (residues 215–222), and α9 (residues 237–252). Ile32 and Pro242 introduce ~90° kinks into the first and last helices, respectively. This domain resembles the Rossman fold, which is commonly found in enzymes.
FIG. 1. Structure of BioH.
A, the overall folding of BioH molecule as viewed along the b-sheet. The α-helices and the β-strands are numbered, and the termini are labeled. The catalytic, α/b/a domain (Rossman fold) consists of a seven strand β sheet (yellow) surrounded by α-helices (magenta). The auxiliary, α only domain consists of four α-helices (helices 4–7). The rest of the molecule is shown in green. Also shown as a thick wire model are the residues of the catalytic triad and parts of the inhibitor PMSF. B, the auxiliary domain viewed from above the molecule. Four α-helices of the V-shaped double bend and the catalytic residues Ser82, His235, and Asp207 are shown along with disordered inhibitor PMSF, Trp22, and disordered Phe143. The blue spheres represent solvent molecules around the catalytic site. The orientations in A and B are related by ~90° rotation around the horizontal axes on the paper.
A small auxiliary domain is formed by the C-terminal segment of the polypeptide chain (Cys110–Asp187) and is inserted into the catalytic domain. The auxiliary domain contains four α-helices, residues 122–134 (α4), 136–145 (α5), 155–166 (α6), and 173–185 (α7), that create a bundle of two V-shaped bends (Fig. 1B). The two domains are connected by a hinge region near Cys110 and Asp187. The interface between domains is stabilized by multiple hydrophobic interactions including helices α6 and α7 that run across the surface of catalytic domain and intramolecular hydrogen bonds between the carbonyl of Pro109 and the nitrogen of Leu188 and two hydrogen bonds between Asp187 and Arg189.
Automated Structural Bioinformatics Reveals a Ser-His-Asp Catalytic Triad
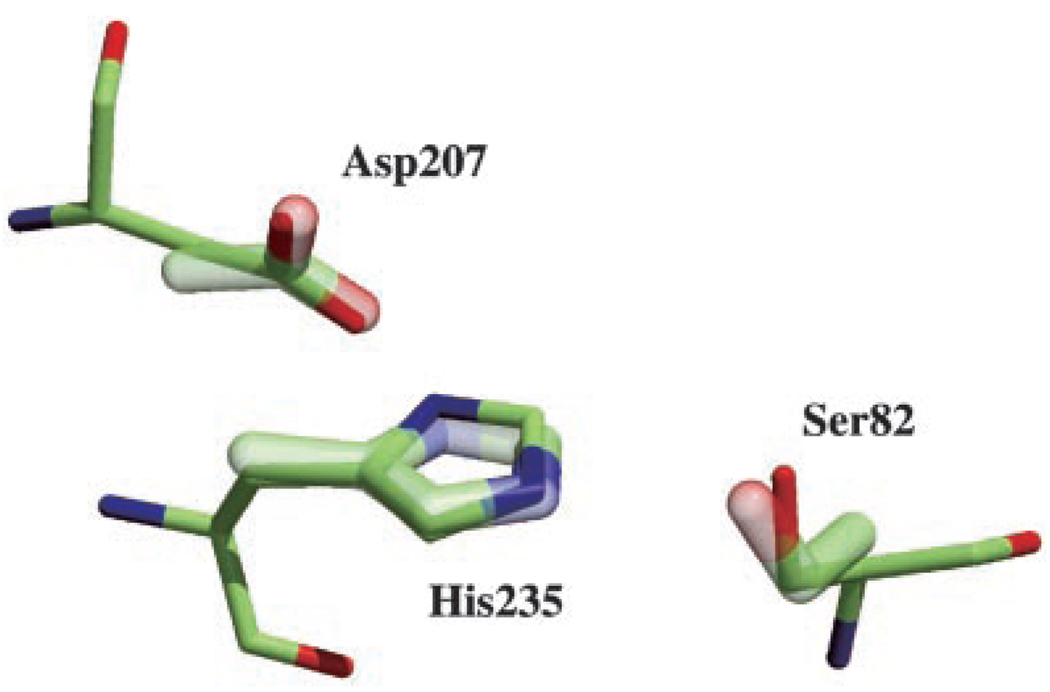
One of the aims of structural proteomics is to perform more comprehensive automated analysis of protein structures to reduce the level of time-intensive human intervention. To screen new structures for potential catalytic function, we have created a data base of ~189 three-dimensional enzyme active site structural templates.1 The BioH structure was scanned against this data base of using the TESS program (3). This automated search gave a close match of BioH to the Ser-His-Asp catalytic triad of lipases (21) (EC 3.1.1.3). The BioH residues involved (Ser82, His235, and Asp207) matched the template with a root mean square deviation of 0.28 Å for the overlaid side chains (Fig. 2). This is well within the cut-off of 1.2 Å used for discriminating true from false matches for this template. The presence of the catalytic triad suggested that BioH might possess lipase, protease, or esterase activity. Furthermore, the serine nucleophile (Ser82) is located within one of the two earlier identified Gly-Xaa-Ser-Xaa-Gly motifs (22), which is typical for acyltransferases and thioesterases.
