Abstract
High-throughput (HT) protein crystallography is severely impeded by the relatively low success rate of protein crystallization. Proteins whose structures are not solved in the HT pipeline owing to attrition in any phase of the project are referred to as the high-hanging fruit, in contrast to those proteins that yielded good-quality crystals and crystal structures, which are referred to as low-hanging fruit. It has previously been shown that proteins that do not crystallize in the wild-type form can have their surfaces engineered by site-directed mutagenesis in order to create patches of low conformational entropy that are conducive to forming intermolecular interactions. The application of this method to selected proteins from the Bacillus subtilis genome which failed to crystallize in the HT mode is now reported. In this paper, the crystal structure of the product of the YdeN gene is reported. Of three prepared double mutants, i.e. E124A/K127A, E167A/E169A and K88A/Q89A, the latter gave high-quality crystals and the crystal structure was solved by SAD at 1.8 Å resolution. The protein is a canonical α/β hydrolase, with an active site that is accessible to solvent.
1. Introduction
Structural genomics, which synergistically involves both NMR and X-ray crystallography, is a global effort aimed at the determination of three-dimensional protein structures on a genomic scale in a high-throughput mode (Berman et al., 2000). Of particular interest in this early phase of the program is the determination of structures of proteins believed to represent new or barely known families. Every new structure deposited in the PDB increases the accuracy of homology modeling. This in turn allows better understanding of the structure–function relationships in those proteins for which structures cannot be directly determined experimentally (Stevens, 2000). However, the effectiveness of high-throughput (HT) crystallographic analysis is impeded by several bottlenecks, including the relatively low success rate of crystallization. It is estimated that of all proteins expressed in a soluble state, only between 10 and 30% form X-ray quality crystals (Claverie et al., 2002; Sulzenbacher et al., 2002). The most recent statistics available from the Midwest Structural Genomics Center (http://www.mcsg.anl.gov) show that of all successfully purified soluble proteins, less than 40% produced crystals, of which less than 60% were X-ray grade crystals that yielded useful diffraction. Thus, the first sweep of the structural genomics program is likely to harvest only the ‘low-hanging fruit’, i.e. readily expressed, stable and easily crystallizable proteins. Unfortunately, this may leave many biologically important proteins ‘hanging high and dry’.
To circumvent one of the bottlenecks, i.e. the crystallization problem, we proposed a novel approach to crystallization of proteins based on rational surface mutagenesis to create patches with low overall conformational entropy in order to facilitate the formation of crystal contacts (Derewenda, 2004). This approach proved successful in a model system of human RhoGDI and allowed the crystallization and crystal structure determination of new proteins (Longenecker, Garrard et al., 2001; Longenecker, Lewis et al., 2001; Mateja et al., 2002). We and others have also shown that the method is useful to generate crystal forms that diffract to much higher resolution than the wild-type protein, which may prove to be of importance in drug design (Mateja et al., 2002; Munshi et al., 2003).
We are now applying this strategy to those selected targets of the Bacillus subtilis structural genomics effort which failed to crystallize in the HT pipeline. This paper describes the crystallization by surface modification and structure determination of the product of the YdeN gene from B. subtilis (Midwest Centre for Structural Genomics code APC 1086). The protein was originally thought to have a unique sequence and potentially a novel fold. Subsequent tertiary-structure prediction revealed that it shows similarities to α/β hydrolases and this information was used to design three double mutants, including the K88A/Q89A mutant, in which residues with high conformational entropy located on putative surface loops were targeted. The K88A/Q89A double mutant produced crystals of high quality that diffracted to 1.8 Å. The structure revealed that the protein is a member of the ubiquitous α/β hydrolase family and structural considerations suggest that it is a hydrolase active on a soluble ester, possibly a thioester, but is not an interfacially activated lipase. The molecular model, including H atoms and anisotropic displacement parameters, was refined to a conventional R factor of 12.4%.
