Abstract
The crystal structure of a SlyA transcriptional regulator at 1.6 Å resolution is presented, and structural relationships between members of the MarR/SlyA family are discussed. The SlyA family, which includes SlyA, Rap, Hor, and RovA proteins, is widely distributed in bacterial and archaeal genomes. Current evidence suggests that SlyA-like factors act as repressors, activators, and modulators of gene transcription. These proteins have been shown to up-regulate the expression of molecular chaperones, acid-resistance proteins, and cytolysin, and down-regulate several biosynthetic enzymes. The structure of SlyA from Enterococcus faecalis, determined as a part of an ongoing structural genomics initiative (www.mcsg.anl.gov), revealed the same winged helix DNA-binding motif that was recently found in the MarR repressor from Escherichia coli and the MexR repressor from Pseudomonas aeruginosa, a sequence homologue of MarR. Phylogenetic analysis of the MarR/SlyA family suggests that Sly is placed between the SlyA and MarR subfamilies and shows significant sequence similarity to members of both subfamilies.
Comparisons of amino acid sequences and sequence patterns in prokaryotic genomes have identified many putative DNA-binding proteins and transcription factors. A sequence pattern common to several families of Escherichia coli regulatory proteins (GalR/LacI, DeoR, IclR, GntR, Crp, MarR, MerR, and AsnC) has been identified (1), and the structures of several transcriptional regulators have been determined (2). Many of these proteins contain the classic helix-turn-helix (HTH) DNA-binding motif and exhibit an associated sequence pattern.
A large number of the putative transcription factors, however, reveal new DNA-binding motifs (2, 3). The MarR/SlyA family is one example. Members of this large family modulate gene expression and are widely distributed. The COG data base includes 149 members of the MarR (multi-antibiotic resistance) family, among them 23 MarR homologues from Bacillus subtilis, MprA from E. coli, PecS from Erwinia chrysanthemi, and Hpr from B. subtilis (4, 5). The crystal structure of MarR revealed a new winged-helix (WH)1 DNA-binding motif (6). This structural motif was recently found in the MexR repressor from Pseudomonas aeruginosa, a sequence homologue of MarR (7).
SlyA-like proteins were found in 118 species of bacteria and 15 species of archaea, with 16 SlyA homologues in E. coli alone (http://www.ncbi.nlm.nih.gov:80/cgi-bin/Entrez/blink). This family includes such proteins as SlyA of Salmonella typhimurium and E. coli, and the Rap, Hor, and RovA proteins from Serratia marcescens, Erwinia carotovora, and Yersinia enterocolitica, respectively (8) (Fig. 1). In E. coli, SlyA regulates expression of cytolysin A, a cytotoxic protein that forms, stable, cation-selective transmembrane pores (9, 10).
FIG. 1. Multiple sequence alignment and phylogenetic relationships among SlyA homologues.
The threonine residue identical in all sequences is shown in dark gray; highly conserved amino acids (identical amino acids present in E. faecalis SlyA and at least 12 other proteins) and amino acids with similar properties are shown in gray. The amino acid sequence alignment was created using ClustalX, and the neighbor-joining tree was created using PAUP. Bootstrap values (1000 trials) of >50% are shown.
The slyA gene is implicated in virulence and environmental adaptation of S. typhimurium. It has been shown to up-regulate the expression of molecular chaperones, acid-resistance proteins, and cytolysin and down-regulate several biosynthetic enzymes (11). It has also been suggested that regulation of gene expression by SlyA is crucial for intracellular survival and/or replication of both enteroinvasive E. coli and Salmonella enterica serovar Typhimurium in phagocytic host cells.
RovA, whose amino acid sequence is 77% identical to that of the S. typhimurium SlyA, plays a role in regulation of the invasion of mammalian cells by Yersinia and mediates regulation of invasin in response to environmental signals (12). Invasin is a primary factor that allows efficient internalization of Yersinia pseudotuberculosis into eukaryotic cells. All the current evidence shows that SlyA-like factors act as repressors, activators, or modulators of gene transcription.