FIG. 2. Superposition of the Ser82, His235, and Asp207 residues onto the catalytic triad template.
The template side chains are depicted by the thicker, transparent bonds, whereas the BioH residues are represented by the thinner, solid bonds and include the main chain atoms. The root mean square deviation between equivalent atoms in the template and matched side chains is 0.28 Å.
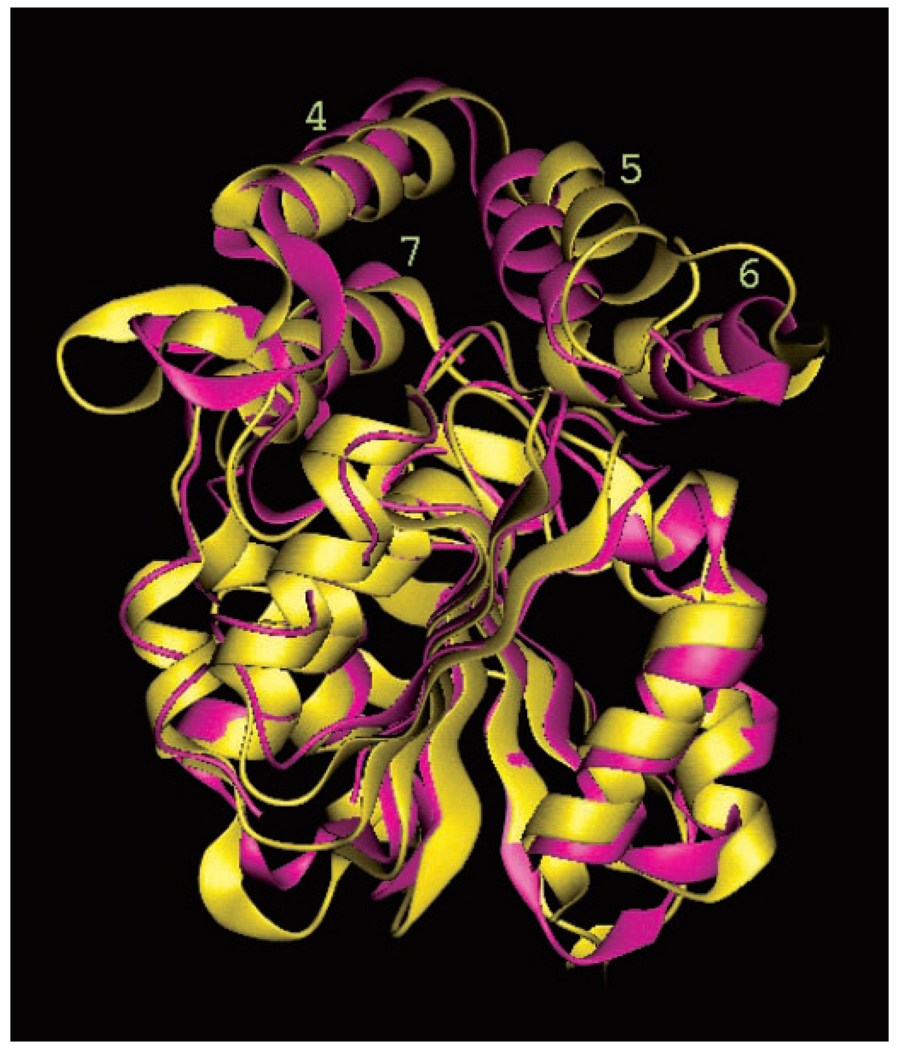
The structure of BioH was also compared with all other known structures using conventional methods such as the DALI algorithm (23). The results from the DALI search revealed structural homology to a large number of proteins with a broad range of enzymatic functions. The closest matches with strong structural similarities include a bromoperoxidase (EC 1.11.1.10; Z score, 22.6; Protein Data Bank code 1brt), an aminopeptidase (EC 3.4.11.5; Z score, 21.1; Protein Data Bank code 1qtr), two epoxide hydrolases (EC 3.3.2.3; Z scores, 20.5 and 18.2; Protein Data Bank codes 1ehy and 1cr6, respectively), two haloalkane dehalogenases (EC 3.8.1.5; Z scores, 20.2 and 16.2; Protein Data Bank codes 1bn6 and 1b6g, respectively), and a lyase (EC 4.2.1.39; Z score, 17.2; Protein Data Bank code qj4). A comparison of BioH with a chloroperoxidase (EC 1.11.1.10) is shown in Fig. 3. The sequence identities between BioH and these proteins range from 15 to 25% and therefore do not suggest a specific catalytic function for BioH. Further manual analysis of these enzymes and literature review would have revealed to the expert that each contains a Ser-His-Asp catalytic triad in their active sites.