2. Materials and methods
2.1. Design of mutants
The pMCSG7 expression vector containing the YdeN gene with a cleavable N-terminal hexa-His tag was obtained from the Midwest Center for Structural Genomics. Initial sequence-homology searches carried out at the beginning of the project indicated that the protein has no homologues of known structures and no identifiable domains. However, further analysis, including secondary- and tertiary-structure prediction using the Polish Bioinformatics Site Meta Server (Ginalski et al., 2003; Ginalski & Rychlewski, 2003; von Grotthuss et al., 2003), yielded results that strongly suggested that the protein belongs to the α/β hydrolase family. Despite the lack of identifiable sequence similarities, the fold-recognition algorithm generated an amino-acid alignment with bromoperoxidase A1, chloroperoxidase F, chloroperoxidase T (Hofmann et al., 1998) and 2-hydroxyl-6-oxo-6-phenylhexa-2,4-dienoic acid (HPDA) hydrolase (BphD enzyme) from Rhodococcus sp. (Nandhagopal et al., 2001). This alignment revealed the presence of three conserved canonical residues forming a putative catalytic triad, i.e. Ser, Asp and His, lending credibility to the result. As this work was in progress, similar annotation appeared in the NCBI Blast search.
Using a canonical model of an α/β hydrolase, three double mutants were designed so that the mutation sites were expected to be on solvent-exposed loops. The mutants were E124A/K127A, E167A/E169A and K88A/Q89A. The Quik-Change mutagenesis kit (Stratagene) was used to introduce the designed mutations into the expression plasmid.
2.2. Protein expression and purification
The wild-type and mutated proteins were expressed in the Escherichia coli BL21(DE3) strain. Cells were grown in LB medium containing 100 μg ml−1 ampicillin at 310 K until the OD600 reached 1.2. Cultures were induced with 1 mM IPTG. Expression was performed at 293 K for 17 h. Cells were harvested by centrifugation at 6000 rev min−1 for 30 min, resuspended in buffer A (50 mM Tris pH 8.5, 300 mM NaCl and 10 mM imidazole) and subsequently lysed by sonication. The soluble fraction was obtained by centrifugation at 14 000 rev min−1 for 40 min. The protein was purified using Ni-affinity chromatography. The supernatant was incubated with 10 ml Ni–NTA agarose for 2 h to allow His6-tagged protein to bind to the resin. After collecting the flowthrough, the column was washed with 1.5 l buffer A. The protein was eluted in an imidazole gradient (75–150 mM) and dialyzed against 50 mM Tris pH 8.0, 1.0 mM EDTA, 0.5 mM DTT buffer for protease cleavage. The His6 tag was removed during incubation with rTEV protease for 48 h at 283 K. The cut protein was loaded onto an Ni–NTA agarose column to separate it from uncut protein, rTEV and His6 tag. The protein was dialyzed against 20 mM Tris pH 8.0 and concentrated to 15 mg ml−1 (Bradford assay, BioRad). The SeMet-labeled protein was purified in the same way as the native protein and finally dialyzed against 20 mM Tris pH 8.0, 200 mM KH2PO4 buffer and concentrated to 15 mg ml−1.
2.3. Crystallization
The preliminary screens with the wild-type and mutated protein samples were carried out using a screen including 96 solutions, a subset of the 108 best crystallization conditions reported by the Joint Center for Structural Genomics (Page et al., 2003). The sitting-drop method was used throughout. No crystals were obtained for the wild-type protein. The optimized crystallization conditions of the K88A/Q89A mutant involved mixing 1 μl protein solution and 1 μl reservoir solution containing 30% PEG 1500 and 0.2 M KH2PO4 pH 5.6. Crystals appeared overnight. For the SeMet-containing crystals, the well solution was 20% PEG 8000 and 0.05 M KH2PO4 pH 4.6. Crystals were frozen using the harvesting solution containing 20% PEG 8000, 0.05 M KH2PO4 pH 4.6 and 10% ethylene glycol.
2.4. Data collection
Data were collected from a single SeMet-substituted crystal at the SER-CAT (Southeast Regional Collaborative Access Team) beamline 22-ID at the Advanced Photon Source, Argonne National Laboratory at nominal wavelengths of 0.97920 Å (peak), 0.97546 Å (remote) and 0.97931 Å (edge). All data were processed and scaled using HKL2000 (Otwinowski & Minor, 1997). The unit cell was identified as P212121, with unit-cell parameters a = 36.2, b = 54.1, c = 93.2 Å. The Matthews coefficient is 2.1 Å3 Da−1, assuming one polypep-tide per asymmetric unit.