The SlyA protein from a pathogen, Enterococcus faecalis (SlyA-Ef), was selected for this study as part of an ongoing structural genomics initiative (www.mcsg.anl.gov). The specific biochemical function, the signal molecule, and the DNA-binding properties of this protein are unknown. Phylogenetic analysis of the MarR/SlyA family suggests that SlyA-Ef is placed between the SlyA and MarR subfamilies. It shows significant sequence similarity to members of both subfamilies (Fig. 1). In this report we present the crystal structure of a SlyA transcriptional regulator at 1.6 Å resolution and discuss structural relationships between members of the MarR/SlyA family.
MATERIALS AND METHODS
The E. faecalis SlyA open reading frame was amplified by polymerase chain reaction, with the NdeI and BamHI sites engineered at the translation start codon and immediately downstream of the translation stop codon, respectively, and cloned between the NdeI and BamHI sites of the pET15b vector (Novagen) in-frame with the His-tag and the rTEV cleavage site. Expression of the His-tagged fusion protein in E. coli strain BL21(DE3) carrying the pMAGIC vector was induced with isopropyl-β-d-thiogalactoside. Cells were harvested after 4 h of culture at 37 °C; suspended in 50 mm phosphate buffer, pH 8.0, 300 mm NaCl, 10 mm imidazole, 10 mm β-mercaptoethanol, and 10% glycerol; and lysed by sonication. The fusion protein was purified by affinity chromatography using nickel-nitrilotriacetic Superflow resin (Qiagen). The His-tag was removed by digestion with rTEV, and the resulting protein was purified by affinity chromatography using nickel-nitrilotriacetic Superflow resin (Qiagen). In this design, three amino acid residues were added at the N terminus of SlyA. The cleaved protein was further purified on an SP-Sepharose Fast Flow column (Amersham Biosciences) using 0.5 and 1.0 m NaCl two-step elution and concentrated with simultaneous buffer exchange using Centriplus-3 (Amicon) (3-kDa cutoff). A 2 mm protein stock solution in 10 mm Tris-HCl, pH 7.4, 20 mm NaCl, and 1 mm dithiothreitol was used for crystallization. Selenomethionine-labeled SlyA protein was prepared using the methionine biosynthesis inhibition method (13).
Equal volumes of SlyA protein stock solution and buffers were mixed in hanging droplets and equilibrated against 1-ml aliquots of solutions from the Hampton Research sparse matrix crystallization screening kit. SlyA was crystallized from 30% polyethylene glycol (PEG) 3350, 20% glycerol and 100 mm Hepes-sodium (pH 7.6) at 283 K. Crystals (0.2 × 0.02 × 0.4 mm) were rinsed in cryoprotectant solution consisting of 25% glycerol in the crystallization solution and flash-frozen in liquid nitrogen. Diffraction data were collected at 100 K at the 19BM beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory.
Crystals of native SlyA protein and its selenomethionine derivative diffracted to 1.8 and 2.0 Å, respectively. The space group was P212121 with cell dimensions of a = 43.512, b = 48.429, and c = 95.911. MAD data were collected to 2.0 Å of resolution from a single crystal containing selenomethionine-labeled protein at three different x-ray wavelengths near the selenium edge. Inverse beam strategy was used. The absorption edge was determined by a fluorescent scan of the crystal as described previously (13). The data were processed using HKL2000 suite (14). Crystal characteristics and data collection statistics are presented in Table I.
TABLE I.
Summary of Crystal and MAD data
| Crystal Data | |||
|---|---|---|---|
| Unit cell |
a = 43.512 Å, b = 48.429 Å, c = 95.911 Å, α = β = γ = 90° |
||
| Space group | P21 21 21 | ||
| Molecular mass (Da) (145 residues) |
15,950 | ||
| Molecules (AU)a | 2 | ||
| Selenomethionine (AU)a | 4 | ||
| MAD data collection | Edge | Peak | Remote |
| Wavelength (Å) | 0.9795 | 0.9793 | 0.95354 |
| Resolution range (Å) | 1.6 | 1.7 | 1.6 |
| No. of unique reflections | 35,499 | 30,018 | 32,997 |
| Completeness (%) | 98.1 | 99.1 | 91.1 |
| R merge (%) | 7.3 | 7.1 | 7.3 |
AU, asymmetric unit.