FIG. 3. Superposition of the E. coli BioH (magenta) and Streptromyces aureofaciens chloroperoxidase (yellow) structures; superposition of the catalytic domains, including the loops connecting secondary structure elements.
The overall architecture of the auxiliary domains is the same for both proteins, although the placement of the helices, especially of α5 and α6, relative to the catalytic domain differs.
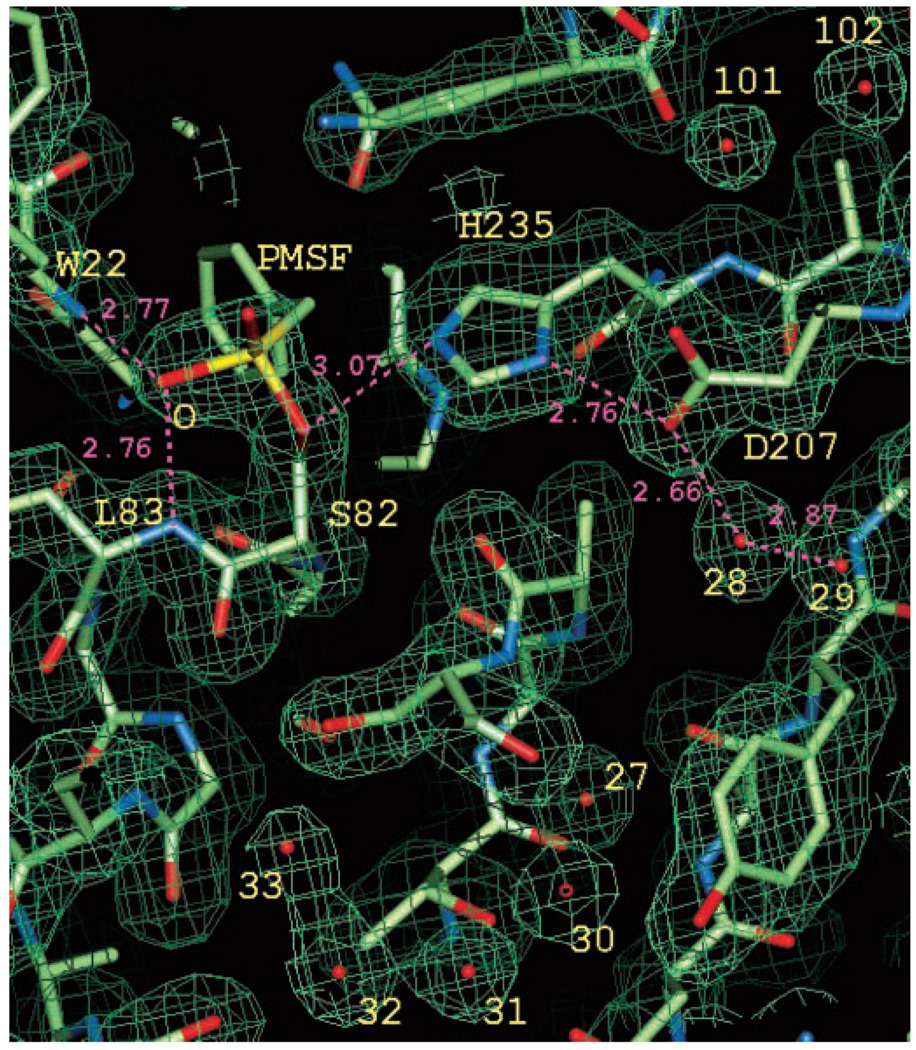
Ser82 Is Covalently Modified by a Hydrolase Inhibitor
The structural informatics provided initial evidence for the location of the BioH catalytic site. The experimental density maps also showed an unusual feature that extended from the side chain of Ser82 (Fig. 4). The shape of the density and its environment, insinuated that the corresponding compound was covalently attached to the Oγ atom of Ser82 and formed hydrogen bonds with the backbone nitrogens of Trp22 and Leu83. To investigate the properties of the Ser82 modification, we analyzed the full length and trypsinized BioH with mass spectroscopy. Under denaturing conditions, two major peaks were observed with molecular masses of 29,152 Da (corresponding to the full-length protein) and 29,306 Da with similar intensity. Treatment of the protein with mild base caused the peak at 29,306 Da to disappear over time and the peak at 29,152 Da to increase in relative intensity. In addition, a new peak was detected with mass of 172 Da, interpreted as singly hydrated 154-Da molecule (see below). We also examined the mass of the tryptic fragment of BioH that contains Ser82. When the tryptic digestion was done under slightly acidic conditions and examined by both MALDI and ESI-MS, only the Ser82-containing fragment showed a 154-Da adduct. Therefore the catalytic potential of Ser82 seems responsible for observed additional mass attached to Ser82.