2.5. Structure solution
Analysis of the MAD data using XPREP (Bruker AXS, Madison, USA) revealed incompatibilities with the assumed wavelengths, suggesting that the real wavelengths differed from the nominal values. The SAD method was used instead, based on the absorption-peak data. The anomalous substructure was solved with SHELXD (Schneider & Sheldrick, 2002). The two Se sites were fed into the autoSHARP procedure incorporating SHARP (de La Fortelle & Bricogne, 1997), SOLOMON (Abrahams & Leslie, 1996) and the CCP4 suite (Collaborative Computational Project, Number 4, 1994). The model was built automatically in ARP/wARP (Perrakis et al., 1999), which was able to build 184 out of 190 residues. The program O (Jones et al., 1991) was used for subsequent manual model improvement. Refinement using all data to 1.8 Å resolution was performed with REFMAC5 (Murshudov et al., 1997). The refinement of the model utilizing individual isotropic displacement parameters (B factors) converged with an R factor of 14.0% (Rfree = 18.4%). In the final stages of the refinement, H atoms were added to the model and anisotropic displacement parameters were used. This improved the agreement factors to 12.5 and 18.0%, respectively. MOLPROBITY (Lovell et al., 2003) and PROCHECK (Laskowski et al., 1993) were used as validation tools during refinement process. Relevant crystallographic data are shown in Table 1.
Table 1.
Data-collection and refinement statistics.
| Values in parentheses correspond to the last shell. | |
|---|---|
| Data-collection statistics | |
| Space group | P212121 |
| Unit cell (Å) |
a = 36.2, b = 54.1, c = 93.2 |
| Wavelength (Å) | 0.97920 |
| Resolution (Å) | 50.0–1.70 (1.76–1.70) |
| Total reflections | 109985 |
| Unique reflections† | 31784 |
| Redundancy | 3.5 (1.4) |
| Completeness (%) | 99.8 (16.3) |
| Rsym‡(%) | 8.2 (31.0) |
| I/σ(I) | 13.9 (1.9) |
| Phasing statistics | |
| Anomalous phasing power§ | 0.660 |
| Anomalous Rcullis¶ | 0.919 |
| Figure of merit (acentric) | 0.233 |
| After density modification | 0.790 |
| Refinement statistics | |
| Resolution (Å) | 50.0–1.80 |
| Reflections (working) | 15495 |
| Reflections (test) | 1069 |
| Rwork†† (%) | 12.5 |
| Rfree†† (%) | 18.0 |
| No. protein atoms | 1513 |
| No. waters | 292 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.018 |
| Bond angles (°) | 1.55 |
| Average B factor (Å2) | |
| Main chain | 11.8 |
| Side chain | 15.1 |
| Waters | 32.1 |
This number accounts for separation of Friedel mates.
Rsym = ∑ |Ii −⟨I⟩|/∑ I, where Ii is the intensity of the ith observation and ⟨I⟩ is the mean intensity of the reflections. The values are for unmerged Friedel pairs.
Phasing power = r.m.s.(|Fh|/E), where |Fh| is the heavy-atom structure-factor amplitude and E is the residual lack-of-closure error.
for acentric reflections, where |Fh, obs| is the observed heavy-atom structure-factor amplitude and |Fh, calc| is the calculated heavy-atom structure-factor amplitude.
, the crystallographic R factor, and where all reflections belong to a test set of randomly selected data.