The structure of SlyA was determined using the MAD phasing method. All four Se sites were found in the asymmetric unit. MAD phases were calculated using the CNS suite (15) phases and improved using the density modification (DM) method as implemented by the CNS. Electron density maps were high quality and allowed autotracing of the amino acid chains using the wARP program (16). The procedure provided an initial model that included the majority of amino acid residues. The model was refined using 1.6 Å data and the CNS suite. Manual adjustment and model building was performed using the program QUANTA (17). The final R-factor and R-free are 24.8% and 27.5%, respectively, with excellent stereochemistry (Table II). The final structure included 145 amino acid residues and 179 water molecules. The PROCHECK analysis showed that all residues are in the allowed region of the Ramachandran plot (18). The electron density for the main chain is high quality, except for the N-terminal Met and the five C-terminal residues.
TABLE II.
Crystallographic statistics
| Phasing | |||||||
|---|---|---|---|---|---|---|---|
| Resolution range (Å) | Centric | Acentric | All | ||||
| FOMa | Phasing power | FOMa | Phasing power | No. | FOMa | Phasing power | |
| 30.0-1.8 | 0.8308 | 2.5720 | 0.6452 | 2.4129 | 42969 | 0.6571 | 2.4245 |
| Density modification | 0.80826 | ||||||
| Refinement | |||||||
| Resolution range (Å) | 30-1.6 | ||||||
| No. of reflections | 32997 | ||||||
| σ cutoff | 0.0 | ||||||
| R-value (%) | 24.8 | ||||||
| Free R-value (%) | 27.5 (2819) | ||||||
| Root mean square deviations from ideal geometry | |||||||
| Bond length (1–2) (Å) | 0.009 | ||||||
| Angle (°) | 1.00 | ||||||
| Dihedral (°) | 17.3 | ||||||
| Improper (°) | 0.67 | ||||||
| No. of atoms | |||||||
| Protein | 2331 | ||||||
| Water | 289 | ||||||
| Mean B-factor (Å2) for all atoms | 21.0 | ||||||
| Ramachandran plot statistics (%) | |||||||
| Residues in most favored regions | 99.2 | ||||||
| Residues in additional allowed regions | 0.8 | ||||||
| Residues in disallowed region | 0.0 | ||||||
FOM, figure of merit.
RESULTS AND DISCUSSION
Structure of SlyA
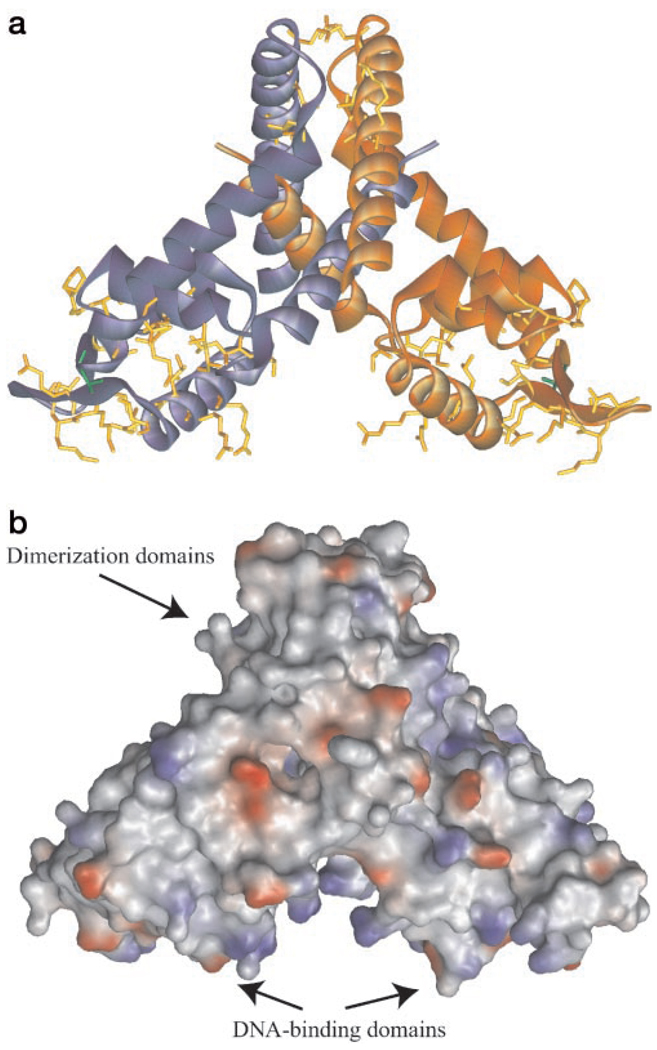
The crystal of unliganded SlyA-Ef contains two monomers in an asymmetric unit related by a pseudo-2-fold dyad. The 2-fold symmetry of the DNA-binding domains in the crystal structure (Fig. 2) is consistent with the palindromic character of the DNA sequences recognized by members of the SlyA family (9). The SlyA dimer appears to have a “Swiss cheese”-like structure with numerous channels, cavities, and voids. We suggest that these features are used for interaction with as yet unknown signal molecules and other cofactors working together in transcription regulation (Fig. 2b).