FIG. 4. Experimental electron density maps after density modification.
Residues of the catalytic triad, Ser82, Asp207, and His235 of the refined model are labeled along with Trp22, Leu83, and several solvent molecules (labeled with numbers only). Note the additional electron density extending from the Oγ atom of Ser82. According to biochemical data, this density was interpreted as PMSF, but only parts of it are visible because of disorder and partial occupancy. Several functionally important hydrogen bonds, including those between the sulfonate oxygen of the inhibitor and backbone nitrogens of Trp22 and Leu83 (oxyanion hole) are shown with magenta dashed lines and numbers.
For crystallographic experiments and initial MALDI and ESI-MS, the BioH protein was purified in the presence of protease inhibitor phenylmethylsulfonyl fluoride (PMSF), which is known to react with the catalytic serine in hydrolases (24) and form a stable covalent adduct. Therefore it appears that BioH was modified during purification The protein purified in the absence of PMSF did not reveal this modification. These results strongly suggest that the modification corresponds to the addition of PMSF (expected Δm = 154) at Ser82 and that the serine possesses nucleophilic properties.
BioH Is a New Carboxylesterase in E. coli
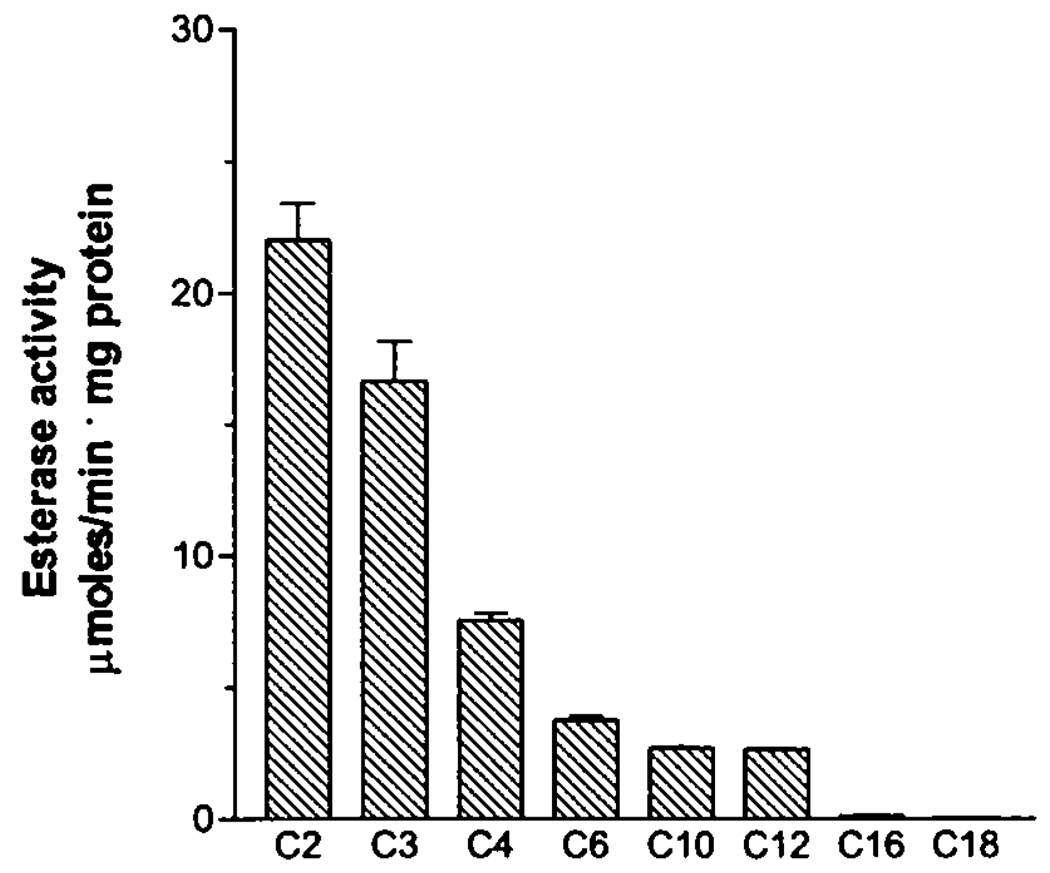
BioH purified in the absence of PMSF was subjected to several enzymatic assays that focused on hydrolase function including carboxylesterase, lipase, thioesterase, phosphatase, endopeptidase, aminopeptidase, and bromoperoxidase. BioH demonstrated significant carboxylesterase activity (Table III) (EC 3.1.1.1) and hydrolyzed p-nitrophenyl esters