3. Results and discussion
3.1. The quality of the model
The refined model of YdeN includes residues 2–187 out of a total of 190 in the single polypeptide chain. The quality of the model was verified with PROCHECK (Laskowski et al., 1993), which showed that 88.0% of non-glycine residues are in the favored regions and 11.4% are in the allowed regions of the Ramachandran plot. Notably, Ser71 lies outside this range (φ = 55, ψ = −117°). It is located within a tight turn and its strained conformation (ε region in the Ramachandran plot) is a characteristic feature of all α/β hydrolases (Derewenda & Derewenda, 1991; Ollis et al., 1992). One other residue, His18, also shows the ε secondary structure (φ = 58, ψ = −134°) and the electron density for this residue is equally well resolved. The strained conformation of this buried residue is stabilized by both van der Waals and hydrogen bonds, including a close packing against the adjacent Trp19. The side chain of Arg87 is not visible in the σA-weighted 2mFobs – DFcalc electron-density map contoured at 1σ and it was consequently excluded from refinement. Six side chains (Asn41, Ser55, Gln58, Glu63, Glu124 and Asp147) have at least two conformers and the occupancies were adjusted to correspond to the electron density.
3.2. Overall structure
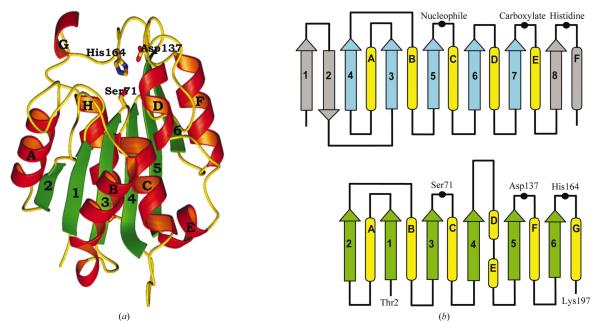
The refined YdeN model reveals the canonical tertiary fold of α/β hydrolases, albeit lacking the typically observed first two antiparallel strands (Fig. 1). Consequently, the molecule consists of six central parallel β-strands with the topology 213456 and eight α-helices. The strand–turn–helix motif following β3 has an architecture which is consistent with the nucleophilic elbow motif that is typical of all α/β hydrolases and is also observed in an unrelated enzyme, malonyl-CoA acyl transferase (Serre et al., 1995). The typical amino-acid sequence around the nucleophilic Ser in these proteins is Gly-X1-Ser-X2-Gly. The glycines are found in this motif because of the acute angle formed by the strand and the helix, which brings the side chains in these two positions into collision (Ollis et al., 1992). However, the first glycine in the elbow of YdeN is replaced with an Ala. This is not unusual; such departures from a canonical sequence have been observed before and are made possible by a slight opening of the elbow (Lawson et al., 1994; Uppenberg et al., 1994). The fingerprint sequence of the nucleophilic elbow of YdeN is similar to that of lipases from filamentous fungi, where X1 and X2 are also His and Leu, respectively (Derewenda et al., 1994).
Figure 1.
(a) Ribbon diagram of the YdeN molecule. The strands and helices are numbered and the three catalytic residues within the triad are shown in full. (b) Comparison of a canonical α/β hydrolase (upper panel) with the YdeN fold (lower panel).
3.3. Comparison with other α/β hydrolases
Members of the ubiquitous α/β hydrolase superfamily share a common architecture consisting of a central predominantly parallel β-sheet motif connected by loops and helices. The β-sheets create a half-barrel in which the first and the last strand are twisted with respect to each other by approximately 90°. Insertions containing long Ω loops or even large subdomains frequently occur after strands β3, β4, β6, β7 or β8. These insertions typically participate in the formation of a substrate-binding site and define the enzyme’s specificity (Heikinheimo et al., 1999). In many cases, such as those of the pancreatic lipase (van Tilbeurgh et al., 1993), bromoperoxidase A2 (Hofmann et al., 1998) or the 2-hydroxyl-6-oxo-6-phenylhexa-2,4-dienoic acid (HPDA) hydrolase (Nandhagopal et al., 2001), the insertion constitutes a ‘lid’ which undergoes structural rearrangement during the catalytic process. Smaller enzymes in this family, such as carboxylesterase (Kim et al., 1997) or cutinase (Martinez et al., 1991; Nicolas et al., 1996), have solvent-accessible active sites and are active against water-soluble small esters or thioesters.