FIG. 2. Structure of SlyA-Ef.
a, SlyA-Ef dimer with subunits labeled in shades of blue and orange. Threonine residue present in all members of the MarR/SlyA family is shown in green, and other conserved residues are shown in yellow. b, SlyA-Ef dimer showing the solvent-accessible surface (drawn using 1.4 Å radius) and electrostatic potential. The SlyA-Ef reveals several channels and cavities filled with ordered water molecules. The positively charged surface associated with the HLH motif is likely to bind to a DNA duplex. Drawings were prepared with WebLabViewerPro.
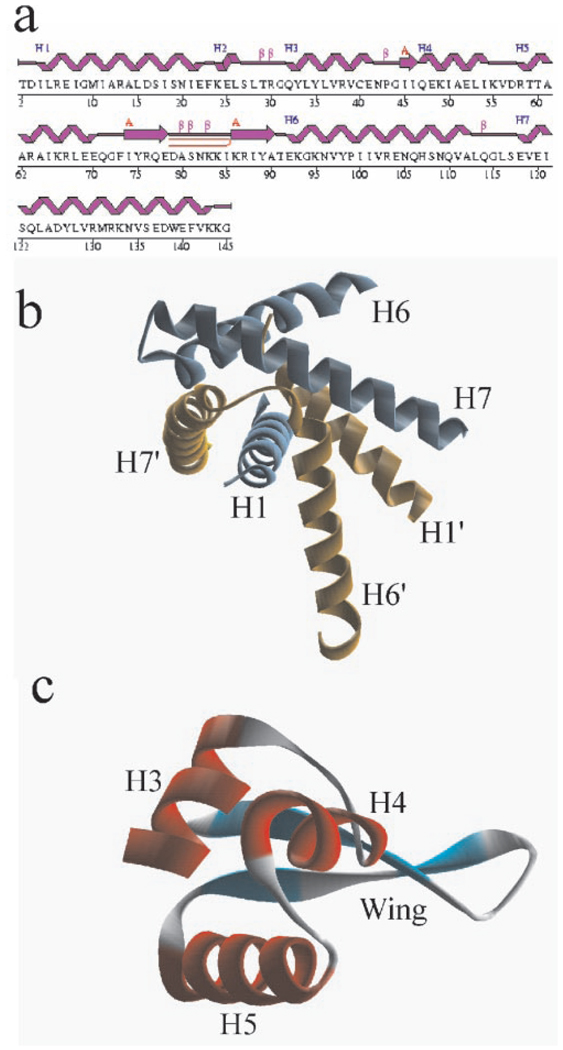
The protein dimer consists of three function-related structural elements, i.e. a dimerization unit composed of two domains and two independent DNA-binding domains. The dimerization domain includes an N-terminal α-helix, H1, and two C-terminal α-helices, H6 and H7 (Fig. 3). The structure of the dimerization unit reveals domain swapping. Helix H1 (subunit A) is inserted between H6′ and H7′ (subunit B) and forms a coil-coil with H7′. This interaction is mediated by a number of hydrophobic interactions and a salt bridge between conserved Glu-120 and conserved Arg-130′ from the opposite subunits. Arg-130′ also forms a hydrogen bond to Asp-126′. The C terminus of H6 also contributes to the dimer interface. The interaction between subunits is very extensive, with a total surface of 5025 Å2 buried between subunits, a very large area for such a small 150-residue protein.