of fatty acids. The enzyme showed rather narrow pH optimum (8.0–8.5) and broad substrate specificity with a preference for short chain substrates (Fig. 5). The kinetic parameters of BioH were determined for several substrates (Table III). These results demonstrate that although BioH was most active with pNP-acetate, the Km for all C-2–C-6 substrates was essentially the same. In agreement with the results of the mass spectrometry and crystallography, BioH was strongly inhibited by PMSF (10.5% of residual activity after 10 min of incubation with 2 mm PMSF). Purified BioH showed classical Michaelis-Menten kinetics, and linear double reciprocal plots were obtained for all of the pNP substrates tested (data not shown).
TABLE III.
Steady state kinetic parameters for E. coli BioH carboxylesterase activity with various substrates
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | m−1 s−1 | |
| pNP-acetate (C2) | 0.29 ± 0.04 | 18.5 ± 1.7 | 63.8 × 103 |
| pNP-propionate (C3) | 0.35 ± 0.08 | 13.1 ± 1.2 | 37.4 × 103 |
| pNP-butyrate (C4) | 0.33 ± 0.06 | 6.1 ± 0.7 | 18.5 × 103 |
| pNP-caproate (C6) | 0.25 ± 0.02 | 4.0 ± 0.1 | 16.0 × 103 |
| pNP-laurate (C12) | 0.60 ± 0.13 | 1.5 ± 0.2 | 2.5 × 103 |
FIG. 5. Carboxylesterase activity of BioH on p-nitrophenyl esters with various acyl chain lengths.
Equal amounts of protein (0.3 µg) were incubated with saturating concentrations (0.6 mm) of several p-nitrophenyl esters: C2, pNP-acetate; C3, pNP-propionate; C4, pNP-butyrate; C6, pNP-caproate; C10, pNP-caprate; C12, pNP-laurate; C16, pNP-palmitate; C18, pNP-stearate. Each bar represents an average of the results from at least four independent determinations, with standard deviations indicated by error bars.