The YdeN protein is a relatively small α/β hydrolase, smaller than cutinase (Martinez et al., 1991). The extra β-strand at the C-terminus (strand β6) arises from a difference in the tertiary fold in that region. Aside from a distinct similarity to cutinase, the tertiary fold of YdeN is also similar to the haloperoxidase, dienelactone hydrolase, esterase or serine carboxypeptidase subfamilies as defined by Heikinheimo et al. (1999). In common with the other small hydrolases, YdeN has no lid covering its active site, which is consequently solvent-accessible.
An automated comparison using DALI also identified a number of α/β hydrolases with a high level of structural similarity, including bromoperoxidase A2 (Hofmann et al., 1998; r.m.s. difference 2.3 Å over 170 amino acids), thioesterase (Lawson et al., 1994; r.m.s. 2.6 Å over 170 amino acids), dienoate hydrolase (Nandhagopal et al., 2001; r.m.s. 2.5 Å over 167 amino acids) and carboxylesterase (Kim et al., 1997; r.m.s. 2.4 Å over 161 amino acids). A structural comparison of the active centers shows that all the triads are very similar. The r.m.s. differences for all side-chain atoms of the triad residues are 0.56, 0.58, 0.33 and 0.25 Å, respectively.
3.4. The active site
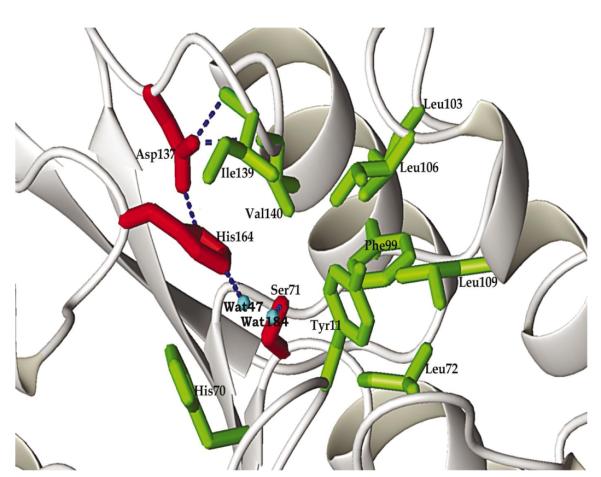
The three catalytic triad residues always occur in the same order along the poly-peptide chain in all α/β hydrolases, i.e. first the nucleophile (Ser, Cys or Asp), then the carboxylic acid (Asp or Glu) and finally histidine. The three residues form an approximate mirror-image of the functionally analogous triad of serine proteases (Ollis et al., 1992; Derewenda & Wei, 1995). In YdeN this sequence is preserved and the three residues involved in the putative catalytic site are Ser71, Asp137 and His164. However, there is no hydrogen bond between the Ser hydroxyl oxygen and the imidazole of histidine because the hydroxyl group is shifted away from His164 Nε2. Instead, the hydroxyl forms a hydrogen bond with an adjacent water molecule (Fig. 2). A comparison of diverse enzymes containing classical triads shows that the geometry of the Ser-His interaction disfavors a strong hydrogen bond in the ground state (Z. S. Derewenda, unpublished work). In several cases, the hydroxyl of the serine is clearly turned away from the His (Ho et al., 1997; Devedjiev et al., 2000). In YdeN, the unpaired hydrogen-bonding potential of His164 Nε2 is satisfied through an interaction with an additional ordered water molecule in the active site. On the other hand, His164 Nε1 donates a typical hydrogen bond to Asp137 Oδ2 (2.74 Å). Asp137 Oδ1 is in turn held in its position by a hydrogen bond with the main-chain amides of Ile139 (2.81 Å) and Val140 (2.97 Å).
Figure 2.
The stereochemical details of the active site. The residues of the triad are shown in red and the hydrogen bonds within this constellation are shown by dotted lines. The residues presumed to participate in substrate binding and selectivity are shown in green.
The function of the nucleophilic triad in triad-containing enzymes is complemented by the oxyanion hole, which stabilizes the incipient oxyanion during the course of the reaction via hydrogen bonds donated by amide groups (Matthews et al., 1975; Nicolas et al., 1996). These hydrogen bonds create a tetrahedrally distorted carbonyl C atom, which becomes susceptible to nucleophilic attack by the serine. The oxyanion hole can be formed by two amide groups, e.g. serine proteases and carboxylesterase (Kim et al., 1997), or three amide groups, as in acetylcholinesterase (Sussman et al., 1991) and the Streptomyces scabies esterase (Wei et al., 1995). In the last two cases, the oxyanion hole may be created by three main-chain amides or by two main-chain amides and one side-chain amide.