FIG. 3. Secondary and domain structure of SlyA-Ef.
A, secondary structure of SlyA-Ef. b, SlyA-Ef dimerization domain showing domain swapping. c, DNA-binding domain with winged helix motif. Drawings prepared with WebLabViewerPro.
The DNA-binding domain is formed by the central part of the polypeptide and includes α-helices H2, H3, H4, and H5, and a β-hairpin (residues 73–91). There is virtually no interaction between DNA-binding domains in the SlyA dimer. This domain contains a DNA-binding motif described previously as a winged helix. In SlyA-Ef, it is composed of H5 and the 19-residue β-hairpin as a wing. This architecture closely resembles the WH motif of the well established major groove DNA-binding domain found in a number of transcriptional regulators (6, 19–22).
The WH motif in SlyA-Ef is preceded by two short α-helices, H3 and H4, with a short β-strand and a small loop between them. The motif is followed by H6, which is a part of the dimerization domain. Two residues conserved in the SlyA family, Thr-91 and Gly-94, are located at the junction between the wing and H6 (Fig. 1). These residues are also conserved in the MarR family and may be important for the positioning of the DNA-binding domain on its DNA target.
Interactions within the DNA-binding domain are mostly hydrophobic and involve several conserved residues (Fig. 1 and Fig. 2). Interactions between the DNA-binding and dimerization domains include the following four regions of the protein: 1) the C terminus of H7 (Trp-139 and Lys-143); 2) the C terminus of H1, short helix H2 (Phe-25), and a loop between H2 and H3; 3) the N terminus of H6 (Tyr-98); and 4) the N terminus of H1 of the opposite subunit, including a salt bridge between Glu-41 and Arg-6′ (these two residues are not strictly conserved in the SlyA family). It has been suggested that conserved glycine residues between secondary structure elements in the WH motif serve as flexible hinges to accommodate DNA targets with different half-site spacings (20). In SlyA-Ef, glycines 31, 44, 72, and 94 may perform this function.
SlyA Family Shows Strong Structural Homology to MarR
The SlyA-Ef has a number of structural homologues identified in the Protein Data Bank (PDB) both by the program available at the DALI server and by the Secondary Structure Matching Program at EMBL-EBI. The closest match identified by the program is the structure of MexR from P. aeruginosa (PDB code 1LNW). 88% of the secondary structure elements match, and the sequences are 17% identical. The root mean square deviation between structurally equivalent Cα positions of the monomer is 2.07 Å.
The next most significant matches (at 75% of the secondary structure elements matched) are to MarR from E. coli (1JGS) and MotA from bacteriophage T4 (1BJA), with sequence identities of 16 and 12% and root mean square deviations of 2.79 and 3.67 Å, respectively. The Swiss cheese characteristic of the SlyA dimer is also apparent in MarR and MexR proteins, although the precise nature of the accessible regions differs among them. This difference may reflect specificity for different signaling molecules or different modes of action.
Interaction with DNA
Crystal structures of the three MarR homologues as well as SlyA were determined without their DNA target or signal molecules bound. The structure of MarR protein includes salicylate, a known inhibitor of DNA binding (6). The structures of MexR and SlyA-Ef discussed here are of unliganded proteins, posing an important question of how these proteins interact with their specific DNA targets and whether these structures can provide new insight into DNA recognition.
The MexR structure shows three different dimers with varied spacing between their WH motifs and high degrees of conformational flexibility (7). The largest space between Cα positions of Arg-73/Arg-73′ was 29.9 Å. In the SlyA-Ef structure, the equivalent spacing (Arg-67/Arg-67′) is shorter (24.63 Å). Analysis of the MarR structure using a program available at the PITA server to generate biological dimers2 indicates that the spacing between the Cα positions of Arg-77/Arg-77′ is 28.1 Å. These key arginine residues exhibit different arrangements in the three structures. In MexR, they are positioned on the DNA binding surface and are expected to point toward the bases in the DNA major groove. In MarR and SlyA, the arginine side chains are also at the DNA binding surface but are expected to point away from the bases and the major groove. These differences suggest that the mode of binding of these proteins to DNA is not identical. Moreover, these data suggest that MarR/SlyA proteins may span a narrow minor groove and may recognize a rather short palindromic DNA sequence (14–16 bp).