Purified BioH showed low enzymatic activities for thioesterase (using palmitoyl-CoA as a substrate; 186.5 ± 18.6 nmol/min/mg protein), lipase (using olive oil; 18.5 ± 1.3 nmol/min/mg protein), and aminopeptidase (using leucine-p-anilide as a substrate; 3.8 nmol/min/mg protein) and showed no detectable enzymatic activity for phosphatase (using p-nitrophenyl phosphate as a substrate), trypsin-like endopeptidase (using benzoyl-arginine-p-nitroanilide as a substrate), or bromoperoxidase (phenol red and monochlorodimedone as potential substrates).
Our data combined with results reported in the literature suggest that BioH represents a novel carboxylesterase in E. coli. E. coli is known to express at least three other proteins with carboxylesterase activity: carboxylesterase YbaC (25), thioesterase TesA (26, 27), and thioesterase TesB (28). BioH shows no significant sequence similarity with these enzymes (data not shown). BioH also possessed different enzymological properties compared with the other enzymes. Compared with BioH, YbaC and TesA exhibit higher affinities for the long chain fatty acids, pNP-octanoate (C8) and pNP-decanoate (C10). The specific activity of BioH for short C2 or C3 substrates was in the same range as for YbaC and at least 10–30 times lower as compared with TesA. Both BioH and TesA also displayed thioesterase activity with palmitoyl-CoA (however ~13 times lower for BioH) but show no activity with acetyl-CoA as a substrate. The ratio of carboxylesterase/thioesterase activities (with pNP-palmitate/palmitoyl-CoA) was 0.3 for TesA and 1.3 for BioH.
The specificity for the short chain fatty acid esters likely arises from the fact that the catalytic site of BioH is buried between two domains (Fig. 1) and is not readily accessible for bulkier compounds. Substrates with acyl chain length of up to 6 carbons (C-2–C-6) could be accommodated within the hydrophobic crevice in the V-shaped cap domain of BioH where the invariant Phe143 (Fig. 1B) can act as a facilitator of binding. In fact, the walls of the active site are quite hydrophobic; therefore binding of acyl substrates to BioH is likely to be mediated mostly by hydrophobic interactions, and the active site is sufficiently large to accommodate short chain substrates with very similar affinities for the C-2–C-6 range. This is consistent with the observation that BioH shows essentially same Km for C-2–C-6 substrates (Table III).
A Possible Role for BioH in Biotin Biosynthesis
In microorganisms and plants, biotin is synthesized from pimeloyl-CoA by the enzymes BioF, BioA, BioD, and BioB in a conserved fourstep reaction (29–31). In the Gram-negative bacteria, such as E. coli, pimeloyl-CoA is produced from l-alanine and/or acetate (32) using the BioC and BioH proteins (33), whose exact biochemical roles have not been elucidated. The bioC gene is widely distributed in bacteria, whereas bioH is not found in many bioC-containing bacterial genomes; in these organisms, bioH appears to be complemented by other genes (bioG and bioK) (34). In some Gram-positive bacteria, such as Bacillus sphaericus and Bacillus subtilis, pimeloyl-CoA is produced from pimelic acid by pimeloyl-CoA synthetase (BioW) (35, 36). Efforts to identify the precursors of pimeloyl-CoA in E. coli using 13C NMR labeling studies have been inconclusive (32, 37) but preclude a pimelic acid intermediate. Most studies support a mechanism based on the condensation of acetyl-CoA or malonyl-CoA moieties into pimeloyl-CoA (38). Consistent with this model, Lemoine et al. (22) identified two Gly-Xaa-Ser-Xaa-Gly motifs in BioH that are characteristic of acyl-transferase and thioesterase proteins. BioH was suggested to transfer pimeloyl units from BioC directly to CoA, and the E. coli BioC protein may function as an acyl-carrier protein involved in pimeloyl-CoA synthesis. The discovery of a BioH-CoA complex (by liquid chromatography-mass spectrometry) (39) supports a role for BioH as a CoA donor to a pimeloyl-acyl-carrier protein (or pimeloyl-BioC), releasing pimeloyl-CoA.