A close analysis of the stereochemistry of the putative active site of YdeN shows that main-chain amides of Tyr11 and Leu72 might serve to form an oxyanion hole. These amino acids correspond to Leu23 and Gln115 in carboxylesterase (Kim et al., 1997). There are no glutamine or asparagine side chains in the proximity of the active site that could contribute to the oxyanion hole.
3.5. YdeN belongs to a novel family of α/β hydrolases
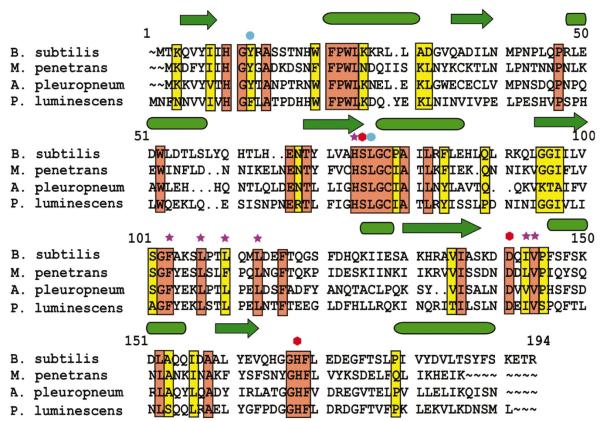
YdeN belongs to a small family of homologous microbial proteins, including a conserved hypothetical protein from Mycoplasma penetrans (43% identity), a predicted esterase from Actinobacillus pleuropneumoniae serovar (40% identity) and a hypothetical protein from Photorhabdus luminescens (36% identity). The sequence alignment (Fig. 3) shows that all the residues that line the specificity pocket are closely conserved in this family, with only minor deviations. This suggests that all proteins may hydrolyze the same or closely related substrates.
Figure 3.
Amino-acid alignment of four known representatives of the YdeN family. Secondary-structure elements are shown above the sequences as arrows (β-strands) or cylinders (α-helices). The active-site residues are denoted by red hexagons, the residues within the putative substrate binding site are denoted by stars and the two residues whose amides are involved in the oxyanion hole are denoted by blue circles.
3.6. The substrate specificity of YdeN
The function of YdeN is not known, but the overall similarity to other α/β hydrolases strongly suggests that the protein is an enzyme with hydrolytic properties. Until very recently it was thought that all α/β hydrolases are enzymes, but the discovery of a non-catalytic protein from Mycobacterium tuberculosis with an analogous tertiary fold has changed this view (Wilson et al., 2004). Nonetheless, in the non-catalytic protein the nucleophile is absent and it is probably safe to assume that the presence of a complete triad is a strong indicator of hydrolytic function. Many α/β hydrolases are lipases, but the absence in YdeN of a lid that would allow interfacial activation suggests that it is active against water-soluble esters, although a thioesterase function cannot be ruled out.
There are a number of hydrophobic amino acids in close proximity to the triad, including Val140, Ile139, Leu109, Leu106, Leu103, Phe99 and His70. Together, they form a small but well defined specificity pocket (Fig. 2). Comparison with carboxylesterase from Pseudomonas fluorescens revealed that Ile139, Leu109, Leu106, His70 in YdeN correspond to residues that build the specificity pocket found in the carboxylesterase, i.e. Val170, Ile70, Met73 and Phe113, respectively. Moreover, all of these residues are conserved among sequence homologues of YdeN. We conclude that it is very likely that YdeN is a carboxylesterase active on a water-soluble ester in which the substrate, possibly the acyl group, has a hydrophobic nature.