The high-degree of flexibility of DNA-binding domains displayed in different crystals provides indirect evidence of the ability of this fold to adapt in order to recognize various DNA targets. Analysis of the crystal structure of the complex bound to DNA will test this hypothesis. It is also evident that the C terminus of SlyA is in position to facilitate an interaction with DNA. The C terminus of SlyA and its homologues contains several conserved positively charged residues that could make contacts with phosphate groups or bases.
In the SlyA-Ef dimer, the WH motifs are positioned to interact with a palindromic recognition sequence, with specific contacts expected to occur predominantly between H5 and the wing and the major groove of DNA. The WH motif in SlyA-Ef is strongly positively charged due to the presence of five arginines and four lysines; however, these residues are not conserved between SlyA and MarR families, suggesting that different DNA targets or different sets of residues in the same DNA target are recognized.
A cavity between DNA-binding domains in the SlyA-Ef dimer and extending to the dimerization domain could serve as a binding site for a signal molecule. In MexR, a C-terminal tail of the C monomer is inserted into this cavity (7). The residues lining the cavity are rather hydrophobic but not conserved, suggesting that these proteins respond to different signals. Interestingly, binding of a single ligand molecule to the dimer is expected to perturb the 2-fold symmetry, allowing the molecule to detect differences in the DNA target sequence. A signal molecule could also control spacing between DNA-binding domains by arranging them differently. Another key difference between SlyA and MarR is that the entrance to the cavity in MarR is closed off by two salt bridges between Asp-67 and Arg-73. These interactions are not present in the SlyA or MexR proteins. Recent evidence suggests that SlyA-dependent regulation may involve other protein cofactors such as the cAMP receptor or nitrate reduction regulator (23), which may involve direct protein-protein interactions.
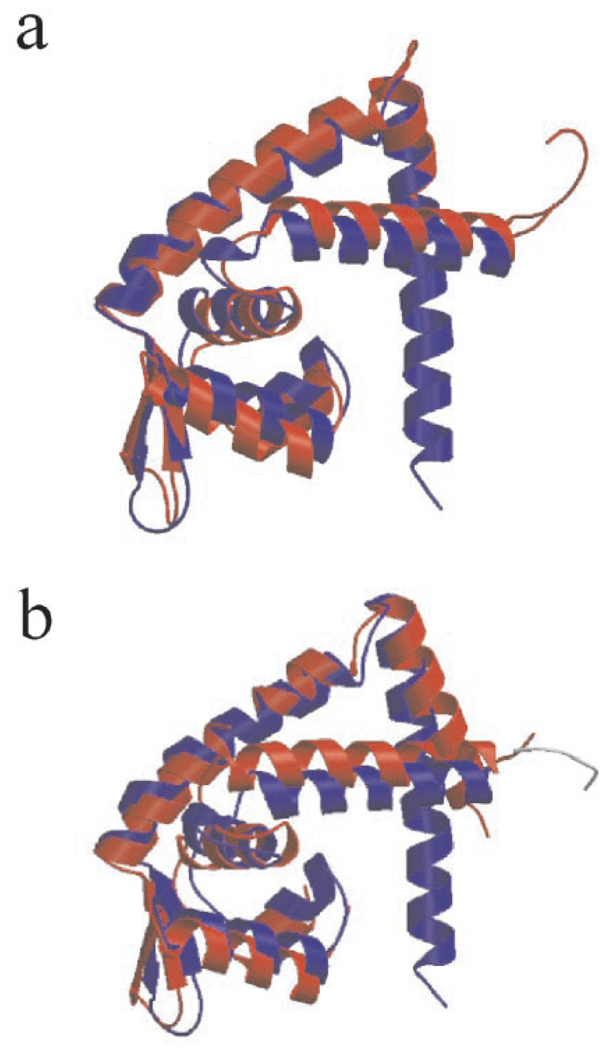
Overall, the SlyA structure is very similar to the structures of MarR and MexR (Fig. 4), showing that the MarR/SlyA family shares a common fold and similar DNA-binding properties despite their low amino acid sequence similarity. However, these proteins show significant local divergence in sequence and structure that apparently allows them to respond to different signal molecules and to bind to diverse DNA targets by using alternative modes of action. The MarR/SlyA structure is an example of a highly adaptable protein fold and provides a plausible explanation for its wide use in transcription regulation in many different organisms.