Our biochemical and structural data are consistent with the current model of the BioH reaction, which proposes that BioH transfers pimeloyl units from pimeloyl-BioC to CoA (22) and therefore should possess both esterase (carboxylesterase or thioesterase) and acyltransferase activities. We demonstrated that purified BioH shows carboxylesterase and low thioesterase activities and that BioH cannot use free pimelic acid for pimeloyl-CoA synthesis. Therefore, we propose that the function of BioH is to condense CoA and pimelic acid into pimeloyl-CoA. Several surface residues, Arg138, Arg142, Arg155, Arg159, and Lys162, which are disordered in the crystal structure but are nevertheless conserved throughout many bacteria, could potentially mediate CoA binding. It is also possible that BioC, which is proposed to function as a specific pimeloyl-acyl-carrier protein in the synthesis of pimeloyl-CoA (22), may interact with BioH and facilitate the delivery of a pimeloyl unit to the BioH catalytic site.
Perspective
Three-dimensional structures are now being generated for many proteins of unknown function. In many cases, such as for BioH, the structural data combined with existing clues in the literature, or even the intuition of an experienced investigator, can point the experimentalist in the right direction to identify and confirm biochemical function. However, as structural proteomics efforts gain momentum, there will be an increase in the number of protein structures for which there is no existing body of literature. The annotation of these proteins will demand methods that do not depend on specialists who are experts in a specific area of biology. The three-dimensional structure of BioH was analyzed using several automated methods for structural comparison and also with a series of generic enzymatic assays. This approach enabled us to rapidly characterize BioH enzymatic activity and reveal a new enzyme in E. coli. The development and refinement of these combined methods will significantly increase the value of structural genomics/proteomics results in the future by assigning biochemical or enzymatic functions to complement the structural information.
Acknowledgments
We thank all members of the Structural Biology Center at Argonne National Laboratory and of the Ontario Centre for Structural Proteomics for help in conducting experiments and James J. De Voss for helpful discussions regarding BioH.
Footnotes
This work was supported by the United States Department of Energy Office of Biological and Environmental Research, the Ontario Research and Development Challenge Fund, and National Institutes of Health Grant GM 62414. This work has been created by the University of Chicago as Operator of Argonne National Laboratory under Contract W-31-109-ENG-38 with the United States Department of Energy.
The atomic coordinates and structure factors (code 1m33) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/ ).
C. Porter, manuscript in preparation.
The abbreviations used are: ESI-MS, electrospray ionization mass spectrometry; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; PMSF, phenylmethylsulfonyl fluoride; pNP, p-nitrophenyl.
REFERENCES
- 1.Stevens RC, Yokoyama S, Wilson IA. Science. 2001;294:89–92. doi: 10.1126/science.1066011. [DOI] [PubMed] [Google Scholar]
- 2.Zarembinski TI, Hung LW, Mueller-Dieckmann HJ, Kim KK, Yokota H, Kim R, Kim S-H. Proc. Natl. Acad. Sci. U. S. A. 1988;95:15189–15193. doi: 10.1073/pnas.95.26.15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace AC, Borkakoti N, Thornton JM. Protein Sci. 1997;6:2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R-G, Skarina T, Kats JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure. 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vorderwülbecke T, Kieslich K, Erdmann H. Enzyme Micriob. Technol. 1992;14:631–639. [Google Scholar]
- 6.Berge RK, Farstad M. Methods Enzymol. 1981;71:234–242. doi: 10.1016/0076-6879(81)71030-9. [DOI] [PubMed] [Google Scholar]