3.7. Crystal contacts
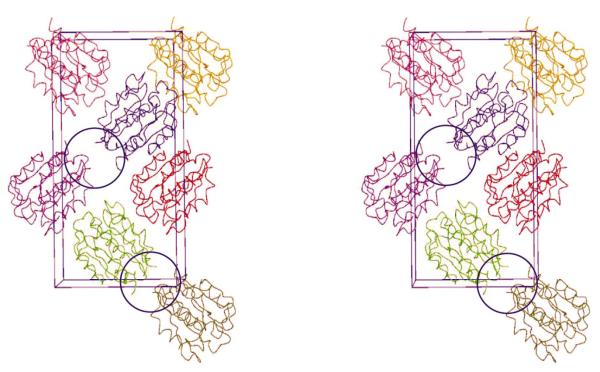
As we predicted on the basis of modeling, the two mutated residues Lys88 and Gln89 are located on a sharp loop connecting helix C to strand 4. The loop is in close proximity to a crystal contact (Fig. 4), but the two alanines are not involved in any direct intermolecular interactions between molecules in the crystal lattice. The wild-type Lys88 would have pointed towards the contact, but it is unlikely that it would seriously interfere with the crystal packing. However, it is noteworthy that along with the preceding Arg87 a lysine in position 88 would contribute to a highly charged surface patch. The loop faces the N-terminus of the adjacent molecule, potentially leading to electrostatic repulsion assuming the wild-type sequence. It is therefore possible that unlike other successful examples of crystallization by surface engineering, crystals of YdeN are not formed becuase of a reduction of excess conformational surface entropy but because of altered electrostatic interactions. If indeed more than one mechanism is at play leading to high-quality crystals, this further increases the potential of surface mutagenesis for crystallization.
Figure 4.
A stereoview of the crystal packing of the YdeN molecules. The crystal contact involving the mutated surface patch is circled.
Acknowledgments
This work was funded by NIGMS (grant GM62615 to ZSD and, in part, grant P-50-GM62414 to AJ and WM). We would like to thank the staff of SER-CAT for help with data collection. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID at the Advanced Photon Source (APS), Argonne National Laboratory. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38.
References
- Abrahams JP, Leslie AGW. Acta Cryst. 1996;D52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. Nature Struct. Biol. 2000;7(Suppl):957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- Claverie JM, Monchois V, Audic S, Poirot O, Abergel C. Comb. Chem. High Throughput Screen. 2002;5:511–522. doi: 10.2174/1386207023330002. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Derewenda U, Swenson L, Green R, Wei Y, Dodson GG, Yamaguchi S, Haas MJ, Derewenda ZS. Nature Struct. Biol. 1994;1:36–47. doi: 10.1038/nsb0194-36. [DOI] [PubMed] [Google Scholar]
- Derewenda ZS. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Derewenda ZS, Derewenda U. Biochem. Cell Biol. 1991;69:842–851. doi: 10.1139/o91-125. [DOI] [PubMed] [Google Scholar]
- Derewenda ZS, Wei Y. J. Am. Chem. Soc. 1995;117:2104–2105. [Google Scholar]
- Devedjiev Y, Dauter Z, Kuznetsov SR, Jones TL, Derewenda ZS. Structure. 2000;8:1137–1146. doi: 10.1016/s0969-2126(00)00529-3. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Rychlewski L. Proteins. 2003;53(Suppl 6):410–417. doi: 10.1002/prot.10548. [DOI] [PubMed] [Google Scholar]
- von Grotthuss M, Pas J, Wyrwicz L, Ginalski K, Rychlewski L. Proteins. 2003;53(Suppl 6):418–423. doi: 10.1002/prot.10547. [DOI] [PubMed] [Google Scholar]
- Heikinheimo P, Goldman A, Jeffries C, Ollis DL. Structure. 1999;7:141–146. doi: 10.1016/s0969-2126(99)80079-3. [DOI] [PubMed] [Google Scholar]
- Ho YS, Swenson L, Derewenda U, Serre L, Wei Y, Dauter Z, Hattori M, Adachi T, Aoki J, Arai H, Inoue K, Derewenda ZS. Nature (London) 1997;384:89–93. doi: 10.1038/385089a0. [DOI] [PubMed] [Google Scholar]