FIG. 4. Comparison of the SlyA-Ef structure with the structures of MarR and MexR.
a, superimposition of SlyA-Ef (blue) on MarR (red) subunits. b, superimposition of the SlyA-Ef (blue) and MexR (red) structures.
Acknowledgments
We thank all members of the Structural Biology Center at Argonne National Laboratory for help in conducting these experiments. We also thank Lindy Keller for help in preparation of this manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM62414 and the United States Department of Energy, Office of Biological and Environmental Research, under contract W-31-109-Eng-38 with the University of Chicago as Operator of Argonne National Laboratory.
The atomic coordinates and structure factors (code 1LJ9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: WH, winged helix; MAD, multiple-wavelength anomalous dispersion; CNS, crystallography NMR software; SlyA-Ef, SlyA protein from Enterococcus faecalis.
H. Ponstingl, manuscript in preparation.
REFERENCES
- 1.Perez-Rueda E, Collado-Vides J. J. Mol. Evol. 2001;53:172–179. doi: 10.1007/s002390010207. [DOI] [PubMed] [Google Scholar]
- 2.Huffman JL, Brennan RG. Curr. Opin. Struct. Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 3.Kenney LJ. Curr. Opin. Microbiol. 2002;5:135–141. doi: 10.1016/s1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- 4.Dehoux P, Cossart P. Mol. Microbiol. 1995;15:591. doi: 10.1111/j.1365-2958.1995.tb02272.x. [DOI] [PubMed] [Google Scholar]
- 5.Thomson NR, Cox A, Bycroft BW, Stewart GS, Williams P, Salmond GP. Mol. Microbiol. 1997;26:531–544. doi: 10.1046/j.1365-2958.1997.5981976.x. [DOI] [PubMed] [Google Scholar]
- 6.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. Nat. Struct. Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 7.Lim D, Poole K, Strynadka NC. J. Biol. Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 8.Revell PA, Miller VL. FEMS Microbiol. Lett. 2001;205:159–164. doi: 10.1111/j.1574-6968.2001.tb10941.x. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig A, Bauer S, Benz R, Bergmann B, Goebel W. Mol. Microbiol. 1999;31:557–567. doi: 10.1046/j.1365-2958.1999.01196.x. [DOI] [PubMed] [Google Scholar]
- 10.Oscarsson J, Westermark M, Lofdahl S, Olsen B, Palmgren H, Mizunoe Y, Wai SN, Uhlin BE. Infect. Immunol. 2002;70:5759–5769. doi: 10.1128/IAI.70.10.5759-5769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spory A, Bosserhoff A, von Rhein C, Goebel W, Ludwig A. J. Bacteriol. 2002;184:3549–3559. doi: 10.1128/JB.184.13.3549-3559.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagel G, Lahrz A, Dersch P. Mol. Microbiol. 2001;41:1249–1269. doi: 10.1046/j.1365-2958.2001.02522.x. [DOI] [PubMed] [Google Scholar]
- 13.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Acta Cryst. D Biol. Cryst. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Perrakis A, Morris R, Lamzin VS. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 17.Accelrys. QUANTA. San Diego, CA: Accelrys; 2000. [Google Scholar]
- 18.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 19.Gajiwala KS, Burley SK. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhang RG, Kim Y, Skarina T, Beasley S, Laskowski R, Arrowsmith C, Edwards A, Joachimiak A, Savchenko A. J. Biol. Chem. 2002;277:19183–19190. doi: 10.1074/jbc.M112171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnin MS, Cicero MP, Davies C, Porter SJ, White SW, Kreuzer KN. EMBO J. 1997;16:1992–2003. doi: 10.1093/emboj/16.8.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littlewood TD, Evan GI. Protein Profile. 1994;1:635–709. [PubMed] [Google Scholar]
- 23.Westermark M, Oscarsson J, Mizunoe Y, Urbonaviciene J, Uhlin BE. J. Bacteriol. 2000;182:6347–6357. doi: 10.1128/jb.182.22.6347-6357.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]