- 7.Nixon M, Chan SHP. Anal. Biochem. 1979;97:403–409. doi: 10.1016/0003-2697(79)90093-9. [DOI] [PubMed] [Google Scholar]
- 8.Bienvenue DL, Mathew RS, Ringe D, Holz RC. J. Biol. Inorg. Chem. 2002;7:129–135. doi: 10.1007/s007750100280. [DOI] [PubMed] [Google Scholar]
- 9.Gan Z, Marquardt RR, Xiao H. Anal. Biochem. 1999;268:151–156. doi: 10.1006/abio.1998.3053. [DOI] [PubMed] [Google Scholar]
- 10.Kuo M-H, Blumenthal HJ. Biochim. Biophys. Acta. 1961;54:101–109. doi: 10.1016/0006-3002(61)90942-8. [DOI] [PubMed] [Google Scholar]
- 11.Pelletier I, Altenbuchner J. Microbiology. 1995;141:459–468. doi: 10.1099/13500872-141-2-459. [DOI] [PubMed] [Google Scholar]
- 12.Evans G, Pettifer RF. J. Appl. Crystallogr. 2001;34:82–86. [Google Scholar]
- 13.Westbrook EM, Naday I. Methods Enzymol. 1997;276:244–268. [PubMed] [Google Scholar]
- 14.Pflugrath JW. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kuunstleve RW, Kuszewski J, Nilges M, Pannu N, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 17.Perrakis A, Morris R, Lamzin VS. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 18.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 19.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project 4. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. [Google Scholar]
- 21.Schrag JD, Li Y, Cygler M, Lang D, Burgdorf T, Hecht HJ, Schmid R, Schomburg D, Rydel TJ, Oliver JD, Strickland LC, Dunaway CM, Larson SB, Day J, McPherson A. Structure. 1997;5:187–202. doi: 10.1016/s0969-2126(97)00178-0. [DOI] [PubMed] [Google Scholar]
- 22.Lemoine Y, Wach A, Jeltsch JM. Mol. Microbiol. 1996;19:645–647. doi: 10.1046/j.1365-2958.1996.t01-4-442924.x. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. Proteins. 1994;19:165–173. doi: 10.1002/prot.340190302. [DOI] [PubMed] [Google Scholar]
- 24.Gold AM. Biochemistry. 1965;4:897–901. doi: 10.1021/bi00881a016. [DOI] [PubMed] [Google Scholar]
- 25.Kanaya S, Koyanagi T, Kanaya E. Biochem. J. 1998;332:75–80. doi: 10.1042/bj3320075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bronner WM, Bloch K. J. Biol. Chem. 1972;247:3123–3133. [PubMed] [Google Scholar]
- 27.Lee Y-L, Chen JC, Shaw J-F. Biochem. Biophys. Res. Commun. 1997;231:452–456. doi: 10.1006/bbrc.1997.5797. [DOI] [PubMed] [Google Scholar]
- 28.Naggert J, Narasimhan ML, DeVeaux L, Cho H, Randhawa ZI, Cronan JE, Jr., Green BN, Smith S. J. Biol. Chem. 1991;266:11044–11050. [PubMed] [Google Scholar]
- 29.Samols D, Thornton CG, Murtif VL, Kumar GK, Haase FC, Wood HG. J. Biol. Chem. 1988;263:6461–6464. [PubMed] [Google Scholar]
- 30.Baldet P, Alban C, Douce R. Methods Enzymol. 1997;279:327–339. doi: 10.1016/s0076-6879(97)79037-2. [DOI] [PubMed] [Google Scholar]
- 31.Demoll E. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanic B, Reznikoff WS, Riley M, Schachter M, Umbarger HE, editors. Washington, D.C.: ASM Press; 1996. pp. 704–709. [Google Scholar]
- 32.Ifuku O, Miyaoka H, Koga N, Kishimoto J, Haze S, Washi Y, Kajiwara M. Eur. J. Biochem. 1994;220:585–591. doi: 10.1111/j.1432-1033.1994.tb18659.x. [DOI] [PubMed] [Google Scholar]
- 33.Barker DF, Campbell AM. J. Bacteriol. 1980;143:789–800. doi: 10.1128/jb.143.2.789-800.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov DA, Mironov AA, Gelfand MS. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gloeckler R, Ohsawa I, Speck D, Ledoux C, Bernard S, Zinsius M, Villeval D, Kisou T, Kamogawa K, Lemoine Y. Gene (Amst.) 1990;87:63–70. doi: 10.1016/0378-1119(90)90496-e. [DOI] [PubMed] [Google Scholar]
- 36.Bower S, Perkins JB, Yocum RR, Howitt CL, Rahaim P, Pero J. J. Bacteriol. 1996;178:4122–4130. doi: 10.1128/jb.178.14.4122-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanyal I, Lee SL, Flint D. J. Am. Chem. Soc. 1994;116:2637–2638. [Google Scholar]
- 38.Lezius A, Ringelman E, Lynen F. Biochem. Z. 1963;336:510–525. [PubMed] [Google Scholar]
- 39.Tomczyk NH, Nettleship JE, Baxter RL, Crichton H, Webster SP, Campopiano DJ. FEBS Lett. 2002;513:299–304. doi: 10.1016/s0014-5793(02)02342-6. [DOI] [PubMed] [Google Scholar]