- Hofmann B, Tolzer S, Pelletier I, Altenbuchner J, van Pee KH, Hecht H-J. J. Mol. Biol. 1998;279:889–900. doi: 10.1006/jmbi.1998.1802. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kim KK, Song HK, Shin DH, Hwang KY, Choe S, Yoo OJ, Suh SW. Structure. 1997;5:1571–1584. doi: 10.1016/s0969-2126(97)00306-7. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, McArthur MW, Moss DS, Thornton JM. J. Appl. Cryst. 1993;26:282–291. [Google Scholar]
- Lawson DM, Derewenda U, Serre L, Ferri S, Szittner R, Wei Y, Meighen EA, Derewenda ZS. Biochemistry. 1994;33:9382–9388. doi: 10.1021/bi00198a003. [DOI] [PubMed] [Google Scholar]
- Longenecker KL, Garrard SM, Sheffield PJ, Derewenda ZS. Acta Cryst. 2001;D57:679–688. doi: 10.1107/s0907444901003122. [DOI] [PubMed] [Google Scholar]
- Longenecker KL, Lewis ME, Chikumi H, Gutkind JS, Derewenda ZS. Structure. 2001;9:559–569. doi: 10.1016/s0969-2126(01)00620-7. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Martinez C, de Geus P, Lauwereys M, Matthyssens G, Cambillau C. Nature (London) 1991;356:615–618. doi: 10.1038/356615a0. [DOI] [PubMed] [Google Scholar]
- Mateja A, Devedjiev Y, Krowarsch D, Longenecker K, Dauter Z, Otlewski J, Derewenda ZS. Acta Cryst. 2002;D58:1983–1991. doi: 10.1107/s090744490201394x. [DOI] [PubMed] [Google Scholar]
- Matthews DA, Alden RA, Birktoft JJ, Freer ST, Kraut J. J. Biol. Chem. 1975;250:7120–7126. [PubMed] [Google Scholar]
- Munshi S, Hall DL, Kornienko M, Darke PL, Kuo LC. Acta Cryst. 2003;D59:1725–1730. doi: 10.1107/s0907444903015415. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nandhagopal N, Yamada A, Hatta T, Masai E, Fukuda M, Mitsui Y, Senda T. J. Mol. Biol. 2001;309:1139–1151. doi: 10.1006/jmbi.2001.4737. [DOI] [PubMed] [Google Scholar]
- Nicolas A, Egmond M, Verrips CT, Longhi S, Cambillau C, Martinez C. Biochemistry. 1996;35:395–410. doi: 10.1021/bi9515578. [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A. Protein Eng. 1992;5:597–611. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Page R, Grzechnik SK, Canaves JM, Spraggon G, Kreusch A, Kuhn P, Stevens RC, Lesley SA. Acta Cryst. 2003;D59:1028–1037. doi: 10.1107/s0907444903007790. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Nature Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Acta Cryst. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Serre L, Verbree EC, Dauter Z, Stuitje AR, Derewenda ZS. J. Biol. Chem. 1995;270:12961–12964. doi: 10.1074/jbc.270.22.12961. [DOI] [PubMed] [Google Scholar]
- Stevens RC. Curr. Opin. Struct. Biol. 2000;10:558–563. doi: 10.1016/s0959-440x(00)00131-7. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G, et al. Acta Cryst. 2002;D58:2109–2115. [Google Scholar]
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- van Tilbeurgh H, Egloff M-P, Martinez C, Rygani N, Verger R, Cambillau C. Nature (London) 1993;362:814–820. doi: 10.1038/362814a0. [DOI] [PubMed] [Google Scholar]
- Uppenberg J, Hansen MT, Patkar S, Jones TA. Structure. 1994;2:293–308. doi: 10.1016/s0969-2126(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Wei Y, Schottel J, Derewenda U, Patkar S, Derewenda ZS. Nature Struct. Biol. 1995;2:218–223. doi: 10.1038/nsb0395-218. [DOI] [PubMed] [Google Scholar]
- Wilson RA, Maughan WN, Kremer L, Besra GS, Futterer K. J. Mol. Biol. 2004;335:519–530. doi: 10.1016/j.jmb.2003.11.001. [DOI] [PubMed] [Google Scholar]